Abstract
Restriction fragment length polymorphisms (RFLPs) were used to study the population genetics and temporal dynamics of the cassava bacterial pathogen Xanthomonas axonopodis pv. manihotis. The population dynamics were addressed by comparing samples collected from 1995 to 1999 from six locations, spanning four different edaphoclimatic zones (ECZs). Forty-five different X. axonopodis pv. manihotis RFLP types or haplotypes were identified between 1995 and 1999. High genetic diversity of the X. axonopodis pv. manihotis strains was evident within most of the fields sampled. In all but one site, diversity decreased over time within fields. Haplotype frequencies significantly differed over the years in all but one location. Studies of the rate of change of X. axonopodis pv. manihotis populations during the cropping cycle in two sites showed significant changes in the haplotype frequencies but not composition. However, variations in pathotype composition were observed from one year to the next at a single site in ECZs 1 and 2 and new pathotypes were described after 1997 in these ECZs, thus revealing the dramatic change in the pathogen population structure of X. axonopodis pv. manihotis. Disease incidence was used to show the progress of cassava bacterial blight in Colombia during the 5-year period in different ecosystems. Low disease incidence values were correlated with low rainfall in 1997 in ECZ 1.
Cassava, Manihot esculenta Crantz, is a starchy root crop that is among the most important tropical foods. About 80% of the cassava produced is consumed in developing countries and constitutes the principal carbohydrate source for more than 500 million people (14). Cassava bacterial blight (CBB) has caused extensive damage to the crop during the past 2 decades (6, 14, 30). The causal agent, Xanthomonas axonopodis pv. manihotis, can induce a wide variety of symptoms, including angular leaf spots, blight, gum exudation, stem cankers, shoot wilt, vascular necrosis, and dieback (15). Resistance, which has been identified in M. esculenta and the wild relative Manihot glaziovii, is thought to be polygenic and additively inherited (9). Molecular markers have been employed to construct a genetic map of the cassava genome consisting of several hundred markers (8). The mapping of CBB resistance has recently been reported both in controlled conditions and in the field (10, 11). Resistance to X. axonopodis pv. manihotis strains is controlled by several quantitative trait loci (QTL) located in different linkage groups, suggesting a specific interaction between the plant and the pathogen. Some of these QTL are specific to one strain, showing the complexity of this host plant-pathogen interaction (10). Since 1983, the entire cassava breeding strategy has been based on improving varieties for different edaphoclimatic zones (ECZs) (1). The ECZs were defined according to the importance of cassava production in that region, climatic conditions, predominant soil type, and pest and disease problems (2). Seven zones exist: ECZ 1, subhumid tropics; ECZ 2, acid soil savannas; ECZ 3, humid tropical lowlands; ECZ 4, midaltitude tropics; ECZ 5, high-altitude tropics; ECZ 6, subtropics; and ECZ 7, semiarid areas. In addition, regions sharing conditions from two ECZs were defined, e.g., ECZ 2-4.
In previous studies, X. axonopodis pv. manihotis populations have been characterized by restriction fragment length polymorphism (RFLP) (24, 27, 28) and virulence variation (20). The RFLP technique, using the pathogenicity gene pthB as a probe, is precise and discriminatory in differentiating among X. axonopodis pv. manihotis strains (28). It permitted the description of basic population genetic parameters and processes like geographical differentiation of the X. axonopodis pv. manihotis subpopulations in ECZs and the migration of strains between ECZs (22, 24). The role of host selection in X. axonopodis pv. manihotis population structure was also reported (18). The study of the virulence variation led to the conclusions that the cassava-X. axonopodis pv. manihotis interaction is pathotype-cultivar specific and that different pathotypes are present in Colombia and Venezuela (20, 22, 29). However, changes in the pathogen population structure over time have not been studied, and until now, little was known regarding the population dynamics of X. axonopodis pv. manihotis.
Understanding the mechanisms by which bacterial populations evolve to overcome host resistance is critical for developing strategies to improve the durability of resistance. This study examines the genetic change in X. axonopodis pv. manihotis populations over time from 1995 to 1999 by using RFLP and virulence assays. Temporal dynamics were studied within and between years. Since changes in pathogen pathotypes have long-term implications in disease management and varietal improvement, the pathotype composition among X. axonopodis pv. manihotis populations was also addressed.
MATERIALS AND METHODS
Sampling.
