The structure of a complex of an endoglucanase with cellotriose was determined with a hexagonal unit cell and showed how the substrate interacts with the enzyme.
Keywords: carbohydrate utilization, catalytic domain, cellulase, molecular interactions
Abstract
The endoglucanase EglA from Piromyces rhizinflata found in cattle stomach belongs to the GH5 family of glycoside hydrolases. The crystal structure of the catalytic domain of EglA shows the (β/α)8-barrel fold typical of GH5 enzymes. Adjacent to the active site of EglA, a loop containing a disulfide bond not found in other similar structures may participate in substrate binding. Because the active site was blocked by the N-terminal His tag of a neighbouring protein molecule in the crystal, enzyme–substrate complexes could not be obtained by soaking but were prepared by cocrystallization. The E154A mutant structure with a cellotriose bound to the −3, −2 and −1 subsites shows an extensive hydrogen-bonding network between the enzyme and the substrate, along with a stacking interaction between Trp44 and the −3 sugar. A possible dimer was observed in the crystal structure, but retention of activity in the E242A mutant suggested that the enzyme probably does not function as a dimer in solution. On the other hand, the first 100 amino acids encoded by the original cDNA fragment are very similar to those in the last third of the (β/α)8-barrel fold, indicating that EglA comprises at least two catalytic domains acting in tandem.
1. Introduction
Cellulose from the plant cell wall is an abundant resource of renewable energy (Dashtban et al., 2009 ▶). It is a linear biopolymer of anhydroglucopyranose molecules joined by β-1,4-glycosidic bonds. Coupling of cellulose chains leads to a crystalline structure that exhibits great tensile strength and low solvent accessibility (Zhang et al., 2006 ▶). The complete degradation of cellulose requires three types of cellulases. Endoglucanase (EC 3.2.1.4) randomly cleaves internal β-1,4-glycosidic bonds, cellobiohydrolase (exoglucanase; EC 3.2.1.91) processively acts on the chain termini to release cellobiose, and β-glucosidase (EC 3.2.1.21) hydrolyzes cellobiose to glucose (Zhang et al., 2009 ▶). Cellulases have found wide applications in the bioconversion of plant matter to fermentative products in many industries (Bhat & Bhat, 1997 ▶; Zhang et al., 2006 ▶).
According to the glycoside hydrolase (GH) classification in the CAZy database (http://www.cazy.org/), the GH5 family encompasses more than 2000 archaeal, bacterial and fungal enzymes, which include endoglucanases, cellobiohydrolases, chitosanases, mannanases, xylanases and xyloglucanases, reflecting the diverse protein sequences. All GH5 enzymes share a common (β/α)8-barrel fold and retain the anomeric configuration at the cleavage site. Two glutamic acid residues at the C-terminal ends of strands βIV and βVII serve as the proton donor and the nucleophile in the double-displacement reaction (Bortoli-German et al., 1995 ▶; Wang et al., 1993 ▶; Tull et al., 1991 ▶). Although there are at least 30 known GH5 structures, significant variations in the surface loops may account for the different substrate specificities.
The ruminal fungal population represents a rich source of cellulases which are very active against crystalline cellulose (Denman et al., 1996 ▶). In a previous study, a cDNA fragment encoding the catalytic domain of a GH5 cellulase, EglA (GenBank accession No. AF094757; 474 amino-acid residues including a linker), was cloned from Piromyces rhizinflata found in cattle stomach (Liu et al., 2001 ▶). As both an endoglucanase and a cellobiohydrolase, EglA efficiently degrades microcrystalline Avicel and filter paper, as well as a broad range of other substrates including barley β-glucan, carboxymethyl cellulose, lichenin and oat-spelt xylan (Liu et al., 2001 ▶). The dual activity of EglA for both glucan-based and xylan-based polysaccharides is unusual for a GH5 enzyme, making it promising for industrial applications.
In this study, the catalytic domain of P. rhizinflata EglA with an N-terminal His tag was expressed in Escherichia coli. The recombinant protein (denoted rEglA) was purified and crystallized and three-dimensional structures were determined for both an unliganded form and a mutant form bound to a substrate molecule. Structural comparison with other homologous enzymes showed an additional disulfide-containing loop adjacent to the active site. The crystal structure also suggested possible dimer formation, which was investigated by site-specific mutation.
