Abstract
Although the precise biochemical function of DJ-1 remains unclear, it has been found to exert cytoprotective activity against oxidative stress. Cys106 is central to this function since it has a distinctly low pKa rendering it extremely susceptible for oxidation. This characteristic, however, also poses a severe hindrance to obtain reduced DJ-1 for in vitro investigation. We have developed an approach to produce recombinant human DJ-1 in its reduced form as a bona fide basis for exploring the redox capacities of the protein. We solved the crystal structure of this DJ-1 at 1.56 Å resolution, allowing us to capture Cys106 in the reduced state for the first time. The dimeric structure reveals one molecule of DJ-1 in its reduced state while the other exhibits the characteristics of a mono-oxygenated cysteine. Comparison with previous structures indicates the absence of redox dependent global conformational changes in DJ-1. The capture of reduced Cys106 is facilitated by stabilization within the putative active site achieved through a glutamate side chain. This side chain is provided by a crystallographic neighbor as part of a ‘Leu-Glu’ motif, which was added to the C-terminus of DJ-1. In the structure this motif binds DJ-1 in close proximity to Cys106 through extended hydrophilic and hydrophobic interactions depicting a distinct binding pocket, which can serve as a basis for compound development targeting DJ-1.
Keywords: oxidative stress, crystal structure of reduced DJ-1, reduced putative active site cysteine, (putative) active site binding, protecting DJ-1 oxidation, compound template
1. DJ-1: Cys106 and reactive oxygen species
Reactive oxygen species (ROS) are formed in cellular processes that regulate metabolism, host defense, and apoptosis (Thannickal and Fanburg, 2000). The machinery of antioxidant enzymes and small molecule scavengers not only detoxify excess ROS and protect against cellular oxidative stress buildup but also maintain intracellular reducing environment (Mathers et al., 2004). Dysregulated production of ROS due to genetic defects and environmental factors have been implicated as a major contributing factor for cancer, neurodegenerative diseases and aging (Migliore and Coppede, 2002).
A small 21 kDa DJ-1 (also known as PARK7) has been implicated in protection against ROS (Takahashi-Niki et al., 2004; Yokota et al., 2003) and is over expressed in multiple cancer settings, including lung, prostate and breast cancer (Hod, 2004; MacKeigan et al., 2003; Nagakubo et al., 1997). Over expression of DJ-1 was shown to effect cell survival and impart cytoprotection against oxidative damage of cancer cell lines by modulating the PI3K/PTEN/AKT pathway (Kim et al., 2005). Additionally, DJ-1 has been implicated as a target protein for certain sperm toxicants and has been assigned an essential role in mouse fertilization (Klinefelter et al., 1997; Klinefelter et al., 2002; Wagenfeld et al., 1998). Genetic mutations in DJ-1 were demonstrated to contribute to the early-onset of familial Parkinson’s disease (Abou-Sleiman et al., 2004; Bekris et al., 2010; Moore et al., 2005). Recently, exposure to herbicides such as rotenone and paraquat were implicated to cause sporadic form of Parkinson’s disease by affecting DJ-1 oxidative status in dopaminergic neuronal cells (Menzies et al., 2005; Meulener et al., 2005). These studies, despite the fact that the precise biochemical function of the protein still remains unclear, suggest that DJ-1 is an oxidative stress response protein (Hao et al., 2010; Lev et al., 2006; McCoy and Cookson, 2011).
Structurally, human DJ-1 folds into a three-layered α-β-α sandwich architecture, which assembles to form a homodimer (Tao and Tong, 2003; Wilson et al., 2003). The overall fold of DJ-1 is conserved from bacteria to humans and is structurally classified into the class I glutamine amidotransferase family.
Human DJ-1 contains three cysteine residues (Cys46, Cys53 and Cys106). The most evolutionarily conserved cysteine, Cys106, is critical for DJ-1’s cytoprotective function and is situated at the tip of a sharp turn between a β-strand and α-helix, which commonly bears the catalytic nucleophile in α/β hydrolases (Heikinheimo et al., 1999; Wilson, 2011). The most notable feature of DJ-1 Cys106 is its depressed pKa value of 5.4 rendering its thiol moiety highly susceptible for oxidative modification (Witt et al., 2008). This is exemplified by previous high-resolution crystal structures of DJ-1 that commonly show oxidation of Cys106 to its sulfinic acid form (Blackinton et al., 2009; Canet-Aviles et al., 2004). The high susceptibly of DJ-1 to oxidation furthermore causes severe difficulties in obtaining reduced DJ-1 as basis to study DJ-1 function and drug compound development targeting the DJ-1 putative active site.