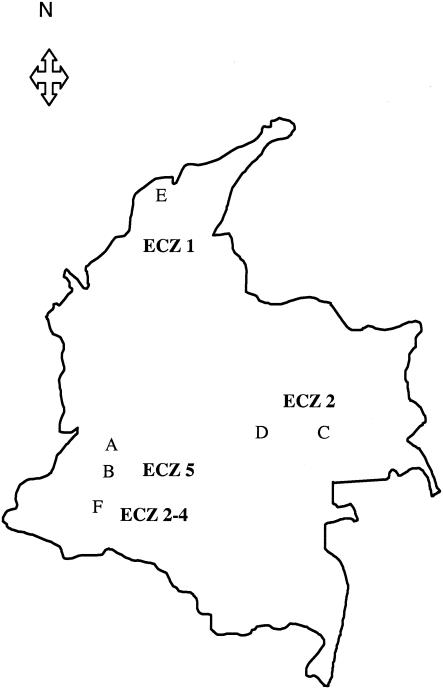
All strains were isolated from stem or leaf samples of cassava plants between 1995 and 1999. Six sites located throughout four ECZs in Colombia were frequently visited: Villavicencio and Carimagua (ECZ 2), Pivijay (ECZ 1), Mondomo and Cajibio (ECZ 5), and Santander de Quilichao (ECZ 2-4) (Fig. 1 and Table 1). ECZ 2-4 corresponds to an ECZ showing conditions and constraints found in both ECZ 2 and ECZ 4 (F. Calle, personal. communication). In a previous study (23) ECZ 2-4 was mistakenly classified as ECZ 2-5. In all, 736 strains were collected. At each site, two to three fields were sampled intensively at random and along diagonals. The number of samples collected from each site depended on the severity of the disease at the time of collection. All sites were sampled only once during the cropping cycle with the exception of Mondomo (ECZ 5) and Pivijay (ECZ 1) fields, which were visited twice during the cropping year, 3 and 7 months after plantings. Samples were collected from 14 improved cultivars at Mondomo and 27 cultivars at Pivijay. Samples were collected from the same cultivars at the beginning and the end of the crop cycle. For most of the fields, the same cultivars were planted over the years. However, a high number of new M. esculenta cultivars were planted in Mondomo and Villavicencio in 1997.
FIG. 1.
Geographical distribution of the six cassava fields sampled in Colombia. In each ECZ, strains were collected at different sites. A, Cajibio; B, Mondomo; C, Carimagua; D, Villavicencio; E, Pivijay; F, Santander de Quilichao.
TABLE 1.
Number of isolates, genetic diversities, disease incidence, number of haplotypes, and chi-square analysis of haplotypic frequencies for X. axonopodis pv. manihotis populations collected at six sites in four ECZs in Colombia
| ECZ | Sitea | Yrb | Incidencec | Nid | Haplotype
|
Hsiteg | χ2h | |
|---|---|---|---|---|---|---|---|---|
| Nhe | HYf | |||||||
| 1 | Pivijay | 1995 | 3.5 | 30 | 4 | 0.58 | ||
| 1996 | 3 | 9 | 3 | 0.56 | ||||
| 1997 | 1 | 48 | 4 | 0.56 | ||||
| 1998 | 3.5 | 39 | 9 | 0.52 | ||||
| 1999 | 4.5 | 39 | 4 | 0.46 | 0.69 | 118.8 (0.001) | ||
| 2 | Carimagua | 1995 | 4 | 36 | 8 | 0.83 | ||
| 1996 | 4 | 18 | 8 | 0.83 | ||||
| 1997 | 4 | 26 | 6 | 0.78 | 0.87 | 113.5 (0.001) | ||
| Villavicencio | 1995 | 4 | 24 | 10 | 0.81 | |||
| 1996 | 4 | 13 | 5 | 0.91 | ||||
| 1997 | 2 | 39 | 7 | 0.64 | ||||
| 1998 | 4 | 55 | 9 | 0.62 | ||||
| 1999 | 4 | 88 | 10 | 0.7 | 0.80 | 201.1 (0.001) | ||
| 5 | Mondomo | 1996 | 4.4 | 52 | 6 | 0.46 | ||
| 1997 | 3.7 | 49 | 4 | 0.65 | ||||
| 1998 | 4 | 21 | 4 | 0.69 | ||||
| 1999 | 2 | 35 | 4 | 0.69 | 0.73 | 120.1 (0.001) | ||
| Cajibio | 1995 | 3 | 22 | 3 | 0.26 | |||
| 1996 | 3.5 | 27 | 3 | 0.15 | 0.22 | 2.7 (0.446) | ||
| 2-4 | Santander de Quilichao | 1996 | 2 | 6 | 4 | 0.30 | ||
| 1997 | 3.5 | 60 | 3 | 0.26 | 0.58 | 66 (0.001) | ||
Site of collection of the isolates; six sites were visited in four ECZs.
Year of collection of the isolates.
Estimates of disease incidence were made in each field during the sampling, as recommended by Boher and Agboli (5).
Number of isolates collected in each site and each year.
Number of RFLP haplotypes detected in each site and each year.
Genetic diversity of the X. axonopodis pv. manihotis strains collected in 1 year.
Genetic diversity of the X. axonopodis pv. manihotis strains collected in one site in all years.
The chi-square value was significant at the 5% level.
Estimates of disease incidence were made in each field during the sampling, as recommended by Boher and Agboli (5). Disease incidence was recorded by randomly selecting 15 plants of each cultivar for each field and rating them on a 1 to 5 scale as follows: 1, no disease symptoms; 2, presence of angular leaf spots; 3, wilting of leaves and exudates; 4, leaf wilting, exudates, and wilting of some shoots (dieback); and 5, dieback of all shoots. Other parameters recorded included rainfall (millimeters) and minimum and maximum day-night temperatures (degrees Celsius). These were continuously monitored from 1995 to 1999 in each location and provided by IDEAM (Instituto de Hidrologia y Meteorologia y Estudios Ambientales, Bogotá, Colombia).
Characterization of isolates. (i) RFLP with pthB as probe.
Genomic DNA extractions, digestion, electrophoresis, and Southern blotting were performed as previously described (24). Membranes were hybridized with a plasmid probe, designated pthB, corresponding to a 5.6-kb EcoRI fragment. DNA sequencing of this fragment showed that the pthB gene is a member of the Xanthomonas avr-pth gene family harboring 12.5 internal 102-bp repeats and flanked by near-identical 980-bp direct repeats. The nucleotide sequence of the pthB gene is available in the GenBank database (accession number AF012325). Conditions of hybridization and washes were as previously described (24).
