The complex of MoPrP(120–232) and Fab POM1 has been crystallized (space group C2, unit-cell parameters a = 83.68, b = 106.9, c = 76.25 Å, β = 95.6°). Diffraction data to 2.30 Å resolution have been collected using synchrotron radiation.
Keywords: prions, antibodies, POM1
Abstract
Prion diseases are neurodegenerative diseases that are characterized by the conversion of the cellular prion protein PrPc to the pathogenic isoform PrPsc. Several antibodies are known to interact with the cellular prion protein and to inhibit this transition. An antibody Fab fragment, Fab POM1, was produced that recognizes a structural motif of the C-terminal domain of mouse prion protein. To study the mechanism by which Fab POM1 recognizes and binds the prion molecule, the complex between Fab POM1 and the C-terminal domain of mouse prion (residues 120–232) was prepared and crystallized. Crystals of this binary complex belonged to the monoclinic space group C2, with unit-cell parameters a = 83.68, b = 106.9, c = 76.25 Å, β = 95.6°.
1. Introduction
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs), are a group of fatal progressive neurodegenerative disorders including bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in cervids, scrapie in sheep and goats, and Creutzfeldt–Jakob disease (CJD) in humans (Aguzzi & Polymenidou, 2004 ▶). Several other forms of human prion disease are also known; these include kuru, variant Creutzfeldt–Jakob disease (vCJD), Gerstmann–Straussler–Scheinker (GSS) disease and fatal familial insomnia (FFI) (Collinge, 2001 ▶). These diseases are associated with the conversion of normal cellular prion protein (PrPc) to a pathogenic conformation (PrPsc) that possesses abnormal physiochemical properties such as insolubility, protease resistance and the propensity to polymerize into amyloid-like fibrils (Prusiner, 1991 ▶). According to the ‘protein-only’ hypothesis, TSE transmission is mediated only by PrPsc itself; PrPsc acts as a template for its self-propagation through recruiting PrPc molecules and the cycle continues, leading to the formation of amyloid fibrils. The hallmark of prion diseases is the accumulation of amyloid fibrils in the brain that are associated with neuronal degradation characterized by nerve-cell loss and spongiosis.
To date, there is no treatment available for prion disease; however, any interruption to the conversion of PrPc to PrPsc in neurons could potentially delay disease progression or even cure the disease (White et al., 2003 ▶). Targeting cell-surface PrPc through monoclonal antibodies appears to be a novel therapeutic strategy that could reduce PrPsc accumulation, presumably by disrupting the PrPc–PrPsc interaction (Antonyuk et al., 2009 ▶). Furthermore, PrPc is converted to different conformations during prion propagation; thus, any molecule that binds specifically to PrPc should stabilize the native fold, thereby inhibiting changes in conformation and consequently prion propagation. In several in vitro and in vivo studies antiprion monoclonal antibodies have been shown to reduce the number of scrapie prions, and the development of prion disease has also been found to be delayed in the murine model after an inoculation of antiprion antibody (White et al., 2003 ▶; Féraudet et al., 2005 ▶). Here, we report the crystallization of a complex between a Fab fragment of the antibody POM1 (Polymenidou et al., 2008 ▶) and the C-terminal part of mouse prion protein.
2. Experimental procedure and results
2.1. Mouse prion expression and purification
The pET15b plasmid (Novagen) containing the C-terminal structured domain (residues 120–232) of the mouse prion gene was transformed into Escherichia coli BL21 (DE3) Codon Plus (Stratagene) cells. The cells were grown at 310 K in rich medium containing 100 µg ml−1 ampicillin and the protein [MoPrP(120–232)] was mainly expressed in inclusion bodies. The inclusion bodies were sonicated (4 × 30 s with 60 s intervals at 50% amplitude), pelleted by centrifugation and extensively washed. The inclusion bodies were then incubated in a denaturing buffer consisting of 6 M guanidinium hydrochloride, 10 mM Tris–HCl, 100 mM NaH2PO4, 5 mM imidazole pH 8.0 for 1 h at room temperature with constant stirring. The extracted denatured proteins were loaded onto an Ni–NTA agarose column (Qiagen) at a flow rate of 1 ml min−1 after the addition of 10 mM reduced glutathione. The MoPrP(120–232) protein was refolded on the column by a gradient application of buffer A (denaturing buffer) and buffer B (10 mM Tris–HCl, 100 mM NaH2PO4, 5 mM imidazole, pH 8.0) as described by Yin et al. (2003 ▶). MoPrP(120–232) was then eluted with 300 mM imidazole in buffer B and exchanged with distilled water using Amicon Ultra centrifugal filters (3 kDa molecular-weight cutoff, Millipore). The purity of the protein was confirmed by SDS–PAGE and the concentration was measured by the Bradford method (Bradford, 1976 ▶).
