Abstract
Using epifluorescence microscopy and image analysis, we have quantitatively described the frequency, size, and spatial distribution of bacterial aggregates on leaf surfaces of greenhouse-grown bean plants inoculated with the plant-pathogenic bacterium Pseudomonas syringae pv. syringae strain B728a. Bacterial cells were not randomly distributed on the leaf surface but occurred in a wide range of cluster sizes, ranging from single cells to over 104 cells per aggregate. The average cluster size increased through time, and aggregates were more numerous and larger when plants were maintained under conditions of high relative humidity levels than under dry conditions. The large majority of aggregates observed were small (less than 100 cells), and aggregate sizes exhibited a strong right-hand-skewed frequency distribution. While large aggregates are not frequent on a given leaf, they often accounted for the majority of cells present. We observed that up to 50% of cells present on a leaf were located in aggregates containing 103 cells or more. Aggregates were associated with several different anatomical features of the leaf surface but not with stomates. Aggregates were preferentially associated with glandular trichomes and veins. The biological and ecological significance of aggregate formation by epiphytic bacteria is discussed.
Leaf surfaces can support the development of large populations of diverse bacteria, and most plant-pathogenic bacteria multiply on the leaf surface before they initiate disease (19). Pseudomonas syringae is a prominent example of such epiphytic bacteria in that it can cause disease on many plant species and can incite frost damage because of its ice nucleation activity (20). This species often establishes large epiphytic populations on host and nonhost plants (for a review, see reference 20), and studies have shown that the establishment of large epiphytic populations is a prerequisite for disease or frost occurrence (29, 44). The epiphytic bacterial population of entire leaves is usually estimated, and quantitative analysis of epiphytes on leaves at smaller spatial scales is lacking. A few studies have reported large variations in population sizes of individual bacterial species between leaves or between individual 9-mm2 leaf segments (23), strongly suggesting that bacterial populations are highly variable in size even at small spatial scales.
The patchy recovery of bacteria from leaves in leaf imprint studies suggests that bacteria do not occur in a uniform pattern across leaf surfaces but tend to be localized in particular sites (27). The nature of these sites has been qualitatively studied using scanning electron microscopy, and the most-common sites of bacterial colonization reported were at the base of trichomes, at stomates, and in epidermal cell grooves along veins (31, 33, 43, 46). While most of these observations suggest a relatively scattered colonization, large aggregates embedded in an exopolymeric matrix have been reported on leaf surfaces of different plant species (17, 18, 37). However, such reports do not provide enough information as to the ubiquity or importance of such aggregated cells.
Bacterial aggregates observed on leaf surfaces have characteristics similar to those of cells in biofilms that have been described in aquatic and medical environments (13). Cells in biofilms are encased in a spatially structured exopolymeric matrix. The presence of an exopolymeric matrix may lead to the creation of physical barriers and chemical gradients that can concentrate nutrients from dilute sources, provide protection against predators, and shield cells from lytic enzymes, antibiotics, or other inhibitory compounds (for reviews, see references 13, 14, 36, and 42). A localized high level of density of cells may foster genetic and metabolic exchange. The epiphytic survival of bacteria during desiccation is likely enhanced when they are embedded in a hydrated polymer. The physiology and phenotypes of cells within biofilms have also been reported to be markedly different from those of planktonic cells of the same species. Aggregates may also locally facilitate coordinated bacterial population responses for traits expressed in a density-dependent manner through quorum sensing. Cha et al. (11) have found that many plant-pathogenic bacteria produce autoinducers, suggesting that density-dependent expression of traits (such as virulence factors) involved in interactions with plants is also common on leaf surfaces (15). Since the leaf surface is an unsaturated (i.e., water-limited) environment with constantly changing conditions during the course of the day, some of the properties of bacterial aggregates on leaves might differ from those of biofilms observed in aquatic habitats.
An initial attempt to measure aggregate abundance on leaves (38) revealed that aggregated populations on field-grown broad-leaved endive and parsley leaves constitute between 10 and 40% of the total bacterial population. However, the authors used a semidestructive method that combined leaf washing, filtration, and ultrasonication which did not allow them to determine aggregate sizes or their locations on the leaf surface or to account for cell disintegration or aggregation that might have occurred during removal. With the use of P. syringae pv. syringae strain B728a on bean leaves as a model, the objective of our study was to quantitatively describe the frequency, size, and spatial distribution of bacterial aggregates on leaf surfaces. It has been reported that the majority of the cells of P. syringae associated with healthy bean leaves are present on the plant surface (49), and our preliminary microscopic observations revealed the presence of large aggregates on bean leaf surfaces within a few days after inoculation of plants with P. syringae pv. syringae strain B728a. Our approach consisted of analyzing captured digital images of bacteria (visualized by epifluorescence microscopy) directly on the leaf surface after staining with acridine orange. We demonstrate that bacterial aggregates occur in a wide range of sizes and are not randomly distributed on the leaf surface, reflecting the variability of the leaf surface environment, and discuss the ecological significance of the large aggregates formed on leaves and factors fostering their formation.
MATERIALS AND METHODS
Bacterial strain and culture media.
Pseudomonas syringae pv. syringae strain B728a is a spontaneous rifampin-resistant derivative of a strain isolated from a bean leaflet. Its characteristics have been previously reported (5, 20, 30). Cultures were stored at −80°C in 15% (vol/vol) glycerol in 10 mM potassium phosphate buffer (PB) (pH 7.0) and routinely grown on King's medium B (22) (KB). Bacteria were cultured for 24 h at 28°C on KB containing rifampin (100 μg/ml) for inoculum production. The cells were harvested from plates with a sterile loop and suspended in 1 mM PB. The cell concentration was estimated by spectrophotometry and adjusted by dilution with PB to the desired concentration.
Plant inoculation.
All experiments were conducted with 2-week-old bean plants (Phaseolus vulgaris cv. Bush Blue Lake 274) grown in a greenhouse and incubated in the laboratory under controlled conditions similar to those of other studies (5). The bacteria were applied by immersing the plants in a suspension of P. syringae pv. syringae strain B728a (105 CFU/ml) for about 3 s. The plants were then kept at 22°C in a moist chamber maintained at close to 100% relative humidity (RH) (12-h photoperiod). After 2 days, half of the plants were transferred to a growth chamber and maintained at 22°C (12-h photoperiod) under conditions of a low level of RH (<50% RH) for the duration of the experiments. Plants of the same genotype were also grown for 40 days in a field site located in Berkeley, California.
Estimation of bacterial population sizes.