(ii) Virulence patterns.
Virulence patterns of 96 strains representing the 45 different RFLP haplotypes described in ECZs 1, 2, 2-4, and 5 from 1995 to 1999 were determined. Strains were inoculated as previously described on a set of differential cultivars previously selected for ECZs 1 and 2 (20). Briefly, plants were arranged in the greenhouse according to a randomized complete block design with five replicates. Noninoculated plants were included in each experiment as controls. Cassava stems were inoculated by inserting a toothpick contaminated by passage through a 24-h-old culture of the appropriate isolate. Symptoms were monitored 7, 14, and 30 days after inoculation, and disease was rated according to the following scale: 0, healthy plant, no reaction observed; 1, dark area or necrosis around the inoculation point; 2, gum exudates on the stem; 3, wilting of one or two leaves and exudates; 4, more than two leaves wilted; and 5, complete wilting and dieback. Cultivars with at least one rating of 4 or higher in a replicate were considered susceptible (20).
The cassava differential cultivars were the same as previously used for the characterization of pathotypes in ECZs 1 and 2 (20). For ECZ 5, we used three varieties previously studied (20) and included a larger set of cassava cultivars which had been selected for their adaptation to ECZ 5 (CM7436-7, CM7438-1, MBRA383, MCOL2061, SM1846-12, SM524-1, SM616-22, and CM7595-1). For ECZ 2-4, nine cultivars were selected based on their adaptation to the ECZ conditions (CM523-7, CM2177-2, CM6740-3, SM1219-9, CM7951-5, CM3306-4, CM7514-7, and CM849-1).
Statistical analysis.
Each distinct RFLP banding pattern, obtained after hybridization with probe pthB, was regarded as a haplotype. The haplotype diversity was estimated using the Nei and Tajima index (16), H = [n/(n − 1)](1 − ΣXi2), where Xi is the proportion of the ith distinct pthB haplotype and n is the number of strains. Significance testing for differences in haplotype frequencies over years in each site or ECZ was performed by a chi-square analysis.
Three analyses of molecular variance (AMOVAs) (Arlequin version 1.1; S. Schneider, J. M. Kruffer, D. Roessli, and L. Excoffier, Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland [http://lgb.unige.ch/arlequin/]) were performed. First, AMOVA was used to estimate variance components for RFLP-pthB haplotypes, partitioning the variation among ECZs and fields within ECZs and among fields. A second AMOVA was performed to study changes of X. axonopodis pv. manihotis population structure over a cropping cycle. The variation was partitioned into two populations, Mondomo and Pivijay, each including the two samplings performed at the beginning and at the end of the crop cycle. A third AMOVA was performed in order to assess if the genetic structure of X. axonopodis pv. manihotis had changed over the 5 years under study; strains collected within a year were considered a single population. The analysis was repeated for each ECZ. Genetic distance, calculated as Euclidean metric distance, was computed between all strains and between locations. The Euclidean metric distance between two individuals is equivalent to their total number of observed band differences for RFLP-pthB analysis.
RESULTS
From 1995 to 1999, 45 different RFLP types or haplotypes were detected among 736 strains in Colombia. Ten different haplotypes were detected in ECZ 1, 26 were detected in ECZ 2, 8 were detected in ECZ5, and 7 were detected in ECZ 2-4. A total of 29 bands, ranging from 2.8 to 32 kb, were scored, and one to nine distinct bands were observed per strain (data not shown). In general, in each ECZ a distinct group of haplotypes was detected, confirming the geographical differentiation of the X. axonopodis pv. manihotis strains in Colombia (24). There were four exceptions, with haplotype C13 being found in fields of different ECZs (ECZs 2, 5, and 2-4), C8 being present in ECZ 5 and ECZ 2-4, C32 being present in ECZs 2 and 5, and C28 and C29 being present in ECZ 2 and ECZ 2-4.
The analysis of X. axonopodis pv. manihotis strains collected from 1995 to 1999 revealed extensive genetic variability among subpopulations of X. axonopodis pv. manihotis in different ECZs. Genetic diversities calculated for each site were highest for Carimagua and Villavicencio (ECZ 2) and lowest for Cajibio (ECZ 5) and Santander de Quilichao (ECZ 2-4) (Table 1). Except in Mondomo, genetic diversities decreased over time (Table 1). The AMOVA showed that 55.4% of the haplotype diversity was within locations, 34.8% was among ECZs, and 9.8% was among locations within ECZs. In order to determine if the population structure changed over time, an AMOVA was performed taking all isolates collected in 1 year in an ECZ as a population. More variation was ascribed to differences within the years than to differences among the years for all the ECZs, indicating that genetic diversity was maintained from year to year. The percentages of variation due to differences among and within years of collection were 13.6 and 86.4 for ECZ 2, 17.7 and 82.3 for ECZ 1, 41.6 and 58.4 for ECZ 2-4, and 42.2 and 57.8 for ECZ 5, respectively.