Here we present a method for obtaining reduced DJ-1 as well as the crystal structure of DJ-1 with Cys106 preserved in its reduced state by an artificial ‘Leu-Glu’ motif revealing new means of cysteine stabilization and a basis for targeting the putative active site of DJ-1.
2. Details of expression, purification, mass spectrometry, and crystal structure determination
Full-length human DJ-1 (189 aa) was cloned into NdeI and XhoI sites of pET29a, which encodes a tag-less DJ-1 with vector-derived Leu-Glu residues at the carboxy-terminus (191 aa). The DJ-1 was expressed in BL21(DE3) cells by induction with 0.2 mM IPTG at 20°C overnight. Bacterial cell pellets were suspended in 10 ml lysis buffer (50 mM MES pH 5, 1mM TCEP, 300U DNaseI, 3.5 mM MgCl2, 0.4 mg/ml lysozyme) and the cells were disrupted by sonication. Following centrifugation, the supernatant was passed through a 0.45 μm filter and DEAE resin (50 mM MES, 1mM TCEP, pH 5.0). The DEAE unbound material was loaded on superdex 200 column (10 mM MES, 1mM TCEP, pH 5.0), yielding a highly enriched DJ-1 protein fraction (Fig. 1A).
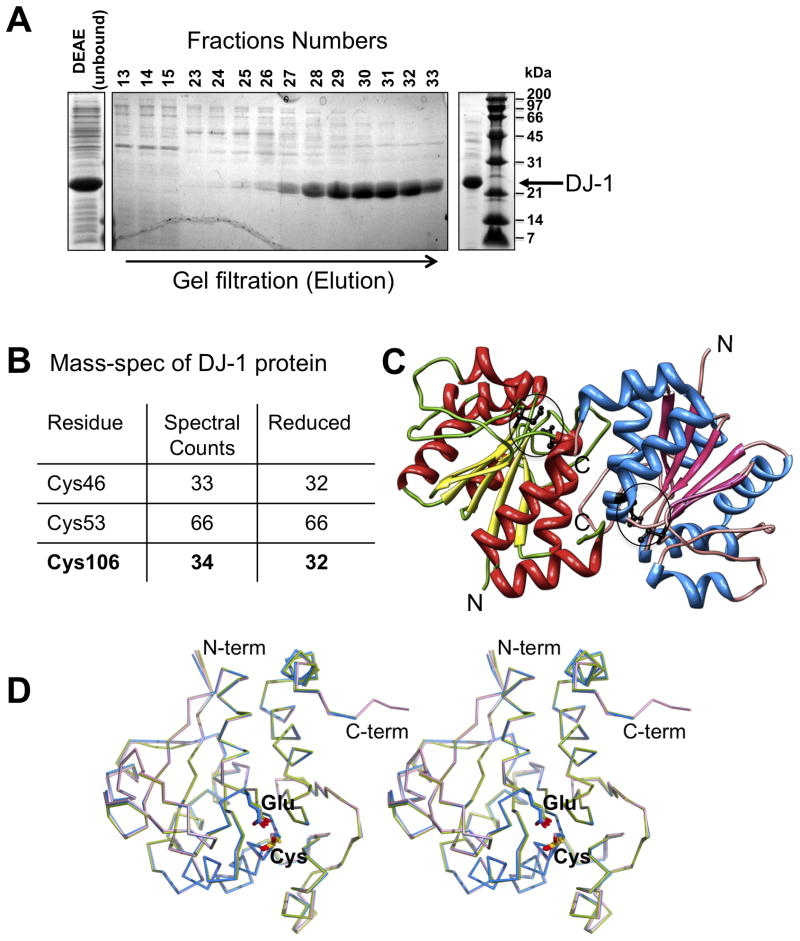
Figure 1. Purification and crystal structure of DJ-1.