Culturable bacterial population sizes on symptomless primary leaves randomly sampled from bean plants, as well as on individual leaf segments, were estimated by dilution plating of leaf washings. At each sampling time, 15 leaves were individually placed in 20 ml of washing buffer (100 mM PB [pH 7.0] containing 0.1% Bacto Peptone; Difco) and sonicated for 7 min in an ultrasonic cleaning bath to recover bacterial cells as described for other studies (39). The weight and surface area of each individual leaf sampled were also measured. To quantify the distribution of population sizes across a leaf, individual leaves were cut into 44-mm2 leaf segments; population sizes were estimated separately on individual segments. The total number of segments per leaf ranged from 54 to 72. For each leaf, segments were cut using a flame-sterilized razor blade, placed in individual sterile Eppendorf microcentrifuge tubes containing 500 μl of washing buffer, and sonicated for 7 min. Appropriate dilutions of leaf or leaf segment washings were plated using a spiral plater (Spiral Systems Inc., Cincinnati, Ohio) on KB and on KB containing rifampin, cycloheximide (Sigma Co., St. Louis, Mo.) (100 μg/ml), and benomyl (Dupont, Wilmington, Del.) (50 μl/mg). The population sizes were estimated from colony counts on plates after 2 to 3 days of incubation at 28°C.
Visualization of cells by microscopy.
The frequency, size, and localization of bacterial aggregates were determined at the microscale by analysis of digital images of micrographs of epiphytic bacteria before and immediately after inoculation and at 2, 7, and 8 days following inoculation of P. syringae pv. syringae strain B728a onto leaf surfaces. At each sampling time, three leaves were randomly selected and 5 to 10 segments (1 by 1 cm) were randomly cut per leaf. Each segment was then individually dipped in a solution of acridine orange (0.1%, pH 7.0) for 2 min to allow visualization of both the bacterial cells and the anatomical structures of the leaf surface (adapted from reference 37). A 1% solution of water agar was prepared and set aside in liquid phase in a water bath at 50°C Leaf segments were then air dried, placed on top of 100 μl of molten water agar on a microscope slide to ensure a flat surface for microscopic observations, and observed after solidification of the agar (i.e., after about 20 s). The upper surface of primary leaves was observed in all experiments. Samples were viewed by epifluorescence microscopy with an Axiophot microscope equipped with a 20×, 1.30-numerical-aperture Plan Neofluar objective (Zeiss Inc., Oberkochen, Germany). A long-pass filter for fluorescein was used in combination with a short-pass filter (Melles-Griot, Irvine, Calif.) (650 nm) to partially block the red autofluorescence of the leaf but not the acridine orange fluorescence. A total of 5 to 10 random fields of view were selected per leaf segment, and the corresponding images were captured with an Optronics DEI750 video camera.
Viability of bacterial cells was determined using a Live-Dead staining kit (Molecular Probes Inc., Eugene, Oreg.). Cells removed from leaf surfaces by sonication were stained and filtered through black polycarbonate filters (Millipore Co., Bedford, Maine) (0.2-μm-pore-size diameter). The filters were then placed between a microscope slide and a coverslip and were immediately observed by epifluorescence microscopy. At least 20 random fields of view (containing a total of at least 400 cells) were observed per slide, and the total numbers of dead (red) and living (green) cells were enumerated.
Our samples were also observed using environmental scanning electron microscopy (ESEM), a process that does not require any sample preparation involving liquids. Leaf segments were mounted on metal supports with double-coated tape, placed in the microscope chamber, and immediately observed. Micrographs were collected using an Electroscan E3 ESEM operated at an accelerating voltage of 5.0 keV.
Frequency and size of aggregates.
The frequency and size of bacterial aggregates on leaf surfaces were determined by image analysis of micrographs obtained by epifluorescence microscopy. To represent measurements in micrometers, the microscope and video camera were calibrated using a micrograded slide having parallel stripes separated by known distances. Captured images were transferred to a personal computer platform and individually processed with Corel Photopaint software (Corel Co., Ottawa, Canada) following a four-step sequence. (i) Due to the irregular topography of the leaf surface, some areas of the image captured were out of focus and were deleted manually. (ii) Bacteria (which fluoresced bright orange) were separated from the dark-brown background fluorescence corresponding to the leaf surface by selecting the appropriate threshold value of pixel intensity. (iii) The few fluorescent objects that were not bacterial cells remaining after the thresholding step were deleted manually. (iv) To facilitate measurements, the resulting image was converted into a binary image with bacteria represented in white and the leaf surface represented in black. Aggregate size and frequency measurements were conducted using IPLab software (Scanalytics Inc., Fairfax, Va.), which allowed automatic measurements of the number of individual objects in an image as well as their corresponding areas. The total number of cells per aggregate was then estimated based on the assumption that aggregates consisted of dense monolayers of bacteria with a surface area of 1.5 μm2 each. This procedure gave us an underestimation of the total number of aggregated bacterial cells. The area of leaf surface observed was deduced for each image by summing the areas of the leaf covered by bacteria (white pixels) and the areas of the leaf not covered by bacteria (black pixels). For ease of interpretation and to facilitate the creation of a visual image of the aggregates observed, aggregate sizes were expressed in values reflecting estimated numbers of cells instead of areas in square millimeters. Since large aggregates may have comprised several layers of bacterial cells as observed under field conditions (37), this convention may have led to an underestimation of the number of cells in such aggregates. On the other hand, the use of a low-magnification objective to maximize the leaf surface area visualized and in focus in a field of view might result in an overestimation of the total number of cells in small (1- to 4-cell) aggregates since the strong fluorescent signals of stained bacteria can form a fluorescent halo around the cells, thus leading to overestimation of their sizes.
Localization of aggregates.
To associate an aggregate with a particular leaf surface feature, the leaf surface area was partitioned into discrete variables and defined as the sum of areas covered by the following anatomical structures: glandular trichomes, hooked trichomes, stomates, undifferentiated epidermal plant cells, and veins. Due to the large number of aggregates observed, only aggregates covering an area of 150 μm2 or more (e.g., 100 bacterial cells) were analyzed. Aggregate sizes were determined using processed images, and the corresponding unprocessed images were used to define the location of a given aggregate. When more than one large aggregate was present in the same image, the x and y coordinates of the center of each were obtained using IPLab software. To determine the relative areas occupied by the different anatomical structures on a leaf, 1-cm-wide transects were cut crosswise from the midvein to the margin at the base, midsection, and tip of a noninoculated primary bean leaf. Transects were then stained with acridine orange (0.1%, pH 7.0) and prepared for epifluorescence microscopy observation as described previously. For each transect, overlapping images were taken from the midvein to the leaf margin. To minimize areas out of focus, images were taken using a 10×, 0.30-numerical-aperture Plan Neofluar objective (Zeiss Inc.); when necessary, several images of the same field of view were taken at different focal planes. Overlapping sections of the captured images were manually deleted before measurements were performed. For each image, masks corresponding to individual glandular trichomes, hooked trichomes, stomates, or veins were manually drawn and their corresponding areas were measured using IPLab software. The area covered by undifferentiated epidermal plant cells was deducted by subtracting the sum of areas covered by the different anatomical structures from the total area measured.