2. Materials and methods
2.1. Production of the rEglA crystals and data collection
The glucanase-encoding gene was amplified from pNZJ068/egla (Liu et al., 2005 ▶) by polymerase chain reaction and cloned into pET46 Ek/LIC (Novagen), which also encoded the N-terminal His tag MAHHHHHHVDDDDK. The E154A and E242A mutations were introduced using the QuikChange kit (Stratagene). The recombinant and mutant proteins were expressed in E. coli BL21 (DE3) cells by induction with isopropyl β-d-1-thiogalactopyranoside and were purified using an Ni–NTA column. Detailed procedures for protein expression and activity measurements are described in the supplementary material1.
Crystals of wild-type rEglA and the E154A mutant in the apo form and in complex with cellobiose or cellotetraose were prepared at room temperature using sitting-drop crystallization kits from Hampton Research (Laguna Niguel, California, USA). The best crystals were produced using a reservoir consisting of 1.5 M sodium citrate. Fig. 1 ▶ shows some typical rEglA crystals. The E154A–substrate complex crystal was produced by incorporating 15 mM cellobiose or 6 mM cellotetraose into the protein solution. The diffraction data were collected on beamline BL13B1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan). For the substrate-free crystal, reservoir solution containing 20% glycerol was used as a cryoprotectant. For the complex crystals, no cryoprotectant was necessary. The data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
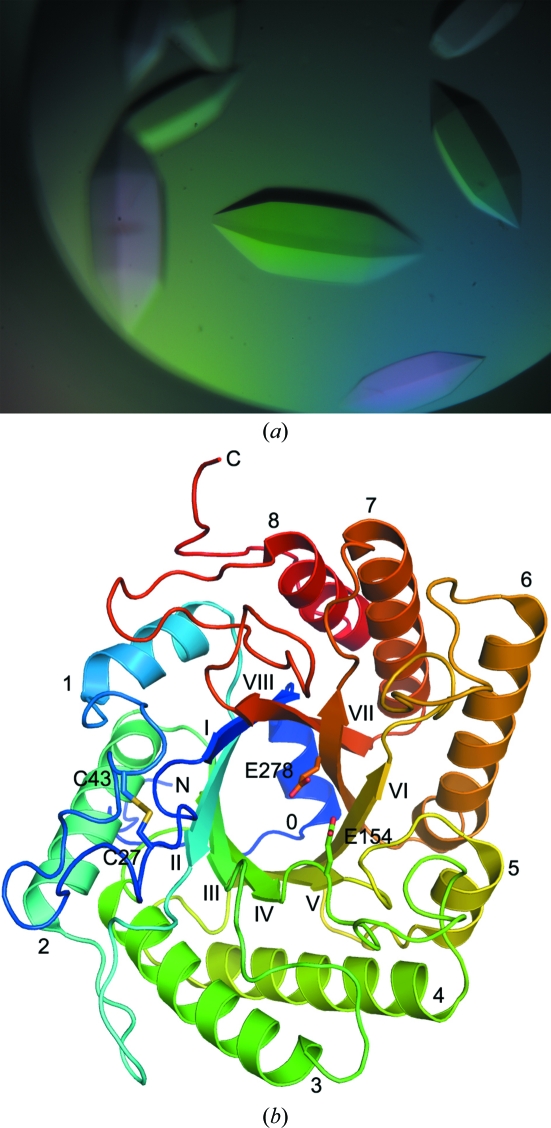
Figure 1.
The unliganded crystal structure of rEglA. (a) The physical dimensions of the crystal in the centre of the picture were 0.5 × 0.2 × 0.2 mm. The E154A mutant–substrate complex crystals had a similar appearance. (b) The structure shows a typical (β/α)8 fold. The protein model is rainbow-coloured from blue to red from the N-terminus to the C-terminus. The α-helices are labelled from 0 to 8 near their N-termini and the β-strands from I to VIII near their C-termini. The side chains of the catalytic glutamate and disulfide-forming cysteine residues are shown as stick models.