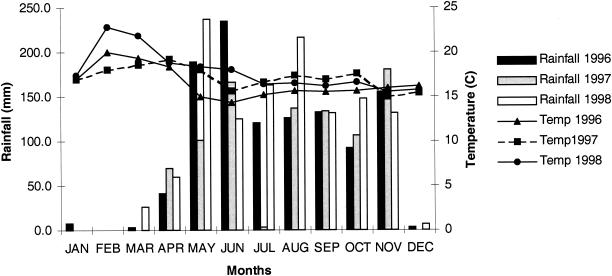
In some cases, disease incidence index was the average of data collected during various samplings in the same field. Disease severity varied from year to year in each ECZ. The most dramatic variation was observed in ECZ 1. In 1997 disease symptoms were rarely observed and disease severity was very low (disease severity of 1) compared to 1996 (disease severity of 3) and 1999 (disease severity of 4,5) (Table 1). The origin of this variation is not known and is difficult to estimate. However, this variation in the disease severity index in ECZ 1 can be correlated with the climatic conditions observed in ECZ 1 in 1997. Differences between minimum and maximum temperatures were low at the beginning of the year, and the amount of rainfall in July was significantly lower, almost nonexistent, compared to 1996 and 1998 (Fig. 2).
FIG. 2.
Rainfall and difference between minimum and maximum temperatures at Pivijay (ECZ 1) in 1996, 1997, and 1998.
Two fields were further selected to study the genetic change of the X. axonopodis pv. manihotis population during one crop cycle. During the first sampling, conducted 3 months after plantings, 36 X. axonopodis pv. manihotis strains were isolated from infected tissues at Mondomo and 37 X. axonopodis pv. manihotis strains at Pivijay. During the second sampling, performed 7 months after plantings, we isolated 17 X. axonopodis pv. manihotis strains at Mondomo and 26 X. axonopodis pv. manihotis strains at Pivijay. Also, four haplotypes were described in the first sampling at Mondomo (C8, C9, C36, and C37) and eight were described at Pivijay (C6, C16, C18, C33, C40, C42, C43, and C44) (Table 2). The haplotypes most frequently found at Mondomo were C8 (41.7%) and C9 (38.9%). At Pivijay the frequency of haplotype C18 was relatively high (62.2%). The other haplotypes occurred at frequencies of less than 14%. In the second sampling, carried out 7 months after planting, the same four haplotypes were detected at Mondomo, while at Pivijay only three haplotypes were detected (C6, C18, and C42) (Table 2). Haplotype frequencies at Mondomo were 37.5% for both C8 and C37 and 25% for both C9 and C36. At Pivijay, frequencies were 62.2% for C18, 34.6% for C42, and 3.2% for C6. Chi-square analysis was used to compare haplotype frequencies over the crop cycle, and results show that they are statistically significant (Table 2). Genetic diversity (H) for each field was 0.71 (Mondomo) and 0.58 (Pivijay). The genetic diversity values showed that diversity increased at Mondomo during a cropping cycle while diversity at Pivijay was lower at the end of the cropping cycle. For the AMOVA, two groups were defined: group 1 comprised Pivijay sampling 1 and Pivijay sampling 2 while group 2 comprised Mondomo sampling 1 and Mondomo sampling 2. Of the variation observed, 31% was among populations, 61% was within samplings, and only 8% was among samplings within populations.
TABLE 2.
Haplotypes and genetic diversities (H) of the strains of X. axonopodis pv. manihotis populations collected in two samplings during a cycle of production
| Site | Samplinga | pthB haplotypes | χ2b | Genetic diversity (H)c |
|---|---|---|---|---|
| Mondomo | 1 | C8, C9, C36, C37 | 0.67 | |
| 2 | C8, C9, C36, C37 | 14.095 (0.003) | 0.74 | |
| Pivijay | 1 | C6, C16, C18, C33, C40, C42, C43, C44 | 0.60 | |
| 2 | C6, C18, C42 | 18.293 (0.011) | 0.52 |
First or second sampling during the cropping cycle. The first sampling was performed 4 months after planting, and the second one was 7 months after planting.
The chi-square value was significant at the 5% level.
Diversity index, based on the RFLP haplotypes, was calculated as described by Nei and Tajima (16).
In all, 96 strains were tested for virulence on a set of differential cultivars adapted to each ECZ. Pathotypes present in ECZs 1, 2, 5, and 2-4 and from 1995 to 1999 are described in Tables 3 to 6, respectively. Nine different pathotypes were defined among the ECZ 1 strains, based on the reactions following inoculation on six cassava cultivars adapted to this ECZ (Table 3). The five cultivars adapted to ECZ 2 were able to distinguish 13 pathotypes among the set of X. axonopodis pv. manihotis strains tested (Table 4). New pathotypes that were not detected in a previous study (20) were described in ECZs 1 and 2 (Tables 3 and 4). In ECZ 5 four pathotypes were identified (Table 5). For ECZ 2-4, a set of cassava differentials was proposed allowing the characterization of three pathotypes in this ECZ (Table 6). All pathotypes from ECZs 5 and 2-4 were highly virulent, producing susceptible reactions on almost all cultivars.
TABLE 3.
X. axonopodis pv. manihotis pathotypes defined in ECZ 1
| Pathotypea | Haplotype(s) | Resistance of cassava cultivarb
|
|||||
|---|---|---|---|---|---|---|---|
| MBRA685 | MCOL22 | MVEN25 | MCOL1505 | MBRA12 | MNGA2 | ||
| C1-1 | C10, C16, C15, C6, C18, C42 | S | S | S | S | S | S |
| C1-2 | C11, C5, C16, C40 | R | S | S | S | S | R |
| C1-3 | C14, C16, C33, C18, C43, C44, C6, C15 | R | S | S | S | S | S |
| C1-4 | C18 | S | S | S | R | S | R |
| C1-5 | C6 | R | R | S | R | R | R |
| C1-6 | C33, C6 | R | S | S | S | R | R |
| C1-7 | C6 | S | S | S | S | R | S |
| C1-8 | C42 | R | R | S | S | S | R |
| C1-9 | C18 | S | S | S | S | S | R |
Pathotypes defined after the analysis of the reactions between strains and cultivars originated from the same ECZ.