Data transformation and statistical analysis.
Estimations of the total number of cells per aggregate and frequency distributions were obtained using Microsoft Excel software (Microsoft Co., Redmond, Wash.). Descriptive statistic determinations, a Kolmogorov-Smirnov test for normality of nontransformed and log-transformed data, Duncan's multiple range test, a chi-square goodness of fit test, and calculation of the cutoff size values and slopes of the truncated power laws (TPL) were all performed with Statistica software (Statsoft Inc., Tulsa, Okla.).
RESULTS
Combination of acridine orange staining and image analysis.
The approach used in this study allowed us to visualize bacteria and to measure the frequency and size of bacterial aggregates directly on the leaf surface. While the spatial patterns of bacterial colonization of bean leaves in the field and in the greenhouse were qualitatively similar, the leaves of plants grown in the field were partially covered with autofluorescent soil particles or plant debris (which was difficult to differentiate from bacterial aggregates) and thus were not suitable for an extensive and accurate quantification of the number and size of bacterial aggregates. Image analysis was therefore performed on bean plants grown in the greenhouse following inoculation with P. syringae pv. syringae strain B728a. The spatial patterns of colonization of bean leaves were qualitatively similar on the abaxial and adaxial leaf surfaces. Characterization of bacterial aggregates was therefore performed on the upper leaf surface, since it has a more even topography as well as a lower density of long autofluorescent hooked trichomes. The relatively low magnification (×250) used allowed us to observe large leaf surface areas and to minimize the percentage of out-of-focus areas on captured images and yet allowed us to visualize individual bacterial cells. The use of a 650-nm short-pass filter was necessary for the visualization of individual cells and small aggregates that would otherwise be masked by the strong red autofluorescence of the leaf. Qualitative observations (using ESEM) of our samples revealed the same colonization pattern of bacteria on leaves. No bacterial cells were observed associated with stomates, and the presence of dark spots of reduced electron reflection, probably corresponding to a localized difference in chemical composition, was observed at the base of trichomes and along veins.
Use of ESEM confirmed that aggregates were preferentially associated with glandular trichomes or veins or were occasionally observed along leaf margins or associated with a wounded epidermal cell, indicating that acridine staining did not redistribute cells. While the spatial distribution of aggregated cells on field-grown plants followed the same colonization pattern, we were not generally able to visualize solitary cells. Samples were observed at a relatively low level of magnification that may not have allowed us to detect individual cells, due to the presence of strongly autofluorescent soil particles or plant debris and also because the size of bacteria grown on leaves is significantly smaller than that of cultured cells (34). While the use of acridine orange did not allow differentiation between P. syringae cells and cells of other bacterial species, the data reported in this study were very consistent with those of experiments performed using green fluorescent protein-tagged P. syringae cells (unpublished data). As determined on the basis of plate counts of culturable populations, in addition, P. syringae cells represented over 99% of the cells present on the leaf surfaces of bean plants.
Quantification of bacterial aggregate sizes.
To obtain a sample of leaf surface bacterial populations sufficiently representative to allow projection across an entire leaf, several square millimeters of leaf surfaces containing between 12,000 and 65,000 aggregates were measured at each sampling time (Table 1). At 0, 2, and 8 days of incubation after inoculation with a dilute P. syringae cell suspension, bacteria covered 2.2% ± 0.4%, 4.8% ± 0.1%, and 12.0% ± 2.5% of the leaf surface, respectively. After incubation for several days, cells were not randomly scattered over the leaf surface but occurred in a wide array of cluster sizes ranging from single cells to aggregates with over 104 cells each. While after 2 days total bacterial populations on leaves had increased little after achieving a population size of ca. 106 CFU/cm2, the average aggregate size increased with time and the largest aggregates observed at 0, 2, and 8 days after inoculation consisted of approximately 200, 2,500, and 11,000 bacterial cells, respectively (Table 1). Aggregates observed on leaf surfaces of plants maintained under conditions of low RH were less numerous and smaller than aggregates observed on leaf surfaces of plants maintained under conducive conditions for the same period of time (data not shown). The proportion of living cells on leaves was less than 1% after 8 days under dry conditions; when plants where kept under conducive conditions, the proportion of living cells remained stable (at ca. 90%) during the course of the experiments. While data are not reported, the same experiments were performed on several additional leaves and the data obtained showed a similar pattern.
TABLE 1.
Number and size of bacterial aggregates observed on bean leaf surfaces through time following inoculation of P. syringae strain B728a
| Days since inoculation | Leaf | Total leaf area observed (mm2) | Bacterial aggregates
|
Area of leaf covered (%) | Cells/cm2 (106) | Variance/mean ratio | ||
|---|---|---|---|---|---|---|---|---|
| No. observed | No. of cells
|
|||||||
| Mean ± SE | Rangea | |||||||
| Preinoculation | 1 | 0.669 | 2,279 | 6.7 ± 0.2 | 108 | 0.98 | 0.6 | 11 |
| 2 | 0.632 | 531 | 7.5 ± 0.6 | 131 | 0.27 | 0.2 | 23 | |
| 3 | 0.810 | 2,949 | 6.3 ± 0.1 | 118 | 1.00 | 0.6 | 9 | |
| 0 | 1 | 0.668 | 6,268 | 7.8 ± 0.1 | 201 | 3.07 | 1.8 | 15 |
| 2 | 0.430 | 2,446 | 6.8 ± 0.2 | 135 | 1.65 | 1.0 | 16 | |
| 3 | 0.570 | 3,764 | 7.1 ± 0.2 | 178 | 2.00 | 1.2 | 14 | |
| 2 | 1 | 0.438 | 5,326 | 10.0 ± 0.4 | 644 | 4.99 | 3.0 | 73 |
| 2 | 0.519 | 6,148 | 10.0 ± 0.4 | 1,375 | 4.88 | 3.0 | 87 | |
| 3 | 0.330 | 3,147 | 11.5 ± 1.0 | 2,522 | 4.54 | 2.7 | 268 | |
| 8 | 1 | 0.351 | 5,177 | 12.5 ± 2.2 | 11,306 | 7.39 | 4.6 | 2,057 |
| 2 | 0.441 | 5,534 | 27.4 ± 2.7 | 7,960 | 12.95 | 8.6 | 1,489 | |
| 3 | 0.462 | 6,354 | 31.3 ± 3.5 | 10,203 | 15.71 | 10.8 | 2,502 | |
Range of bacterial aggregate sizes (number of cells), which also corresponds to the total number of cells observed in the largest aggregates.
Frequency of bacterial aggregates.