2.2. Fab production
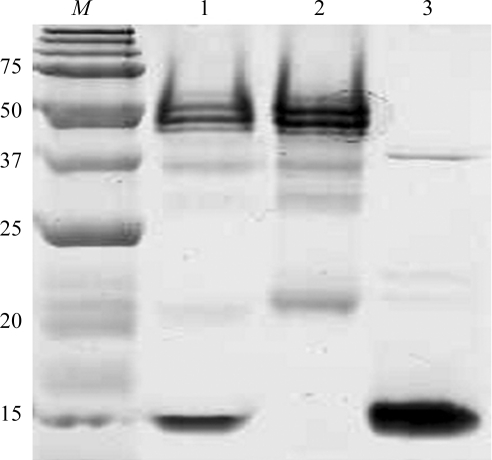
The IgG1 POM1 hybridoma was prepared as described by Polymenidou et al. (2008 ▶). The hybridoma cell-culture supernatant was loaded onto a protein G Sepharose column and the POM1 antibody was eluted with 0.1 M glycine pH 2.8. For Fab production, the IgG1 POM1 (1 mg ml−1) was digested with papain at a POM1:papain ratio of 1:0.02(w:w) in 50 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA and 2 mM cysteine (Andrew & Titus, 2001 ▶). After 5 h incubation at 310 K in a water bath, papain was inactivated by adding iodoacetamide to a final concentration of 3 mM. The digest was then concentrated, exchanged with protein A IgG-binding buffer and loaded onto a Protein A Sepharose column (Pierce). The Fc fragment and the undigested IgG1 POM1 were bound to the protein A column, whereas the POM1 Fab fragments were collected in the flowthrough. The Fab fractions were then assessed for homogeneity by Coomassie Brilliant Blue staining after separation by SDS–PAGE (Fig. 1 ▶).
Figure 1.
SDS–PAGE analysis of purified Fab POM1 and its complex with MoPrP(120–232). Lane M, molecular-weight marker (kDa); lane 1, Fab POM1–MoPrP(120–232) complex; lane 2, Fab POM1; lane 3, MoPrP(120–232).
2.3. Protein preparation and crystallization
Fab POM1 and MoPrP(120–232) were mixed in an equimolar ratio in order to form the protein complex and the complex was purified by Superdex G-75 (GE Healthcare) size-exclusion chromatography in a buffer solution consisting of 50 mM Tris pH 7.0, 100 mM NaCl and 1 mM NaN3. The purified protein complex was then concentrated to 10 mg ml−1 for crystallization studies. Screening of crystallization conditions for the Fab POM1–MoPrP(120–232) complex was carried out using several commercial screening solutions from Hampton Research in 96-well Intelli-Plates (Hampton Research) set up by a crystallization robot (Hydra-Plus-One, Robbins Scientific). Crystallization trays were set up using the sitting-drop vapour-diffusion method, in which 0.4 µl protein sample was mixed with an equal volume of screening solution. An initial crystallization hit was found in a saturating solution of 25% PEG 3350, 0.1 M Bis-Tris pH 6.5 and 0.2 M lithium sulfate (condition G3 of Hampton Research Index screen). After several optimization steps, crystals of approximate dimensions 0.6 × 0.1 × 0.1 mm were obtained after 7 d (Fig. 2 ▶).
Figure 2.
Crystals of the complex between Fab POM1 and MoPrP(120–232) grown using the sitting-drop vapour-diffusion procedure at room temperature.
3. Results and discussion
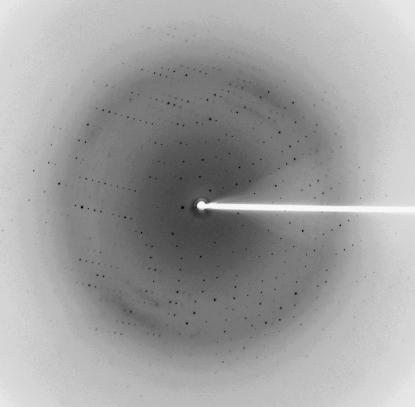
A complete X-ray diffraction data set was collected from a single crystal of the Fab POM1–MoPrP(120–232) complex cryocooled in liquid nitrogen using reservoir solution containing 20% glycerol as a cryoprotectant (Fig. 3 ▶). The synchrotron data were indexed and scaled with the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶). Data-collection statistics are shown in Table 1 ▶. Diffraction data were collected from the crystals of the Fab POM1–MoPrP(120–232) complex on the 08ID-1 beamline, Canadian Macromolecular Crystallography Facility using a 300 mm CCD detector.