A. Purification of reduced DJ-1 at pH 5.0 visualized by SDS-PAGE depicting crude extract (left), gel filtration elution profile (middle) and the DJ-1 preparation used for MS analysis and crystallization (right). B. MS analysis depicting reduced DJ-1. The total number of spectral counts for peptides containing assigned cysteines (“residues”) is displayed as well as the number of these counts referring to alkylated cysteines in these peptides indicating reactive (”reduced”) state of the cysteines. C. Overall structure of the asymmetric unit. The two independent copies of DJ-1 observed in the crystallographic asymmetric unit forming the characteristic DJ-1 dimer are colored separately, with the location of Cys106 and Glu18 indicated by circles D. Structural comparison of DJ-1 in different oxidation states. Structural overlay of two independent copies of DJ-1 reported here showing molecule 1 (with reduced Cys106; blue), molecule 2 (with mono-oxygenized Cys106; magenta) along with a DJ-1 structure determined previously in which Cys106 is oxidized to its sulfinic acid form (green; PDB Id: 1SOA).
Cysteine oxidation status of the purified DJ-1 was analyzed by NanoLC/MS/MS. Essentially, DJ-1 in 10 mM MES, 100 mM NaCl and 1mM TCEP, pH 5.0 was directly transferred to boiling 10% SDS, prior to mixing with SDS-PAGE sample buffer. After electrophoresis and coomassie staining, gel bands were reduced and alkylated with 5 mM DTT and 15 mM iodoacetamide followed by in-gel tryptic digestion. Resulting peptides were concentrated, desalted and then analyzed by automated NanoLC-LTQ MS/MS (see supplement for additional detail).
Diffraction quality DJ-1 crystals were grown at 293K by sitting-drop vapor diffusion method by mixing 2.5 μl of DJ-1 (~12mg/ml) and 2 μl of precipitant solution (45% (v/v) polyproylene glycol 400, 0.1 M Bis-Tris, pH 6.5, 1 mM TCEP) and equilibrating with 50 μl of reservoir solution (precipitant solution containing 10mM TCEP). DJ-1 crystal was flash-frozen in liquid nitrogen after briefly rinsing it with precipitant solution. X-ray diffraction data at 1.54 Å were collected on a Rigaku R-AXIS HTC image plate system mounted on a Rigaku FR-E SuperBright Ultra high-intensity rotating anode X-ray generator. Data were integrated in XDS (Kabsch, 2010), analyzed and converted to MTZ in Pointless (Evans, 2006) and scaled in Scala of CCP4 suite (CCP4, 1994). The structure was determined by molecular replacement with Phaser (McCoy et al., 2007) using monomeric DJ-1 as search model (PDB ID 1PDV). After initial refinement in REFMAC5 (Murshudov et al., 1997) and manual rebuilding in COOT (Emsley and Cowtan, 2004), automated model building was carried using PHENIX Autobuild (Adams et al., 2010). Final refinement cycles involving TLS were performed in PHENIX-refine and REFMAC5. Stereochemical quality of the final model was assessed using AutoDepInputTool (Yang et al., 2004), MolProbity (Davis et al., 2004) and SFcheck 4.0 (Vaguine et al., 1999). A summary of the data processing and refinement statistics is presented in Table 1.
Table 1.
Summary of data collection, and refinement statistics.
| Space group | C2 |
|---|---|
| Unit cell parameters | a=141.88Å, b=41.83Å, c=63.34Å β=115.79° |
| Data Collection Wavelength (Å) | 1.54178 |
| Resolution range (Å) | 20 – 1.53 (1.62 – 1.53) a |
| Number of observations | 230, 297 |
| Number of unique reflections | 47, 218 |
| Mean I/σ(I) | 19.0 (4.7) a |
| Rmerge(%) | 4.4 (25.2) a |
| Multiplicity | 4.9 (4.6) a |
| Model and refinement statistics | |
| Resolution range (Å) | 50 – 1.56 (1.59 – 1.56) a |
| No. of unique reflections | 44, 319 |
| Completeness | 94.4 (89.4) a |
| Average B-factor (Å2) | 25.7 |
| Rwork (%) | 17.2 |
| Rfree (%) | 21.1 |
| RMS Bond lengths (Å) | 0.013 |
| RMS Bond angles (°) | 1.42 |
| Protein residues/all atoms | 378/3,167 |
| φ/Ψ deviations | 98.9 favored |
| 0.8 allowed | |
Highest resolution shell in parentheses
Rwork = Σ||Fobs|−|Fcalc||/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rwork, but for 5% of the total reflections chosen at random and omitted from refinement.