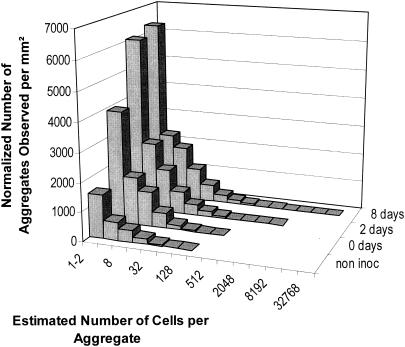
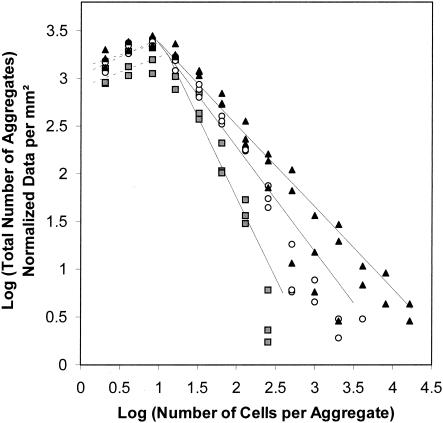
The large majority of bacterial cell aggregates observed on leaves were relatively small, and aggregate sizes exhibited a strong right-hand-skewed frequency distribution (Fig. 1). Aggregates consisting of 64 cells or fewer represented 99.9, 99.3, and 98.1% of the total number of aggregates observed after 0, 2, and 8 days, respectively. The distribution of aggregate sizes was not well described by the log-normal value. To compare frequency distribution of aggregate sizes as a function of time, the TPL, consisting of two power functions separated by a cutoff size (C), were applied to our data. The cutoff sizes and slopes were calculated after plotting the log of the total number of aggregates against the log of the number of cells per aggregate. The slope of the second TPL functions decreased significantly with time, indicating that the number of larger aggregates increased with increases in incubation time (Fig. 2). No significant differences were observed between the slopes of the first power functions which corresponded to small aggregates (less than 12 cells/aggregate).
FIG. 1.
Frequency distribution of bacterial aggregates on leaf surfaces before inoculation and 0, 2, and 8 days following inoculation with P. syringae pv. syringae strain B728a. The total number of aggregates was determined for three leaves at each sampling time, and the data were combined. The characteristics of the total surface of the leaf observed at each sampling time were slightly different, and the total number of aggregates observed was normalized; values are expressed in square millimeters. The number of cells per aggregate was estimated from the surface area of each aggregate; it was assumed that each aggregate consisted of a dense monolayer of bacterial cells with an average surface area of 1.5 μm2.
FIG. 2.
Frequency distribution of aggregate sizes at 0 (gray-shaded squares), 2 (open circles), and 8 (filled triangles) days following inoculation with P. syringae pv. syringae strain B728a on bean leaf surfaces. Three leaves were observed at each sampling time. The TPL were used to compare aggregation patterns as a function of time. The cutoff sizes were 12, 19, and 10 cells per aggregate after 0, 2, and 8 days, respectively. The slope of the second TPL functions significantly (P < 0.001) decreased through time, reaching −1.67 (R2 = 0.88), −1.09 (R2 = 0.89), and −0.86 (R2 = 0.86) after 0, 2, and 8 days, respectively. No significant differences were observed between the slopes of the first power functions corresponding to the smaller aggregates.
Relative percentages of solitary and aggregated cells on leaf surfaces.
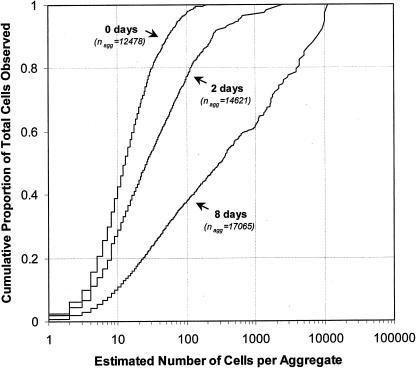
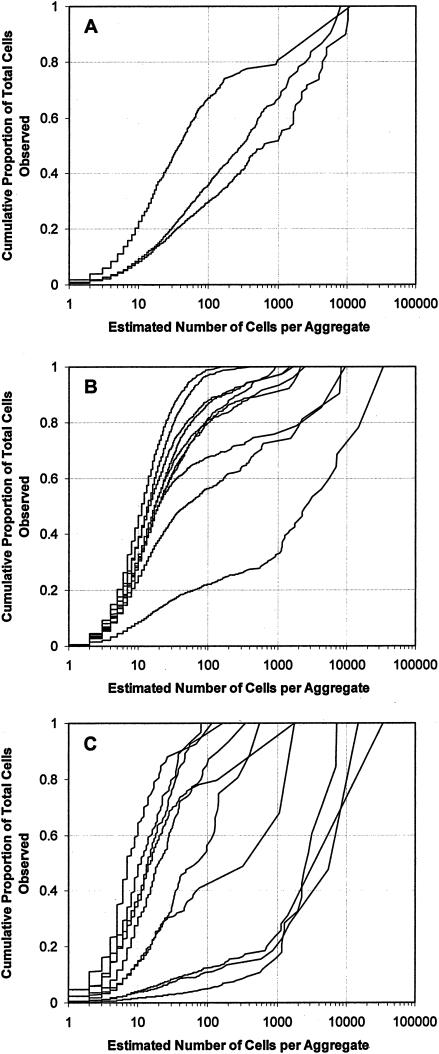
While large aggregates are not frequent on a given leaf, they can account for the majority of the cells present. We also observed that the proportion of the total number of cells that were located in larger aggregates increased with incubation time (Fig. 3). Immediately after inoculation, only 2.7% of the cells were located in aggregates containing 100 cells or more. This proportion dramatically increased with incubation time and, when considered over several leaves, reached (on average) 22.7 and 62.2% after 2 and 8 days, respectively. At 8 days after inoculation, half of the cells were located in aggregates containing 300 cells or more. A great deal of variation in the degree of aggregation was observed between leaves and between leaf segments as well as within individual leaf segments. For example, the proportions of cells located in aggregates containing 100 cells or more for each of three leaves sampled 7 days after inoculation were 32.8, 64.1, and 71.4%, respectively, and the proportions of cells located in aggregates containing 1,000 cells or more were 19.5, 33.7, and 48.8% (Fig. 4A). Observation (at 7 days after inoculation) of 10 different segments within the same leaf revealed that the percentages of cells located in large aggregates differed greatly among leaf segments; we observed that within this collection of leaf segments, the proportions of cells located in aggregates containing 100 cells or more ranged from 1.3 to 78.9% and that the proportions of cells located in aggregates containing 1,000 cells or more ranged from 0.0 to 67.3% (Fig. 4B). In different fields of view taken within a given leaf segment, the proportions of cells located in aggregates containing 100 cells or more ranged from 0.0 to 94.9% and the proportions of cells located in aggregates containing 1,000 cells or more ranged from 0.0 to 85.4% (Fig. 4C).