Figure 3.
A representative 1° oscillation image of data collected from a crystal of the complex between Fab POM1 and MoPrP(120–232).
Table 1. Data-collection and structure-solution statistics.
Values in parentheses are for the highest resolution shell.
| Crystal dimensions (mm) | 0.6 × 0.1 × 0.1 |
| Matthews coefficient VM (Å3 Da−1) | 2.9 |
| Solvent content (%) | 57 |
| Crystal system | Monoclinic |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 83.68, b = 106.9, c = 76.25, α = γ = 90, β = 95.6 |
| No. of molecules in unit cell | 1 |
| Wavelength (Å) | 0.993 |
| Temperature (K) | 100 |
| Resolution range (Å) | 30.0–2.3 (2.38–2.30) |
| No. of unique reflections | 27952 |
| No. of observed reflections | 148536 |
| Completeness (%) | 95.2 (78.7) |
| Multiplicity | 5.3 (4.0) |
| 〈I/σ(I)〉 | 21.0 (4.2) |
| Rmerge† | 0.08 (0.32) |
R
merge = 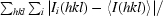
 , where 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
, where 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
The crystals diffracted to 2.3 Å resolution. The crystal was found to belong to space group C2, with unit-cell parameters a = 83.68, b = 106.9, c = 76.25 Å, β = 95.6°. A total of 27 952 unique reflections were measured with an average multiplicity of 5.3. The merged data set was 95.2% complete to 2.3 Å resolution, with an R merge of 8% and a mean I/σ(I) of 21 for all reflections and 4.2 for the highest resolution shell. The calculated Matthews coefficient was 2.9 Å3 Da−1, indicating the presence of one protein complex in the asymmetric unit with a solvent content of 57% (Matthews, 1968 ▶). Structure solution and analysis are currently in progress.
The crystal structure of the Fab POM1–MoPrP(120–232) complex will shed light on the molecular interactions present between the two proteins. This structural information will be helpful in designing therapeutic products against prion diseases.
Acknowledgments
This work was funded by Prionet Canada and AHFMR in grants to MNGJ. The research described in this paper was performed at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada and the University of Saskatchewan. The mouse C-terminal (120–232) prion clone was provided by Dr Trent Bjorndahl of Professor D. Wishart’s group at the University of Alberta.
References
- Aguzzi, A. & Polymenidou, M. (2004). Cell, 116, 313–327. [DOI] [PubMed]
- Andrew, S. M. & Titus, J. A. (2001). Current Protocols in Immunology, ch. 2, Unit 2.8. New York: Wiley. [DOI] [PubMed]
- Antonyuk, S. V., Trevitt, C. R., Strange, R. W., Jackson, G. S., Sangar, D., Batchelor, M., Cooper, S., Fraser, C., Jones, S., Georgiou, T., Khalili-Shirazi, A., Clarke, A. R., Hasnain, S. S. & Collinge, J. (2009). Proc. Natl Acad. Sci. USA, 106, 2554–2558. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Collinge, J. (2001). Annu. Rev. Neurosci. 24, 519–550. [DOI] [PubMed]
- Féraudet, C., Morel, N., Simon, S., Volland, H., Frobert, Y., Créminon, C., Vilette, D., Lehmann, S. & Grassi, J. (2005). J. Biol. Chem. 280, 11247–11258. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Polymenidou, M. et al. (2008). PLoS One, 3, e3872. [DOI] [PMC free article] [PubMed]
- Prusiner, S. B. (1991). Science, 252, 1515–1522. [DOI] [PubMed]
- White, A. R., Enever, P., Tayebi, M., Mushens, R., Linehan, J., Brandner, S., Anstee, D., Collinge, J. & Hawke, S. (2003). Nature (London), 422, 80–83. [DOI] [PubMed]
- Yin, S.-M., Zheng, Y. & Tien, P. (2003). Protein Expr. Purif. 32, 104–109. [DOI] [PubMed]