3. Recombinant production of reduced human DJ-1
The production of recombinant DJ-1 for structural and functional studies is challenged by the high susceptibility of Cys106 to oxidation. While maintained in a reduced environment during bacterial expression, the subsequent oxidative exposure during purification stages bears considerable risks for oxidation of Cys 106, which is increased by alkaline pH and the presence of heavy metal ions (Witt et al., 2008). To bypass these obstacles we took advantage of the massive expression of DJ-1 in E. coli by purifying untagged DJ-1 under acidic conditions (pH 5.0) using a single step size exclusion chromatography (Fig. 1A). This procedure yields high quality DJ-1, which behaves as a dimer in solution (supplemental figure 1) and is suitable for crystallization and future functional studies.
To verify the redox status of DJ-1 Cys106 we subjected the protein material to LC-MS/MS analysis. The MS analysis following protein treatment with iodoacetamide clearly shows substantial carboxyamidomethylation in fragments containing Cys46, Cys53 and Cys106. For the latter 32 out of 33 spectral counts for Cys106 containing peptides (from Lys99 to Lys122) corresponded to alkylation of Cys106 by iodoacetamide hence verifying the reduced state of purified DJ-1 (Fig. 1B, supplementary Table 1).
4. Visualizing reduced DJ-1 in atomic detail
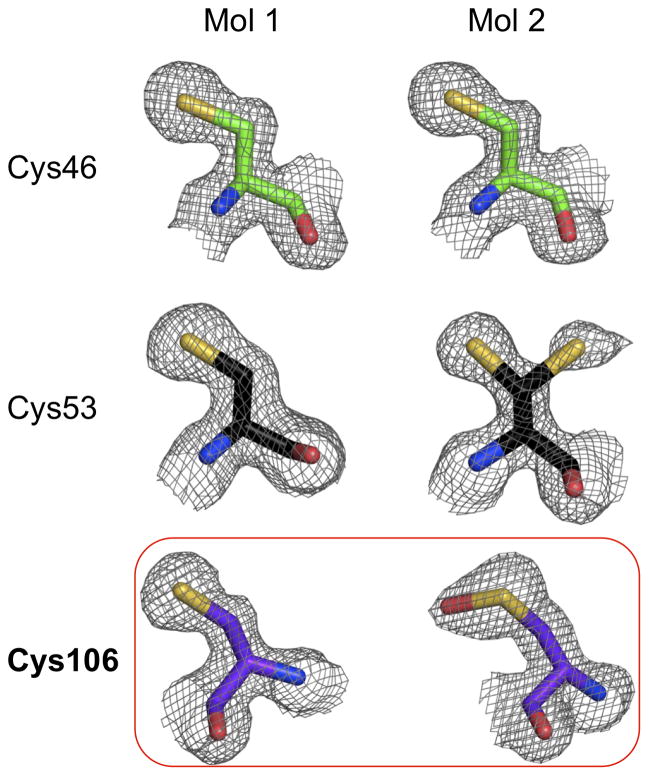
To capture the reduced state of DJ-1, we determined its structure to 1.56 Å resolution from a monoclinic crystal grown at pH 6.5 (Fig. 1C). The crystallographic asymmetric unit comprises two DJ-1 molecules building up the physiological DJ-1 dimer described earlier (Honbou et al., 2003; Lee et al., 2003; Tao and Tong, 2003; Wilson et al., 2003). Overall, interpretable electron density is not observed for the first residue in both DJ-1 molecules and the last three residues at the C-terminus of molecule 1 (aa 189–191). In contrast, molecule 2 shows a clear density for the entire C-terminus including the ‘Leu-Glu’ motif (aa 190–191), which was added during cloning procedures (see section 2 for detail). Analysis of the putative active site Cys106 in molecule 2 shows extra density around its sulfur moiety, which is characteristic of a mono-oxygenation (Fig. 2). Structural comparison shows that the surroundings of the sulfenic acid modified Cys106 are similar to the previous DJ-1 structures bearing a sulfinic acid modified Cys106 (supplemental figure 2). This mono-oxidation of Cys106 likely arises during 4 days of crystal growth despite the presence of 10 mM TCEP during crystallization. Importantly, however molecule 1 reveals the fully reduced state of the Cys106 thiol with no connected additional electron density present (Fig. 2; supplemental figure 3). Structural comparison between molecule 1 (reduced) and 2 (mono-oxidized) or an earlier DJ-1 structure with Cys 106 oxidized to its sulfinic acid form (PDB Id: 1SOA) shows that no global conformational changes or changes in the oligomeric state arise from cysteine oxidation in DJ-1 as evidenced by their near identical Cα-backbone traces (Fig. 1D; supplemental figure 2).