FIG. 3.
Cumulative proportion of the total number of cells observed as a function of the total number of cells per aggregate. Each curve represents the combined data of three individual leaves observed 0, 2, and 8 days following inoculation with P. syringae pv. syringae strain B728a.
FIG. 4.
Cumulative proportion of the total number of cells observed as a function of the total number of cells per aggregate when considered over different spatial scales. Each curve corresponds to the results for an individual leaf (A), an individual leaf segment cut randomly within the same leaf (B), or an individual field of view randomly selected within the same leaf segment (C) observed 7 days following inoculation with P. syringae pv. syringae strain B728a.
Spatial distribution of bacterial aggregates.
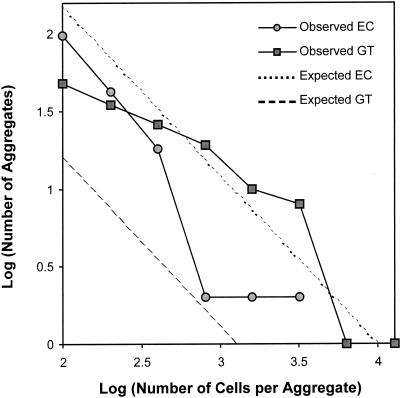
To determine whether bacterial aggregate formation occurred preferentially at particular sites on the leaf surface, we first quantified the relative fraction of the leaf that comprised different anatomical features on the upper surface of a primary bean leaf. Stomates, veins, hooked trichomes, and glandular trichomes accounted for 17.1% ± 0.3%, 7.3% ± 0.4%, 1.2% ± 0.2%, and 0.6% ± 0.1% of the leaf surface, respectively. The remaining 73.8% ± 0.8% was defined as undifferentiated epidermal cells (Table 2). Aggregates were observed associated with all anatomical features of the leaf surface except stomates. Aggregates were observed at the base of glandular and hooked trichomes and in the grooves between cells associated with veins and nondifferentiated epidermal cells. Comparison (by calculation of the chi-square goodness-of-fit values) of the observed and expected (as determined from the relative surfaces of each anatomical feature) numbers of aggregates associated with the different anatomical features demonstrated that aggregates are not randomly distributed on the leaf surface but are preferentially associated with veins and trichomes (P < 0.001). Chi-square goodness-of-fit values were also calculated to test the hypothesis that aggregate sizes are randomly distributed for each anatomical feature. The hypothesis could not be rejected for hooked trichomes (P = 0.81) and veins (P = 0.24). Aggregates associated with glandular trichomes were significantly (P < 0.001) larger and aggregates associated with undifferentiated epidermal cells were significantly (P < 0.005) smaller than those on hooked trichomes and veins (Fig. 5). Bacterial aggregates were most likely to be observed at the base of glandular trichomes where they formed large “pools.” Occasionally, a large aggregate was observed to be associated with a wounded epidermal plant cell.
TABLE 2.
Number and size of bacterial aggregates observed that were associated with the different anatomical features of a bean leaf at 2 and 8 days following inoculation of P. syringae strain B728a
| Bacterial aggregate location | % of the leaf surface | No. and size (no. of cells) of bacterial aggregates on leaves at indicated day after inoculationa
|
|||
|---|---|---|---|---|---|
| 2
|
8
|
||||
| n | Meanb ± SE | n | Mean ± SE | ||
| Glandular trichomes | 0.6 ± 0.1 | 93 | 311.4 ± 38.5 | 114 | 1,092.3 ± 254.1 |
| Veins | 7.3 ± 0.4 | 20 | 301.2 ± 89.8 | 34 | 705.6 ± 214.6 |
| Hooked trichomes | 1.2 ± 0.2 | 10 | 204.1 ± 19.5 | 26 | 409.8 ± 71.7 |
| Nondifferentiated epidermal cells | 73.8 ± 0.8 | 27 | 117.7 ± 2.9 | 163 | 304.1 ± 41.7 |
| Stomates | 17.1 ± 0.3 | 0 | 0 | ||
| Total | 100 | 150 | 268.0 ± 27.3 | 337 | 619.4 ± 92.8 |
Only aggregates with 100 cells or more were included in this study.
Mean aggregate size values represent the inferred number of cells calculated from the area of a leaf covered with a given aggregate and an average cell size of 1.5 μm2.
FIG. 5.
Observed and expected frequency distributions of aggregate sizes associated with glandular trichomes (GT) and nondifferentiated epidermal cells (EC) 8 days following inoculation with P. syringae pv. syringae strain B728a on bean leaf surfaces. Aggregates associated with glandular trichomes were more numerous (P < 0.001) and significantly larger (P < 0.001) than expected. Aggregates associated with nondifferentiated cells were significantly smaller (P < 0.005) and less numerous (P < 0.001) than expected. The same results were observed 2 days after inoculation (data not shown).
Spatial heterogeneity among sampling units and estimation of population sizes.
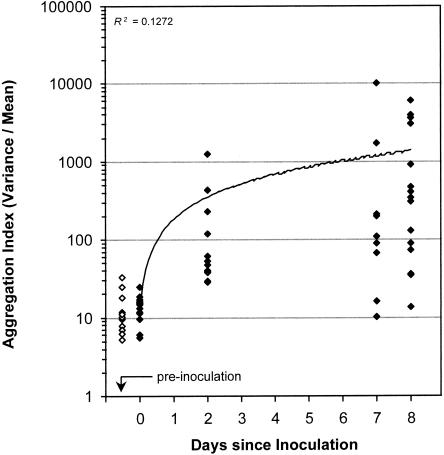
Culturable bacterial-population sizes (determined for entire leaves and leaf segments by plating appropriate dilutions of leaf washings on agar medium) were compared with total population sizes estimated by image analysis. Culturable bacterial-population sizes among the small sampling units employed demonstrated a high level of variability (Fig. 6). The differences between segments in aggregation patterns of total bacteria (as well as the overall increase in the number and size of aggregates through time) are reflected in the large variations observed in mean/variance ratios (used as aggregation indexes) (Fig. 7).
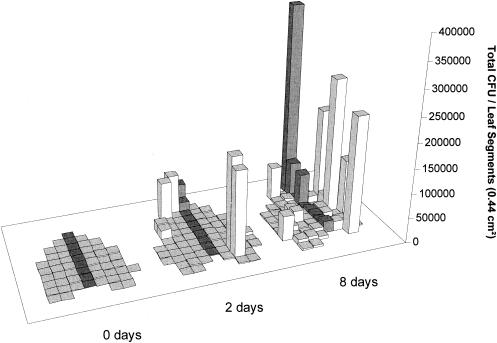
FIG. 6.