2.2. Structure determination and refinement
The structure was determined by molecular-replacement methods using CNS (Brünger et al., 1998 ▶). The search model was PDB entry 1edg (Ducros et al., 1995 ▶). The space group was found to be P6122, with the asymmetric unit containing one protein molecule. The initial R value was 0.52 at 2.5 Å resolution. Prior to further refinement, a randomly selected 5% of reflections were set aside for calculating R free. Although the R value was high, the Fourier map (2F o − F c) showed good electron density for most of the protein molecule, including several surface loops and a disulfide bond. Manual rebuilding using the program O (Jones et al., 1991 ▶) gradually improved the R and R free values to 0.32 and 0.36, respectively.
Despite extensive refinement, the R and R free values could only be reduced to 0.25 and 0.28, respectively, using CNS. However, using REFMAC from CCP4 with TLS (Winn et al., 2001 ▶, 2011 ▶; Collaborative Computational Project, Number 4, 1994 ▶; Potterton et al., 2003 ▶; Murshudov et al., 2011 ▶), the same model yielded R and R free values of 0.18 and 0.22, respectively. The data used in REFMAC were converted directly from those used in CNS to ensure that the same reflections were used in R free calculation. Subsequent refinement also used Coot (Emsley et al., 2010 ▶). Table 1 ▶ shows some statistics for the data and models of the unliganded rEglA and the E154A–substrate crystals. The partially refined E154A–cellobiose model (obtained using CNS) yielded R and R free values of 0.28 and 0.33, respectively, at 2.4 Å resolution. The program PyMOL (DeLano, 2008 ▶) was used to produce figures.
Table 1. Data collection and structural refinement of the rEglA crystals.
The statistics were calculated using the programs HKL-2000, REFMAC5 and Coot. Values in parentheses are for the highest resolution shells.
| Unliganded rEglA | E154A–cellotriose | |
|---|---|---|
| Data collection | ||
| Space group | P6122 | P6122 |
| Unit-cell parameters (Å, °) | a = b = 83.4, c = 225.1, α = β = 90, γ = 120 | a = b = 82.2, c = 221.8, α = β = 90, γ = 120 |
| Resolution range (Å) | 30–2.0 (2.03–2.00) | 30–2.2 (2.24–2.20) |
| Unique reflections | 31439 (1530) | 22798 (1110) |
| Multiplicity | 39.5 (42.8) | 23.1 (25.0) |
| Completeness (%) | 97.9 (97.1) | 97.0 (97.3) |
| Average I/σ(I) | 54.6 (8.8) | 60.7 (8.3) |
| Rmerge (%) | 7.5 (43.7) | 7.6 (61.9) |
| Refinement | ||
| Resolution range (Å) | 30–2.0 (2.05–2.00) | 30–2.2 (2.26–2.20) |
| Positive reflections | 29845 (2240) | 21543 (1629) |
| Rwork (%) | 17.0 (16.4) | 16.3 (18.9) |
| Rfree (%) | 21.6 (22.9) | 21.9 (27.5) |
| R.m.s.d. bond lengths (Å) | 0.020 | 0.018 |
| R.m.s.d. bond angles (°) | 2.1 | 1.9 |
| Dihedral angles in | ||
| Preferred regions (%) | 93.4 | 92.4 |
| Allowed regions (%) | 5.5 | 6.8 |
| Other regions (%) | 1.1 | 0.8 |
| No. of non-H atoms | ||
| Protein | 2971 | 2890 |
| Solvent | 231 | 180 |
| Ligand | 4 | |
| Average B (Å2) | ||
| Protein | 40.3 | 38.3 |
| Solvent | 49.5 | 47.4 |
| Ligand | 34.5 | |
3. Results and discussion
3.1. Overall structure of rEglA
The structure of rEglA, as shown in Fig. 1 ▶, is comprised principally of a (β/α)8 barrel. Near the N-terminus an extended His-tag segment is followed by helix α0, which appears to seal the barrel at the N-terminal side. A disulfide bridge between Cys27 and Cys43 is found in the 25-residue loop connecting strand βI and helix α1. The loop contains a small α-helix and two flanking short turns. Another small helix is located between strand βVI and helix α6. In addition, the ten residues before helix α8 form a β-ribbon structure on the protein surface.