Figure 2. Electron densities observed for cysteine residues in molecule 1 and 2.
The 2mFo-DFc electron density map is displayed around various cysteine residues at a level of 1.0 σcontour.
5. Leu-Glu stabilizes Cys106 in reduced state
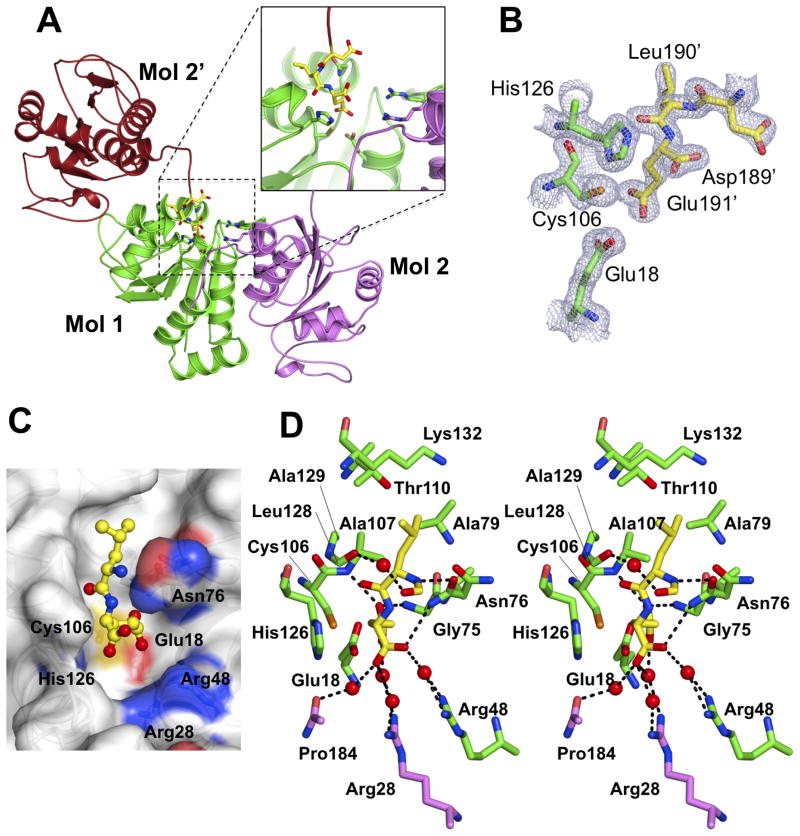
The oxidation status of Cys46, Cys53 and Cys106 for DJ-1 (molecule 1) is shown in Fig. 2. Consistent with previous DJ-1 structures, both Cys46 and Cys53 are unmodified, but with an alternative conformation present for molecule 2 Cys53. As described above, strikingly, Cys106 shows a sufenic acid (-SOH) modification only in molecule 2. The fully reduced Cys106 in molecule 1 is protected through a glutamate residue arising from a ‘Leu-Glu’ motif at the C-terminus of a crystallographic neighbor DJ-1 molecule (Fig. 3A). This protection can either stem from steric blockage of oxidative modification of Cys106 or alteration of the pKa of the thiol caused by the carboxyl group of the glutamate. The ‘Leu-Glu’ motif forms a network of hydrogen bonding and van der Waals interactions with DJ-1 molecule 1 (Fig. 3D). A similar contact is absent in molecule 2 which lacks a protective crystal contact and results in a mono-oxigenized Cys106 which in turn would preclude the interaction with a glutamate motif due to steric and electrostatic hindrances. In the Leu-Glu motif bound reduced DJ-1, the gamma carboxylate of the Glu side chain is positioned within 3.2–3.4 Å from the thiol of Cys106 and forms hydrogen bonding interactions with backbone nitrogen residues of Gly75 and Ala107, and the side chains of Asn76 (Oδ1). The side chain ‘Leu’ is nicely packed within the hydrophobic surroundings of Ala107 (Cβ), Ala129 (Cβ), Ala79 ((Cβ), Thr110 (Cγ2) and Lys132 (Cβ and Cδ) (Fig. 3D) utilizing a defined surface pocket of DJ-1 (Fig. 3C). The ‘Leu-Glu’ interaction is further stabilized by water molecules that bridge the glutamate to Arg48 (molecule 1), as well as Arg28 and Pro184 (molecule 2; Fig. 3D).