Culturable population sizes of P. syringae pv. syringae strain B728a among different leaf segments cut from an individual bean leaf, 0, 2, and 8 days after inoculation. Bacteria were removed from individual 44-mm2 segments at each harvest time. The variance/mean ratios for populations on different segments collected 0, 2, and 8 days after inoculation with P. syringae were 971, 104,434, and 157,812, respectively.
FIG. 7.
Aggregation index (variance/mean) ratios within leaf segments as determined by image analysis as a function of time on noninoculated plants (open diamonds) and following inoculation with P. syringae pv. syringae strain B728a (filled diamonds). Each symbol represents an individual leaf segment.
DISCUSSION
This study provides the first quantitative information on the frequency and size of bacterial aggregates determined directly on leaf surfaces and demonstrates that aggregated bacteria account for the majority of the cells present on a leaf. At 8 days after inoculation of P. syringae on bean leaf surfaces, between 30 and 70% of the total epiphytic bacterial population was located in aggregates containing 100 cells or more. Our data corroborate a previous study (using a method based on the separation of solitary and aggregated cells by filtration of leaf washings) that reported that aggregated cells could account for from 10 to 40% of the total bacterial population on broad-leaf endive and parsley (38). Unpublished data from the same laboratory suggest that on cantaloupe, the aggregated fraction can represent up to 70% of the total epiphytic bacterial population (36). While their approach is a fast and easy way to demonstrate the presence of aggregated cells on individual leaves, it is not as quantitative and may not reflect the original aggregation status of cells on plants or provide information about the size and spatial distribution of aggregates. The direct observation and measurement of bacterial aggregates on the leaf provide more interpretable information on the number of aggregates present and their size frequency distribution and spatial location.
Description of the spatial distribution of epiphytic bacteria at a scale corresponding to their sizes provides biological and ecological information about the processes that lead to successful colonization. Kinkel et al. have shown that bacterial populations are highly variable among leaves and among individual 9-mm2 leaf segments (23). Our data confirm such observations and provide further information about the sites at which bacterial populations are aggregated within individual leaf segments. The variability in spatial aggregation clearly extends even to small regions within a given leaf segment (Fig. 4C). While the variability in population sizes observed at the plant, leaf, or leaf segment level could result from several environmental and biological factors, the variability observed within leaf segments seems to be driven by the presence or absence of microsites conducive to bacterial growth. Cells were not randomly scattered over the leaf surface and occurred in a wide range of cluster sizes, roughly reflecting the spatial heterogeneity of nutrients available on leaf surfaces (28).
Previous qualitative studies (using scanning electron microscopy) revealed that the most common sites of bacterial colonization are at the base of trichomes, at stomates, and at epidermal cell wall junctions, especially in the groove along the veins. Our study revealed that P. syringae pv. syringae strain B728a preferentially formed aggregates at the base of glandular trichomes and in the grooves along the veins, with smaller aggregates found only in the grooves between epidermal cells and at the base of hooked trichomes; we hypothesize that due to a limited amount of nutrients, the latter sites allow limited bacterial growth. For such large aggregates to form, that is, the physical environment must be conducive and an ample supply of carbon-containing nutrients must be present. The use of whole-cell biosensors for sugars on leaves revealed that the sites of ample sugar abundance on a leaf are few and small (28), and the use of such sensors to establish correlation between aggregate formation and the availability of growth substrates should be informative. Aggregates observed on leaf surfaces of plants maintained under conditions of low RH were less numerous and smaller than aggregates observed on leaf surfaces of plants maintained under moist conditions for the same period of time. The lack of free water on leaves may have reduced the rate of nutrient leakage onto leaves and restricted the spatial motility of the nutrients, thereby reducing the rate at which aggregates could grow. We hypothesize that while aggregate formation is favored by the presence of water, aggregates might also contribute to the survival of bacteria periodically exposed to desiccation stress. The repeated exposure of leaves to such stress may be an important factor in causing such a large proportion of cells to be found in such aggregates. In support of such a model, quantification of any differential survival results for solitary and aggregated cells on leaves should prove informative.
Bacterial colonization of plants clearly occurs preferentially at certain features of a leaf. While other studies show that P. syringae pv. syringae can colonize stomates of pear and apple leaves (31, 32), we did not observe any colonization of bean stomates. Small aggregates were sometimes observed associated with stomates under field conditions, but we could not conclude whether such aggregated cells were P. syringae cells or represented other bacterial species. The aggregates that formed along the veins were stretched along the grooves between cells and probably resulted from exudation of nutrients as well as the presence of residual water. Occasionally, a large aggregate was observed in association with a wounded epidermal cell, reinforcing the concept that nutrient availability limits the formation of aggregates on leaf surfaces. Glandular trichomes probably offer optimal conditions for microbial growth due to their ability to retain water droplets (10) and secrete a diversity of chemical compounds, including sugars, proteins, oils, secondary metabolites, and mucilage (2, 40). Aggregates associated with glandular trichomes formed large contiguous pools and were markedly different in shape from aggregates observed near other sites. Their shapes strongly suggest that trichomes leak nutrients onto the leaf surface from a point source and that bacterial growth is promoted by the leakage of these nutrients.
Using different microscopy techniques to observe aggregates on leaf surfaces, Morris et al. have also reported that large aggregates (on field-grown leaves) were often attached to glandular trichomes (37). In a recent study, we have also reported that cells were significantly larger near glandular trichomes (34), suggesting that the physiological state (presumably driven by ample nutrient resources) of the bacteria in such sites is different from that in sites where aggregates are not formed. Different bacterial strains may behave differently, and a given strain may also behave differently on the leaf surfaces of different plant species; however, we have observed that the spatial distributions of Pantoea agglomerans and Pseudomonas fluorescens cells on bean leaf surfaces were similar to that observed for P. syringae (unpublished data). While our result cannot be generalized to other strains, P. syringae pv. syringae strain B728a seems typical of pathogens with an epiphytic phase and other strains such as P. syringae DC3000 seem atypical. As reported by Boureau et al., P. syringae DC3000 is a poor epiphytic colonizer and seems to be an exception to the general P. syringae model (9).