The sequence identity between rEglA and Clostridium cellulolyticum CelCCA (PDB entry 1edg) is 37% (Supplementary Fig. S11). Superposition of the two structures using the program O, with a matching criterion of 2.0 Å, gives a root-mean-square deviation (r.m.s.d.) of 0.79 Å between 282 Cα pairs. In addition to the N- and C-terminal segments, structural differences are found in the shorter loops βIII–α3, βIV–α4, α5–βVI and βVI–α6 of rEglA, but the most significant is in the longer loop βI–α1 (Supplementary Fig. S11). Helix α5 is not homologous in sequence or structure. In CelCCA there are seven cysteines but no disulfide bonds (Ducros et al., 1995 ▶), whereas rEglA contains only a free Cys207. Four of the five variable loops are located on the C-terminal side of the barrel, adjacent to the active site, and probably contribute to the enzyme specificity. The disulfide-containing loop may play an important role in substrate binding (see below).
To avoid confusion, only five other GH5 structures apart from C. cellulolyticum CelCCA were chosen for comparison (Supplementary Table S11). These were Trichoderma reesei β-mannanase (PDB entry 1qnr; Sabini et al., 2000 ▶), Bacillus agaradhaerens Cel5A (PDB entry 1h5v; Varrot et al., 2001 ▶), Paenibacillus pabuli XG5 (PDB entry 2jeq; Gloster et al., 2007 ▶), Thermotoga maritima Cel5A (PDB entry 3mmu; Pereira et al., 2010 ▶) and Clostridium cellulovorans EngD (PDB entry 3ndy; C. M. Bianchetti, R. W. Smith, C. A. Bingman & G. N. Phillips Jr, unpublished work). EngD shares 41% sequence identity with rEglA (Supplementary Table S21). The two Clostridium enzymes CelCCA and EngD have the smallest r.m.s.d. from rEglA (about 1.2 Å for 320 Cα atoms within 4 Å). Tr. reesei β-mannanase and B. agaradhaerens Cel5A differ the most by an r.m.s.d. of 2.1 Å for 195 Cα atoms, despite the Tr. reesei enzyme and rEglA both being of eukaryotic origin (Supplementary Fig. S21). Interestingly, the βI–α1 loop of EngD has the 13-residue segment Cys27–Ala39 in rEglA substituted by a single glycine, making it even shorter than that of CelCCA. This segment is also eight residues shorter in P. pabuli XG5 and is completely absent in Th. maritima Cel5A.
3.2. Enzyme–substrate interactions
Enzyme–substrate complexes were obtained by cocrystallization using the E154A mutant rather than soaking the wild-type crystals with cellobiose or cellotetraose. Both complex crystals were isomorphous to the wild type and each contained a substrate molecule bound to the active site. In the latter, subsequently referred to as E154A–cellotriose for consistency, the electron density clearly showed cellotriose bound to the −3, −2 and −1 subsites at the nonreducing end of the active-site cleft (Fig. 2 ▶). The structure of the bound cellobiose was virtually identical to that of cellotriose in terms of the −2 and −1 sugar units (Supplementary Fig. S31).
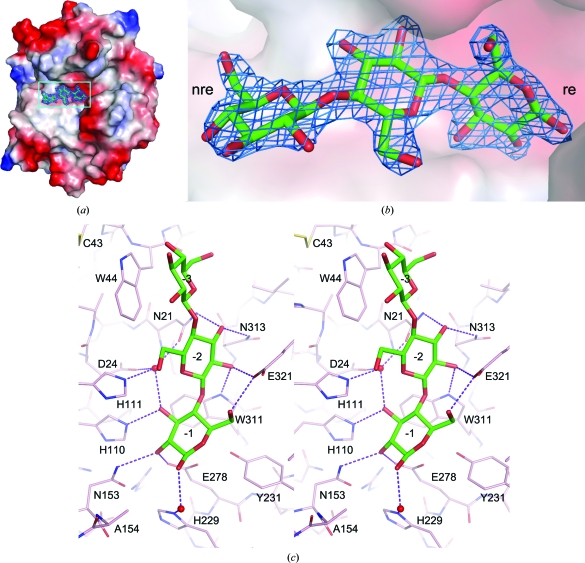
Figure 2.