Figure 3. Leu-Glu interaction conserves reduced DJ-1 Cys106. A. Crystal symmetry related molecule 2 interacts with molecule 1.
The C-terminus of molecule 2 from a crystallographic neighbor molecule (Mol 2′, brown/yellow sticks) binds into the putative active site of DJ-1 molecule 1 (green) though a C-terminal ‘Leu-Glu’ motif. Molecule 2 of the DJ-1 dimer is colored magenta. B. The ‘Leu-Glu’ motif binds in proximity to Cys106 molecule 1. The 2mFo-DFc electron density map is displayed at a level of 1.0 σ contour for Asp- ‘Leu-Glu’ motif (Mol 2′, yellow) and Cys106, Glu18 as well as His 126, which lines the binding pocket (Mol 1, green). C. Surface representation. ‘Leu-Glu’ binding into Cys106 proximal pocket on DJ-1. D. Detailed stereo-view of ‘Leu-Glu’ interaction and the DJ-1 binding pocket. Potential hydrogen bonding interactions are shown in dashed lines. The carboxy-terminal residues ‘Leu-Glu’ from the neighboring molecule 2 (Mol 2′) are shown in yellow sticks. Molecule 1 residues (green sticks) surrounding ‘Leu-Glu’ within 4 Å distance are shown. Also shown is the side chain interaction formed by Arg28 and Pro184 from the biological dimer (Mol 2, magenta).s
6. Reduced DJ-1 - perspective
The production of reduced DJ-1 as verified and described in this work can serve as basis for future biochemical and functional characterization of this enigmatic protein. To this end, a DJ-1 protein purified with the approach presented in this work, but lacking Leu-Glu motif may be more suited. Yet, the structure depicting the ‘Leu-Glu’ motif bound to the putative active site of DJ-1 reveals a binding pocket on DJ-1 (Fig. 3B,C) in which His126 (Chen et al., 2010; Huai et al., 2003) lines one side of the pocket and is contributing to the binding of the C-terminal glutamate. This binding mode may additionally show similarities to the binding of the elusive and at this point putative biochemical substrate(s) of DJ-1 or DJ-1 interacting partners and would notably be hindered already in the mono-oxygenated state of Cys106 as outlined earlier.
The conservation of the reduced state of Cys106 visualized in our structure can furthermore serve as basis to preserve this state in DJ-1 relatives and other proteins/enzyme prone to cysteine oxidation due to a depressed pKa.
In an analogous manner, the ‘Leu-Glu’ “template” depicted in this work can also be exploited in future attempts to develop small molecule compounds targeting DJ-1. This binding pocket can be utilized to either inactivate DJ-1 using a cysteine reactive moiety or preserve the reduced state of its reactive site cysteine as accomplished by the glutamate side chain in our structure. In summary this work represents an important step in the investigation and targeting DJ-1 as a major player in cellular responses to disease and environmental factors.
Supplementary Material
Acknowledgments
We thank Dr. Khatereh Motamedchaboki from the SBMRI proteomics facility for analysis of DJ-1 by mass spectrometry, Dr. Jinghua Yu from our SBMRI X-ray facility for technical assistance, Dr. Andrey Bobkov (protein facility at SBMRI) for AUC measurement and Dr. Guy Salvesen for helpful comments. This work was supported by NIH grant P01 ES016738.