S, susceptible; R, resistant. A cultivar was considered susceptible if it generated symptoms in classes 4 and 5 in at least one out of five plants.
TABLE 6.
X. axonopodis pv. manihotis pathotypes defined in ECZ 2-4
| Pathotypea | Haplotype(s) | Resistance of cassava cultivarb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CM523-7 | CM2177-2 | CM6740-3 | SM1219-9 | CM849-1 | CM7951-5 | CM3306-4 | CM7514-7 | ||
| C2-4-1 | C13, C28, C29, C8 | S | S | S | S | S | S | S | S |
| C2-4-2 | C30 | S | S | S | R | S | S | S | S |
| C2-4-3 | C35 | S | R | S | S | S | S | S | S |
Pathotypes defined after the analysis of the reactions between strains and cultivars originated from the same ECZ.
S, susceptible; R, resistant. A cultivar was considered susceptible if it generated symptoms in classes 4 and 5 in at least one out of five plants.
TABLE 4.
X. axonopodis pv. manihotis pathotypes defined in ECZ 2
| Pathotypea | Haplotype(s) | Resistance of cassava cultivarb
|
||||
|---|---|---|---|---|---|---|
| CM5286-3 | CM2177-2 | MBRA902 | CM6438-14 | CM523-7 | ||
| C2-1 | C1, C2, C13, C12, C34, C17, C29 | S | S | S | R | S |
| C2-2 | C3, C12 | S | S | R | S | S |
| C2-3 | C20, C12 | R | S | S | R | S |
| C2-4 | C4, C21, C12, C27 | S | S | R | R | S |
| C2-5 | C19, C22, C23, C17, C24, C26, C25, C13, C28, C29, C32, C4, C27, C45, C50 | S | S | S | S | S |
| C2-6 | C17 | S | R | S | S | S |
| C2-7 | C17 | S | R | S | R | S |
| C2-8 | C4, C47 | R | S | R | R | R |
| C2-9 | C19, C46 | S | R | R | R | S |
| C2-10 | C23, C26 | S | S | S | R | R |
| C2-11 | C27 | R | R | R | R | R |
| C2-12 | C27 | S | R | S | S | R |
| C2-13 | C51 | R | R | S | R | S |
Pathotypes defined after the analysis of the reactions between strains and cultivars originated from the same ECZ.
S, susceptible; R, resistant. A cultivar was considered susceptible if it generated symptoms in classes 4 and 5 in at least one out of five plants.
TABLE 5.
X. axonopodis pv. manihotis pathotypes defined in ECZ 5
| Pathotypea | Haplotype(s) | Resistance of cassava cultivarb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CG402-11 | MCOL1522 | MCOL2261 | CM7436-7 | CM7438-1 | MBRA383 | MCOI2061 | SM1846-12 | SM616-22 | CM7595-1 | SM524-1 | ||
| C5-1 | C8, C37, C36, C9 | S | S | S | S | S | S | S | S | S | S | S |
| C5-2 | C7 | S | S | S | S | S | S | S | S | S | S | R |
| C5-3 | C36 | S | S | S | S | S | S | S | R | S | S | S |
| C5-4 | C9 | S | S | S | R | S | S | S | R | S | S | R |
Pathotypes defined after the analysis of the reactions between strains and cultivars originated from the same ECZ.
S, susceptible; R, resistant. A cultivar was considered susceptible if it generated symptoms in classes 4 and 5 in at least one out of five plants.
A striking result is that in three ECZs (1, 2) new pathotypes were detected after 1997 (Table 7). Genetic changes in X. axonopodis pv. manihotis populations after 1997 led to the hypothesis either that new pathotypes appeared or that pathotypes already present could be detected because of their increased frequencies. An example can be drawn from ECZ 1. In ECZ 1, after 1997, haplotype C18 became the predominant haplotype and a new highly virulent pathotype (C1-9), belonging to haplotype C18, was detected (Table 7). Some pathotypes were characterized from 1995 to 1999 while others occurred only in a specific year (e.g., C1-6, C1-7, C1-8, and C1-9) (Table 7). In each ECZ, the most frequent pathotype was always the one showing the highest number of compatible interactions (Tables 3 to 6 and 7). These were pathotypes C1-1 in ECZ 1, C2-5 in ECZ 2, C5-1 in ECZ 5, and C2-4-1 in ECZ 2-4. There is no clear correlation between haplotypes and pathotypes, with strains from a specific haplotype belonging to different pathotypes (e.g., haplotype C42 belonging to pathotypes C1-1 and C1-9). One pathotype could also be represented by strains belonging to different haplotypes (e.g., C1-1 grouped C6, C10, C15, C16, C18, and C42) (Table 3). These results must be taken with caution, as we selected a subset of X. axonopodis pv. manihotis strains to be further characterized for their virulence patterns.
TABLE 7.