The bound cellotriose in the E154A mutant. (a) The mutant rEglA protein is shown as a molecular-surface model coloured from red to blue according to the electrostatic potential between −75 and 75 k B T. The bound sugar is shown as a stick model and is superimposed on the F o − F c OMIT map contoured at the 5.0σ level. (b) A close-up view of the bound sugar and the electron density corresponding to the boxed region in (a) is shown with the surface rendered translucent to reveal the hidden part of the sugar. The nonreducing end (nre) and reducing end (re) of cellotriose are indicated. (c) The cellotriose molecule is shown as a bold stick model and the surrounding amino-acid residues are shown as thin sticks. The hydrogen bonds between the substrate and the enzyme are shown as dashed lines. Two mediating water molecules are shown as red spheres. The model is rotated clockwise by approximately 90° from those in (a) and (b).
The r.m.s.d. between the protein models of unliganded rEglA and the E154A–cellotriose crystals was 0.29 Å for 1428 pairs of backbone atoms and 0.79 Å for the side chains. Little conformational change of the enzyme was observed upon substrate binding, but the N-terminal His tag was not visible in the mutant crystal and was probably disordered. There are eight direct hydrogen bonds and at least six water-mediated hydrogen bonds between the −1 and −2 sugars and the enzyme (Fig. 2 ▶). The −3 sugar stacks with the side chain of Trp44 at a distance of 3.5 Å. Only the −1 sugar is entirely confined by protein atoms, whereas some space is available for branched sugar chains around the O3 and O6 atoms of the −2 sugar and even more space is available for the −3 sugar. It is interesting to note that Trp44 is adjacent to Cys43. Upon binding to a substrate branched at O3 of the −3 sugar, or possibly at O6 of the −2 sugar, the Cys27–Cys43 loop should make significant interactions with the substrate.
A comparison of the unliganded rEglA and E154A–cellotriose structures suggests that more than a dozen water molecules in the active site were displaced by the substrate. In fact, seven of these were located in almost identical positions to those where the hydroxyl groups of the cellotriose were bound to the enzyme (Supplementary Fig. S41). Presumably, the displaced water molecules make some entropic contribution to the free energy of substrate binding. The side chain of the superposed acid–base catalytic residue Glu154 will point toward the O1 atom of the −1 sugar at a distance of 1.6 Å. As shown in Fig. 2 ▶, the nucleophile Glu278 makes a hydrogen bond to the −1 sugar and was 3.2 Å from the anomeric C1 atom. This side chain formed a hydrogen bond to the phenolic OH group of the neighbouring Tyr231, another conserved acid–base catalytic residue in the GH5 enzymes. The equivalent residue in Pyrococcus horikoshii endoglucanase is Tyr299 and the Y299F mutant retained only 1–2% residual activity (Kang & Ishikawa, 2007 ▶; Kim & Ishikawa, 2011 ▶).