Like microbial biofilms observed in aquatic environments, aggregates observed in the phyllosphere are characterized by a localized high level of density of cells. Within a few days following inoculation we observed aggregates containing between 104 and 105 cells. The physiology of the aggregated population might be influenced by the high level of cell density. Cells within aquatic biofilms are markedly different phenotypically from planktonic cells of the same species. It has been reported that as many as 30% of the genes of a given species are expressed selectively within biofilms (13), and it has been shown that the expression (including exopolysaccharide production) of several genes is affected in biofilms (41, 48). Although gene expression of bacteria on a leaf surface might be influenced by many factors that differ within the microenvironment encountered on the leaf, several traits important in plant-microbe interactions have been shown to be regulated in a cell density-dependent manner via quorum-sensing mechanisms (4, 12, 15, 26, 47). Several plant-associated bacteria (including Erwinia spp. and Xanthomonas spp.) have been shown to regulate the expression of traits involved in pathogenicity by production of N-acylated homoserine lactone (N-acyl HSL) (4, 26, 47). Several authors have now reported that most plant-pathogenic bacteria (including species with an epiphytic phase, such as Erwinia spp. and Pseudomonas spp.) produce N-acyl HSLs (11, 15, 47). These reports, combined with our observations, suggest that density-dependent expression of traits involved in epiphytic interactions with plants can also occur. Given that P. syringae produces cell-signaling compounds (15, 16, 24, 25), we speculate that cells in aggregates in which such compounds would accumulate such signals might have a different epiphytic fitness than more-solitary cells. Our preliminary results strongly suggest that aggregated cells of P. syringae may benefit from N-acyl HSL production, since mutants of P. syringae pv. syringae strain B728a unable to produce N-acyl HSL exhibit a reduced ability to survive desiccation stress after multiplying on bean plants (unpublished data).
The presence of large clusters of bacteria on leaves might also increase the potential for metabolic and genetic exchange. Bailey et al. have reported that remarkably high rates of transfer of conjugative plasmids can occur on plants (3). In an attempt to demonstrate the role of aggregates in plasmid exchange among epiphytes, Jacques et al. (M.-A. Jacques, C. E. Morris, and M. J. Bailey, Abstr. Proceedings 5th Eur. Meet. Bacterial Gen. Ecol., Nafplion, Greece, vol. 1, abstr. S1, p. 135, 1996) have characterized plasmid distribution among different strains of fluorescent pseudomonads in an individual aggregate isolated from broad-leaved endive and reported that half of the strains contained the same mercury-resistant plasmid. Bjorklof et al. have reported that plasmid transfer was highly efficient when plants were incubated under conditions of high RH (7) but could also occur under conditions of low RH and when bacterial growth had ceased (8). These authors suggested that plasmid transfers occurred in small aggregates observed on the leaf surface. They have also reported that plasmid transfer rates decreased when bacteria were inoculated separately in time and have suggested that the formation of a mucoid matrix might have acted as a barrier for gene transfer by preventing cell-to-cell contact (8). While their observations were not sufficiently detailed to have described the large, but infrequently occurring, aggregates found here, we find it tempting to speculate that the rates of plasmid exchange would be particularly high at such sites. Thus, the patchy distribution of nutrients leading to bacterial aggregates on leaves may provide important breeding grounds for microbial diversity in plant-associated bacteria.
While P. syringae is a plant-pathogenic bacterium with a particularly prominent epiphytic phase (20), our knowledge of how P. syringae is able to survive on leaf surfaces and how disease is initiated is still limited. The results reported in this study provide new insights into its epiphytic life (often viewed as the first step in the infection process). The formation of large aggregates, probably embedded in extracellular polysaccharide (37), might provide an ecological niche for the majority of the cells of P. syringae present on a leaf that is different from that of the more solitary cells which had previously been considered typical of epiphytes. Judging on the basis of the presumption that cells located at the bottom of an aggregate are protected, aggregates might provide a protected site in which bacteria could escape harsh conditions encountered on the leaf surface. While bacterial growth apparently occurs preferentially near potential sources of nutrients, aggregates might also be sites where (because of the large local cell density) phytotoxins (such as syringomycin and syringopeptin) are locally produced, thereby leading to an increased availability of nutrients by increasing plant cell leakiness (6). Aggregates might also be sites where disease is induced because of the selected expression of virulence genes. While there is as yet no published report of N-acyl HSL production on leaf surfaces, P. syringae might express virulence genes in a cell density-dependent manner in epiphytic aggregates.
The objective of our study was to quantitatively describe the spatial distribution of bacterial aggregates on leaf surfaces, specifically during the epiphytic growth phase of P. syringae. Only asymptomatic leaves were sampled, and endophytic colonization was seldom observed during the time course of the different experiments. Endophytic colonization was associated with the different leaf features and occurs in the apoplast between plant cells inside the leaf tissue. No trend regarding correlations between endophytic colonization and any particular leaf feature was observed as Sabaratnam and Beattie have reported (45) for stomates. In their study, the authors considered bacteria resistant to leaf surface sterilization treatment to be endophytic. However, we have reported that a majority of the cells located in aggregates can survive when exposed to environmental stresses such as desiccation (35). Judging on the basis of such observations, and since the majority of bacterial cells are located in large aggregates, we hypothesize that in their study Sabaratnam and Beattie may have underestimated epiphytic populations or overestimated endophytic populations.
In an attempt to link aggregate formation and the disease process, we were able to identify individual necrotic or water-soaked plant cells on young trifoliated leaves and observed that these were heavily colonized by P. syringae cells. However, we were not able to determine whether a given wounded plant cell resulted from the presence of an aggregate formed by this pathogen or whether an aggregate resulted from the presence of a wounded plant cell. While symptoms and endophytic colonization could be observed at a later stage of disease development, the relatively low frequency of occurrence of infections or symptoms on a given leaf (less than 10 per leaf) makes identification of such sites very difficult. The infection process is also probably prolonged and asynchronous, making it very hard to know when to look at and where to look on a leaf to identify the precursors of disease. While it is possible that not all aggregates induce disease, and even if (as is more likely) not all diseases come from aggregates, the goal is to determine the initial site of infection (i.e., that of a single plant cell); however, this will remain a technical challenge.
Aggregate formation among epiphytes has significant ecological implications that must be considered when designing strategies to control plant-pathogenic bacteria. As suggested above, at least some cells in large aggregates may be protected from bactericides applied to plants, especially when the cells are embedded in a mucoid matrix. The progressive decrease in the susceptibility of P. syringae to cupric hydroxide application to leaves with increasing periods of colonization prior to bactericide treatments may reflect the development of protective aggregates (1). Since most antibacterial compounds have been developed on the basis of tests of their activity against planktonic cells in synthetic media, they probably differ in efficacy against aggregated epiphytic populations. In addition, a clustered colonization pattern of the leaf surface (with a majority of the cells located in a few large aggregates) may significantly limit microbial interactions to a few sites as well as providing the “refuges” that have been proposed to account in part for the lack of efficiency of biological control agents applied to leaves (21).
Acknowledgments
This study was supported by grant 99-35303-8633 from the U.S. Department of Agriculture, National Research Initiative, by grant DR-F603-86ER13518 from the U.S. Department of Energy, and by the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego, Calif.