Distribution of pathotypes of X. axonopodis pv. manihotis in Colombia during 1995 to 1999
| ECZ | Pathotype | Result by yra
|
||||
|---|---|---|---|---|---|---|
| 1995 | 1996 | 1997 | 1998 | 1999 | ||
| 1 | C1-1 | + | NT | + | + | + |
| C1-2 | + | NT | ND | + | ND | |
| C1-3 | + | NT | + | + | ND | |
| C1-4 | + | NT | ND | ND | ND | |
| C1-5 | + | NT | ND | ND | ND | |
| C1-6b | ND | NT | ND | + | ND | |
| C1-7b | ND | NT | ND | ND | + | |
| C1-8b | ND | NT | ND | ND | + | |
| C1-9b | ND | NT | ND | ND | + | |
| 2 | C2-1 | + | ND | + | + | ND |
| C2-2 | + | ND | ND | ND | + | |
| C2-3 | + | ND | ND | + | ND | |
| C2-4 | + | ND | ND | ND | + | |
| C2-5 | + | + | + | + | + | |
| C2-6b | ND | ND | + | ND | ND | |
| C2-7b | ND | ND | ND | ND | + | |
| C2-8b | ND | ND | + | + | ND | |
| C2-9b | ND | ND | ND | + | ND | |
| C2-10b | ND | + | ND | ND | ND | |
| C2-11b | ND | ND | + | ND | ND | |
| C2-12b | ND | ND | ND | ND | + | |
| C2-13b | ND | ND | ND | ND | + | |
| 5 | C5-1 | + | NT | + | + | NT |
| C5-2b | + | NT | ND | ND | NT | |
| C5-3b | ND | NT | + | ND | NT | |
| C5-4b | + | NT | ND | ND | NT | |
| 2-4 | C2-4-1b | NT | + | ND | NT | NT |
| C2-4-2b | NT | ND | + | NT | NT | |
| C2-4-3b | NT | ND | + | NT | NT | |
X. axonopodis pv. manihotis strains were not collected in 1995 and 1999 (ECZ 2-4). They were collected but not tested for virulence in 1996 (ECZ 1 and ECZ 5), 1998 (ECZ 2-4), and 1999 (ECZ 5). +, detected; ND, not detected; NT, not tested or X. axonopodis pv. manihotis strains not available.
New pathotype (20).
DISCUSSION
Based on information on the spatial and temporal distribution of the pathogen's genetic diversity, we can develop a picture of the CBB pathogen's population structure and dynamics in Colombia. In previous studies, several molecular markers (repetitive fragment PCR, amplified fragment length polymorphism, and ribotyping) were tested for their utility in characterizing X. axonopodis pv. manihotis populations (3, 19, 23, 24, 28). RFLP was found to be the most informative and discriminative technique for assessing the genetic diversity of X. axonopodis pv. manihotis. For this reason, it was the technique selected to detect spatial and temporal changes in the X. axonopodis pv. manihotis population structure.
In a previous study we identified migrations as a major factor influencing the pathogen population structure and maintaining the high genetic diversity (24). In this study RFLP variation in the X. axonopodis pv. manihotis populations indicated that a high level of variation at the DNA level has remained constant over time and we confirmed that migrations continue to shape the X. axonopodis pv. manihotis population structure. The presence of identical RFLP patterns in several fields of the same or different ECZs indicated pathogen migration. AMOVA confirmed the existence of migrations of strains between locations of the same ECZ. However, in comparing results obtained in 1995 and 1996 (24) with results from this study, it becomes evident that, over time, bacterial populations in sites of the same ECZ become more divergent and populations from different ECZs become less divergent. This change is a consequence of the increasing rate of migration of the pathogen among sites located in different ECZs. If this situation persists, cultivars to be deployed will have to be tested not only with the strains representing the pathotypes found in one ECZ but also with pathotypes from various ECZs.
Genetic changes can be subtle, resulting in simple numerical changes of haplotypes or pathotypes, or be dramatic and affect the pathogen population structure. Contingency chi-square tests showed that differences in haplotype frequencies over the 4 years were statistically significant (Table 1), indicating that X. axonopodis pv. manihotis populations at the site level were unstable. These changes in haplotype frequencies occur rapidly, in less than a year, and during a crop cycle a new population structure is established. In addition, AMOVA showed that genetic variation was higher within a year than among years, confirming how rapidly X. axonopodis pv. manihotis populations are able to evolve. We therefore recommend monitoring the pathogen population each year at the end of the crop cycle.
The influence of host selection or environmental factors on the X. axonopodis pv. manihotis population structure is difficult to estimate. However, as previously mentioned some correlations can be drawn between changes in the population structure and changes in the host population or in the climatic conditions. In fact, in a previous study (20) a correlation was established between host diversity and X. axonopodis pv. manihotis variation. The management and deployment of new cassava resistant genotypes had a significant impact on the population structure of X. axonopodis pv. manihotis over time. In Mondomo (ECZ 5), changes in haplotype frequencies were probably influenced by the change in the host population and the introduction of new cassava genotypes. During our first study in 1995 to 1996, only two varieties were cultivated in farmers' fields. The introduction of new genotypes was followed by the increase in haplotype diversity (Table 1) (20, 24). The same situation occurred in Villavicencio (ECZ 2) (Table 1), where populations changed after the introduction in 1997 of a pair-cross population of 150 individuals derived from an intraspecific cross between two noninbred cassava (M. esculenta Crantz) lines (11). In ECZs 2 and 5, weather conditions were very similar over time (data not shown), and changes in X. axonopodis pv. manihotis population structure may not be explained by changes in the environmental conditions alone.