3.3. Crystal-packing interactions
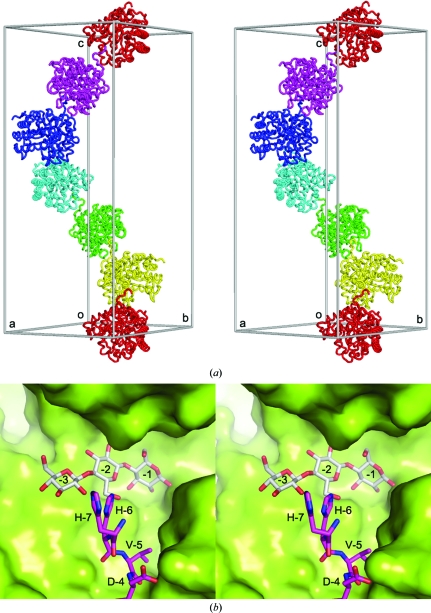
In the unliganded rEglA crystal, the 14-residue N-terminal His tag penetrates into the active site of a neighbouring enzyme molecule related by the 61 symmetry operation (Fig. 3 ▶). Three histidine residues were located adjacent to the −3 and −2 subsites (Fig. 3 ▶). Such an arrangement is likely to block the entrance to the active-site pocket and explains the failure of our initial attempts to obtain complex crystals by soaking. On the other hand, each molecule of rEglA forms a dimer with a crystallographic dyad-related molecule. The dimerization interface area was 1380 Å2 on each protein. The loop βVI–α6 of one protein molecule appeared to be interlocked with two active-site loops βIII–α3 and βIV–α4 of the other (Supplementary Fig. S51). The O1 atom of the −1 sugar was 3.2 Å from the OE2 atom of Glu242* (where the asterisk denotes a residue in the symmetry-related molecule), presumably forming a hydrogen bond. To investigate whether Glu242* plays a role in the hydrolysis of cellotetraose to cellotriose, the mutant E242A was produced and tested. As shown in Table 2 ▶, E242A retains most of the endoglucanase activity, suggesting that the enzyme probably does not function as a dimer in solution.
Figure 3.
His-tag interactions in the unliganded rEglA crystal. (a) Six enzyme molecules related by the P61 screw axis, representing one half of the unit-cell contents of the hexagonal P6122 crystal, are shown in different colours, along with a seventh molecule coloured as the first. The extended N-terminus of each enzyme molecule (e.g. the red molecule) penetrates into the active site of its neighbour (e.g. the yellow molecule). (b) The N-terminal His tag of one molecule is shown as purple sticks and the neighbouring molecule as a green surface model in a close-up view of the contacts between the two 61 symmetry-related rEglA molecules. The bound cellotriose from the E154A complex crystal after superposition of the mutant and the wild-type protein structures is shown as grey sticks.
Table 2. The activity of wild-type and mutant rEglA.
Not detectable.
0.12% of wild-type activity when measured using carboxymethyl cellulose as the substrate.
Comparison with other complex structures suggested that the Trp164 side chain of rEglA, located in loop βIV–α4, should stack with the +1 sugar of cellotetraose. Equivalent residues can be found in C. cellulolyticum CelCCA (Trp180 in PDB entry 1edg), C. cellulovorans EngD (Trp162 in PDB entry 3ndy) and P. pabuli XG5 (Tyr188 in PDB entry 2jeq). The less similar B. agaradhaerens Cel5A (PDB entry 1h5v) also has Trp178 in the corresponding position, but it is in the βV–α5 loop (Supplementary Fig. S61). However, in the E154A–cellotriose crystal the Trp164 side chain stacks with Gly241* and Glu242*, which apparently occupies the +1 subsite. Because the mutant has residual activity (Table 2 ▶), the cellotetraose should have been hydrolyzed (although slowly) to cellotriose before the E154A–cellotriose complex crystallized. The presence of the N-terminal His tag interfered with the strategy to obtain complex crystals by soaking, but it was overcome by cocrystallization. When the active site is occupied by sugars, the His tag can no longer penetrate into it.
4. Concluding remarks
For broad substrate specificity, the substrate-binding region of EglA should also accommodate branched chains, particularly at the nonreducing end. The surface area contributed by the disulfide-containing loop βI–α1 is a possible candidate to interact with branched sugar chains, but the precise binding mode remains to be further investigated. On the other hand, in addition to the catalytic domain of rEglA studied here, the original cDNA sequence also encodes a 110-residue N-terminal region that is highly homologous to the C-terminal region of rEglA, with 93% identity (Supplementary Fig. S71). It spans the secondary-structure elements α6, βVII, α7, βVIII and α8 and implicates another (β/α)8-barrel domain connected to the N-terminus of rEglA via a 30-residue linker. The use of multiple catalytic domains or different enzymes to enhance the efficiency of polymer cleavage is also seen in cellulosomes (Bayer et al., 1998 ▶).