Abbreviations
- TCEP
tris(2-carboxyethyl)phosphine
- DTT
Dithiothreitol
- MES
2-(N-morpholino)ethanesulfonic acid
- Bis-tris
Bis-(2-hydroxy-ethyl)-amino-tris(hydroxymethyl)-methane
- ROS
Reactive oxygen species
Footnotes
Protein Data Bank Accession Code
Coordinates and structure factors have been deposited in the Protein Data Bank under the PDB accession code 3SF8.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Sleiman PM, Healy DG, Wood NW. Causes of Parkinson’s disease: genetics of DJ-1. Cell Tissue Res. 2004;318:185–8. doi: 10.1007/s00441-004-0922-6. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris LM, Mata IF, Zabetian CP. The genetics of Parkinson disease. J Geriatr Psychiatry Neurol. 2010;23:228–42. doi: 10.1177/0891988710383572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–85. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101:9103–8. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen J, Li L, Chin LS. Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum Mol Genet. 2010;19:2395–408. doi: 10.1093/hmg/ddq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–52. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikinheimo P, Goldman A, Jeffries C, Ollis DL. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure. 1999;7:R141–6. doi: 10.1016/s0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- Hod Y. Differential control of apoptosis by DJ-1 in prostate benign and cancer cells. J Cell Biochem. 2004;92:1221–33. doi: 10.1002/jcb.20159. [DOI] [PubMed] [Google Scholar]
- Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, et al. The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J Biol Chem. 2003;278:31380–4. doi: 10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- Huai Q, Sun Y, Wang H, Chin LS, Li L, et al. Crystal structure of DJ-1/RS and implication on familial Parkinson’s disease. FEBS Lett. 2003;549:171–5. doi: 10.1016/s0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–44. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RH, Peters M, Jang Y, Shi W, Pintilie M, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–73. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Klinefelter GR, Laskey JW, Ferrell J, Suarez JD, Roberts NL. Discriminant analysis indicates a single sperm protein (SP22) is predictive of fertility following exposure to epididymal toxicants. J Androl. 1997;18:139–50. [PubMed] [Google Scholar]
- Klinefelter GR, Welch JE, Perreault SD, Moore HD, Zucker RM, et al. Localization of the sperm protein SP22 and inhibition of fertility in vivo and in vitro. J Androl. 2002;23:48–63. doi: 10.1002/jand.2002.23.1.48. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, et al. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J Biol Chem. 2003;278:44552–9. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- Lev N, Roncevic D, Ickowicz D, Melamed E, Offen D. Role of DJ-1 in Parkinson’s disease. J Mol Neurosci. 2006;29:215–25. doi: 10.1385/jmn:29:3:215. [DOI] [PubMed] [Google Scholar]
- MacKeigan JP, Clements CM, Lich JD, Pope RM, Hod Y, et al. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63:6928–34. [PubMed] [Google Scholar]
- Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, et al. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004:157–76. doi: 10.1042/bss0710157. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Cookson MR. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7 doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15:1578–82. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr Biol. 2005;15:1572–7. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Migliore L, Coppede F. Genetic and environmental factors in cancer and neurodegenerative diseases. Mutat Res. 2002;512:135–53. doi: 10.1016/s1383-5742(02)00046-7. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, et al. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum Mol Genet. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–13. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson’s disease patients. Biochem Biophys Res Commun. 2004;320:389–97. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- Tao X, Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J Biol Chem. 2003;278:31372–9. doi: 10.1074/jbc.M304221200. [DOI] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–28. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- Wagenfeld A, Gromoll J, Cooper TG. Molecular cloning and expression of rat contraception associated protein 1 (CAP1), a protein putatively involved in fertilization. Biochem Biophys Res Commun. 1998;251:545–9. doi: 10.1006/bbrc.1998.9512. [DOI] [PubMed] [Google Scholar]
- Wilson MA. The Role of Cysteine Oxidation in DJ-1 Function and Dysfunction. Antioxid Redox Signal. 2011 doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-Å resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci U S A. 2003;100:9256–61. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt AC, Lakshminarasimhan M, Remington BC, Hasim S, Pozharski E, et al. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47:7430–40. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, et al. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–9. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, et al. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312:1342–8. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.