Environmental factors may also have a major influence on disease development and population fluctuations, causing a genetic drift of the population. Temperature and rainfall are the most important factors that affect the severity of CBB (14, 25). The epiphytic phase of X. axonopodis pv. manihotis has important implications in the epidemiology of the disease, and the growth of epiphytical bacterial populations is favored by high humidity and large differences in day-night temperatures (7). In 1997 weather conditions were unfavorable for the development of the disease because of low rainfall as a consequence of the El Niño phenomenon. Consequently a very low percentage of plants showed symptoms at Pivijay (21) and the disease severity index was particularly low. We observed that some haplotypes disappeared while new haplotypes and pathotypes were detected. The predominant haplotype also changed. In Pivijay (ECZ 1) genetic drift had a significant impact on pathogen population structure.
The molecular analysis of diversity showed the potential for X. axonopodis pv. manihotis populations to change. If the haplotypic composition of the population changed during the crop cycles, the pathotypic composition could also change. We showed that the pathotypic composition really changed and the majority of new pathotypes appeared in ECZs 1 and 2 after 1997. As previously stated, a monitoring of the population at the end of the crop cycle and the resistance screening of cassava varieties with the new pathotypes will be the recommended strategy. Instability is the general rule in X. axonopodis pv. manihotis populations, as shown in this study, and genetics had and probably will continue to have a significant impact on the X. axonopodis pv. manihotis population structure. After 1997 in ECZ 1, some haplotype C6 strains that were dominant before 1997 persisted, exhibiting rare pathotypes. Because of this situation we believe that more than one resistance genotype should be deployed at the same time to prevent these rare pathotypes that are potential sources of resistance breakdown from becoming dominant.
In ECZs 5 and 2-4, the high level of virulence showed by the ECZ 5 and 2-4 strains suggests a need for characterizing more suitable sources of resistance. For ECZ 2 more differential cultivars are needed, first because X. axonopodis pv. manihotis populations in ECZ 2 are highly variable and second because, with five differential cultivars, only a limited number of pathotypes can be described. It was the case with ECZ 5, where in a previous study only one pathotype was described (20). By increasing the number of differential cultivars, we characterize new pathotypes that have been present since 1995. In general, results did not show a clear correlation between haplotypes and pathotypes. It could be interesting to know whether the molecular diversity in the avr gene structure of X. axonopodis pv. manihotis may account for the phenotypic diversity observed. The RFLP-EcoRI analysis performed in this study may not detect changes in the copies of the avr genes that are causing a variation in the virulence patterns of the strains. Changes in the avr structure that cause a variation in the virulence pattern of the strain can be due to single mutations at the 3′ terminus of the avrXa7 alleles (4, 26). One strategy could be to sequence the copies of the plasmid-borne avr gene copies in the new pathotypes. It could also be interesting to determine if some of these avr copies in X. axonopodis pv. manihotis have a measurable fitness function.
This study is a part of our continuing effort to understand the population structure of the X. axonopodis pv. manihotis. Knowledge of pathogen population structure and dynamics is useful in deciding the way in which genotypes are deployed in time and space (13). As shown above, a molecular monitoring of the haplotypic composition of the population reflects a change of the pathotypes in each ECZ. If new pathotypes appear in the populations, these must be characterized and included in the set of strains used to screen cultivars to be deployed.
Monitoring the pathogen population requires extensive and repeated surveys. The development and recent application of geographical information systems, geostatistics, and the Kriging technique to plant pathology offer an alternative approach to predict and monitor changes in plant pathogen populations (12, 17). For the future it is important to develop a spatial simulation model to understand the influence of various ecological and genetic factors on the distribution and potential change of X. axonopodis pv. manihotis population structure. Recently we developed a database containing information on pathogen diversity and CBB incidence, host diversity and distribution, and climatic conditions for all the ECZs. This is the first step towards a better understanding of the biology and management of CBB.
Acknowledgments
We thank IDEAM for providing the climatic information for this study. We are grateful to Joe Tohme for his support and to C. Mba and G. Mahuku for critically reading the manuscript.
This research was supported by grants from IRD and CIAT and by a doctoral fellowship awarded to S. Restrepo by the Institut de la Recherche pour le Développement (IRD).
REFERENCES
- 1.Anonymous. 1983. Annual report. Centro Internacional de Agricultura Tropical, Cali, Colombia.
- 2.Anonymous. 1987. Annual report. Centro Internacional de Agricultura Tropical, Cali, Colombia.
- 3.Assigbetsé, K., V. Verdier, K. Wydra, K. Rudolph, and J. P. Geiger. 1998. Genetic variation of the cassava bacterial blight pathogen, Xanthomonas campestris pv. manihotis, originating from different ecoregions in Africa, p. 223-229. In IX International Conference on Plant Pathogenic Bacteria, Madras, India.