Supplementary Material
PDB reference: EglA, 3ayr
PDB reference: E154A EglA–cellotriose, 3ays
Supplementary material file. DOI: 10.1107/S1744309111032428/hv5197sup1.pdf
Acknowledgments
The authors thank the National Synchrotron Radiation Research Center for beam-time allocations. This work was supported by grants from Tianjin Municipal Science and Technology Commission (10ZCKFSY06000 to RTG), the National Basic Research Program of China (2011CB710800 to RTG) and the National Science Council of Taiwan (NSC 98-2313-B-002-033-MY3 to JRL).
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: HV5197).
References
- Bayer, E. A., Shimon, L. J., Shoham, Y. & Lamed, R. (1998). J. Struct. Biol. 124, 221–234. [DOI] [PubMed]
- Bhat, M. K. & Bhat, S. (1997). Biotechnol. Adv. 15, 583–620. [DOI] [PubMed]
- Bortoli-German, I., Haiech, J., Chippaux, M. & Barras, F. (1995). J. Mol. Biol. 246, 82–94. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dashtban, M., Schraft, H. & Qin, W. (2009). Int. J. Biol. Sci. 6, 578–595. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2008). PyMOL http://www.pymol.org.
- Denman, S., Xue, G.-P. & Patel, B. (1996). Appl. Environ. Microbiol. 62, 1889–1896. [DOI] [PMC free article] [PubMed]
- Ducros, V., Czjzek, M., Belaich, A., Gaudin, C., Fierobe, H. P., Belaich, J. P., Davies, G. J. & Haser, R. (1995). Structure, 3, 939–949. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gloster, T. M., Ibatullin, F. M., Macauley, K., Eklöf, J. M., Roberts, S., Turkenburg, J. P., Bjørnvad, M. E., Jørgensen, P. L., Danielsen, S., Johansen, K. S., Borchert, T. V., Wilson, K. S., Brumer, H. & Davies, G. J. (2007). J. Biol. Chem. 282, 19177–19189. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kang, H.-J. & Ishikawa, K. (2007). J. Microbiol. Biotechnol. 17, 1249–1253. [PubMed]
- Kim, H.-W. & Ishikawa, K. (2011). Biochem. J. 437, 220–230.
- Liu, J., Tsai, C., Liu, J., Cheng, K. & Cheng, C. (2001). Enzyme Microb. Technol. 28, 582–589. [DOI] [PubMed]
- Liu, J.-R., Yu, B., Liu, F.-H., Cheng, K.-J. & Zhao, X. (2005). Appl. Environ. Microbiol. 71, 6769–6775. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pereira, J. H., Chen, Z., McAndrew, R. P., Sapra, R., Chhabra, S. R., Sale, K. L., Simmons, B. A. & Adams, P. D. (2010). J. Struct. Biol. 172, 372–379. [DOI] [PubMed]
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Sabini, E., Schubert, H., Murshudov, G., Wilson, K. S., Siika-Aho, M. & Penttilä, M. (2000). Acta Cryst. D56, 3–13. [DOI] [PubMed]
- Tull, D., Withers, S. G., Gilkes, N. R., Kilburn, D. G., Warren, R. A. & Aebersold, R. (1991). J. Biol. Chem. 266, 15621–15625. [PubMed]
- Varrot, A., Schülein, M., Fruchard, S., Driguez, H. & Davies, G. J. (2001). Acta Cryst. D57, 1739–1742. [DOI] [PubMed]
- Wang, Q., Tull, D., Meinke, A., Gilkes, N. R., Warren, R. A., Aebersold, R. & Withers, S. G. (1993). J. Biol. Chem. 268, 14096–14102. [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Zhang, Y.-H. P., Himmel, M. E. & Mielenz, J. R. (2006). Biotechnol. Adv. 24, 452–481. [DOI] [PubMed]
- Zhang, Y.-H. P., Hong, J. & Ye, X. (2009). Methods Mol. Biol. 581, 213–231. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: EglA, 3ayr
PDB reference: E154A EglA–cellotriose, 3ays
Supplementary material file. DOI: 10.1107/S1744309111032428/hv5197sup1.pdf