- 4.Bai, J., S.-H. Choi, G. Ponciano, H. Leung, and J. E. Leach. 2000. Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol. Plant-Microbe Interact. 13:1322-1329. [DOI] [PubMed] [Google Scholar]
- 5.Boher, B., and C. A. Agboli. 1992. La bactériose vasculaire du manioc au Togo: caractérisation du parasite, répartition géographique et sensibilité variétale. Agron. Trop. 46:131-136. [Google Scholar]
- 6.Boher, B., and V. Verdier. 1995. Cassava bacterial blight in Africa: the state of knowledge and implications for designing control strategies. Afr. Crop Sci. J. 2:1-5. [Google Scholar]
- 7.Daniel, J. F., and B. Boher. 1985. Etude des modes de survie de l'agent causal de la bacterie vasculaire vasculaire du manioc, Xanthomonas campestris pathovar manihotis. Agronomie 5:239-246. [Google Scholar]
- 8.Fregene, M., F. Angel, R. Gomez, F. Rodriguez, P. Chavarriaga, W. Roca, J. Tohme, and M. Bonierbale. 1997. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 95:431-441. [Google Scholar]
- 9.Hahn, S. K. 1978. Breeding cassava for resistance to bacterial blight. PANS 24:480-485. [Google Scholar]
- 10.Jorge, V., M. Fregene, M. C. Duque, M. W. Bonierbale, J. Tohme, and V. Verdier. 2000. Genetic mapping of resistance to bacterial blight disease in cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 101:865-872. [Google Scholar]
- 11.Jorge, V., M. Fregene, C. M. Velez, M. C. Duque, J. Tohme, and V. Verdier. 2001. QTL analysis of field resistance to Xanthomonas axonopodis pv. manihotis in cassava. Theor. Appl. Genet. 102:564-571. [Google Scholar]
- 12.Lecoustre, R., D. Fargette, C. Fauquet, and P. De Reffye. 1989. Analysis and mapping of the spatial spread of African cassava mosaic virus using geostatics and the Kriging technique. Phytopathology 79:913-920. [Google Scholar]
- 13.Leung, H., R. J. Nelson, and J. E. Leach. 1993. Population structure of plant pathogenic fungi and bacteria. Adv. Plant Pathol. 10:157-205. [Google Scholar]
- 14.Lozano, J. C. 1986. Cassava bacterial blight: a manageable disease. Plant Dis. 70:1089-1093. [Google Scholar]
- 15.Lozano, J. C., and L. Sequeira. 1974. Bacterial blight of cassava in Colombia: epidemiology and control. Phytopathology 64:83-88. [Google Scholar]
- 16.Nei, M., and F. Tajima. 1981. DNA polymorphism detectable by restriction endonucleases. Genetics 97:145-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, M. N., T. V. Orum, R. Jaime-Garcia, and A. Nadeem. 1999. Applications of geographic information systems and geostatics in plant disease epidemiology and management. Plant Dis. 83:308-319. [DOI] [PubMed] [Google Scholar]
- 18.Restrepo, S. 1999. Etude de la structure des populations de Xanthomonas axonopodis pv. manihotis en Colombie. Ph.D. thesis. Université Paris VI, Paris, France.
- 19.Restrepo, S., M. C. Duque, J. Tohme, and V. Verdier. 1999. AFLP fingerprinting: an efficient technique for detecting genetic variation of Xanthomonas axonopodis pv. manihotis. Microbiology 145:107-114. [DOI] [PubMed] [Google Scholar]
- 20.Restrepo, S., M. C. Duque, and V. Verdier. 2000. Characterization of pathotypes among isolates of Xanthomonas axonopodis pv. manihotis in Colombia. Plant Pathol. 49:680-687. [Google Scholar]
- 21.Restrepo, S., M. C. Duque, and V. Verdier. 2000. Resistance spectrum of selected Manihot esculenta genotypes under field conditions. Field Crops Res. 65:69-77. [Google Scholar]
- 22.Restrepo, S., T. Valle, M. C. Duque, and V. Verdier. 1999. Assessing genetic variability among Brazilian strains of Xanthomonas axonopodis pv. manihotis through RFLP and AFLP analyses. Can. J. Microbiol. 45:754-763. [Google Scholar]
- 23.Restrepo, S., C. M. Velez, and V. Verdier. 2000. Measuring the genetic diversity of Xanthomonas axonopodis pv. manihotis within different fields in Colombia. Phytopathology 90:683-690. [DOI] [PubMed] [Google Scholar]
- 24.Restrepo, S., and V. Verdier. 1997. Geographical differentiation of the population of Xanthomonas axonopodis pv. manihotis in Colombia. Appl. Environ. Microbiol. 63:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takatsu, A., S. Fukuda, and S. Perrin. 1978. Epidemiological aspects of bacterial blight of cassava in Brazil, p. 141-150. In H. Maraite and Y. A. Meyer (ed.), Diseases of tropical food crops. Université Catholique de Louvain, Louvain-la-Neuve, Belgium.
- 26.Vera Cruz, C. M., J. Bai, I. Ona, H. Leung, R. J. Nelson, T.-W. Mew, and J. E. Leach. 2000. Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. USA 97:13500-13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdier, V., B. Boher, H. Maraite, and J. P. Geiger. 1994. Pathological and molecular characterization of Xanthomonas campestris strains causing diseases of cassava (Manihot esculenta). Appl. Environ. Microbiol. 60:4478-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdier, V., P. Dongo, and B. Boher. 1993. Assessment of genetic diversity among strains of Xanthomonas campestris pv. manihotis. J. Gen. Microbiol. 139:2591-2601. [Google Scholar]
- 29.Verdier, V., S. Restrepo, G. Mosquera, M. C. Duque, A. Gerstl, and R. Laberry. 1998. Genetic and pathogenic variation of Xanthomonas axonopodis pv. manihotis in Venezuela. Plant Pathol. 47:601-608. [Google Scholar]
- 30.Wydra, K., and V. Verdier. 2002. Occurrence of cassava diseases in relation to environmental, agronomic and plant characteristics. Agric. Ecosyst. Environ. 93:211-226. [Google Scholar]