We thank Steve Ruzin and Denise Schichnes of the Biological Imaging Facility at the University of California Berkeley for their assistance with microscopy.
REFERENCES
- 1.Andersen, G. L., O. Menkissoglou, and S. E. Lindow. 1991. Occurrence and properties of copper-tolerant strains of Pseudomonas syringae isolated from fruit trees in California [USA]. Phytopathology 81:648-656. [Google Scholar]
- 2.Ascensao, L., and M. S. Pais. 1998. The leaf capitate trichomes of Leonotis leonurus: histochemistry, ultrastructure and secretion. Ann. Bot. 81:263-271. [Google Scholar]
- 3.Bailey, M. J., A. K. Lilley, and J. P. Diaper. 1996. Gene transfer between microorganisms in the phyllosphere, p 103-123. In C. E. Morris, P. C. Nicot, and C. Nguyen-The (ed.), Aerial plant surface microbiology. Plenum Press, New York, N.Y.
- 4.Barber, C. E., J. L. Tang, J. X. Feng, M. Q. Pan, T. J. G. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555-566. [DOI] [PubMed] [Google Scholar]
- 5.Beattie, G. A., and S. E. Lindow. 1994. Comparison of the behavior of epiphytic fitness mutants of Pseudomonas syringae under controlled and field conditions. Appl. Environ. Microbiol. 60:3799-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, C. L., F. Alarcon-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorklof, K., A. Suoniemi, K. Haahtela, and M. Romantschuk. 1995. High frequency of conjugation versus plasmid segregation of RP1 in epiphytic Pseudomonas syringae populations. Microbiology 141:2719-2727. [DOI] [PubMed] [Google Scholar]
- 8.Bjorklof, K., E. L. Nurmiaho-Lassila, N. Klinger, K. Haahtela, and M. Romantschuk. 2000. Colonization strategies and conjugal gene transfer of inoculated Pseudomonas syringae on the leaf surface. J. Appl. Microbiol. 89:423-432. [DOI] [PubMed] [Google Scholar]
- 9.Boureau, T., J. Routtu, E. Roine, S. Taira, and M. Romantschuk. 2002. Localization of hrpA-induced Pseudomonas syringae pv. tomato DC3000 in infected tomato leaves. Mol. Plant Pathol. 3:451-460. [DOI] [PubMed] [Google Scholar]
- 10.Brewer, C. A., W. K. Smith, and T. C. Vogelmann. 1991. Functional interaction between leaf trichomes, leaf wettability and the optical properties of water droplets. Plant Cell Environ. 14:955-962. [Google Scholar]
- 11.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 12.Clough, S. J., K.-E. Lee, M. A. Schell, and T. P. Denny. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179:3639-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, R. Korber, and H. M. Lappinscott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 14.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 15.Dumenyo, C. K., A. Mukherjee, W. Chun, and A. K. Chatterjee. 1998. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur. J. Plant Pathol. 104:569-582. [Google Scholar]
- 16.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fett, W. F. 2000. Naturally occurring biofilms on alfalfa and other types of sprouts. J. Food Prot. 63:625-632. [DOI] [PubMed] [Google Scholar]
- 18.Gras, M., H., C. Druet-Michaud, and O. Cerf. 1994. La flore bactérienne des feuilles de salade fraiche. Sci. Aliments 14:173-188. [Google Scholar]
- 19.Hirano, S. S., and C. D. Upper. 1983. Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21:243-269. [Google Scholar]
- 20.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, K. B. 1994. Dose-response relationships and inundative biological control. Phytopathology 84:780-784. [Google Scholar]
- 22.King, E. O., M. K. Ward, and D. E. Rainey. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 23.Kinkel, L. L., M. Wilson, and S. E. Lindow. 1995. Effects of scales on estimates of epiphytic bacterial populations. Microb. Ecol. 29:283-297. [DOI] [PubMed] [Google Scholar]
- 24.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitten, T., T. G. Kinscherf, J. L. McEvoy, and D. K. Willis. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28:917-929. [DOI] [PubMed] [Google Scholar]
- 26.Koiv, V., and A. Mae. 2001. Quorum sensing controls the synthesis of virulence factors by modulating rsmA gene expression in Erwinia carotovora sp. carotovora. Mol. Genet. Genom. 265:287-292. [DOI] [PubMed] [Google Scholar]
- 27.Leben, C. 1988. Relative humidity and the survival of epiphytic bacteria with buds and leaves of cucumber plants. Phytopathology 78:179-185. [Google Scholar]
- 28.Leveau, J. H. J., and S. E. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 98:3446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindow, S. E. 1983. The role of bacterial ice nucleation in frost injury to plants. Annu. Rev. Phytopathol. 21:363-384. [Google Scholar]
- 30.Loper, J. E., and S. E. Lindow. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449-1454. [Google Scholar]
- 31.Mansvelt, E. L., and M. J. Hattingh. 1987. Scanning electron microscopy of colonization of pear leaves by Pseudomonas syringae pv. syringae. Can. J. Bot. 65:2517-2522. [Google Scholar]
- 32.Mansvelt, E. L., and M. J. Hattingh. 1989. Scanning electron microscopy of invasion of apple leaves and blossoms by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 55:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mew, T. W., and C. M. Vera Cruz. 1986. Epiphytic colonization of host and non-host plants by phytopathogenic bacteria, p. 269-282. In N. J. Fokkema and J. van den Heuvel (ed.), Microbiology of the phyllosphere. Cambridge University Press, New York, N.Y.
- 34.Monier, J.-M., and S. E. Lindow. 2003. Pseudomonas syringae responds to the environment on leaves by cell size reduction. Phytopathology 93:1209-1216. [DOI] [PubMed] [Google Scholar]
- 35.Monier, J.-M., and S. E. Lindow. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 100:15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, C. E., M. B. Barnes, and R. J. C. McLean. 2001. Biofilms on leaf surfaces: implications for the biology, ecology and management of populations of epiphytic bacteria, p138-154. In S. E. Lindow, E. I. Hecht-Poinar, and V. J. Elliot (ed.), Phyllosphere microbiology. APS Press, St. Paul, Minn.
- 37.Morris, C. E., J.-M. Monier, and M.-A. Jacques. 1997. Methods for observing microbial biofilms directly on leaf surfaces and recovering them for isolation of culturable microorganism. Appl. Environ. Microbiol. 63:1570-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris, C. E., J.-M. Monier, and M.-A. Jacques. 1998. A technique to quantify the population size and composition of the biofilm component in communities of bacteria in the phyllosphere. Appl. Environ. Microbiol. 64:4789-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien, R. D., and S. E. Lindow. 1989. Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology 79:619-627. [Google Scholar]
- 40.Olson, D. L., and J. R. Nechols. 1995. Effects of squash leaf trichome exudates and honey on adult feeding, survival, and fecundity of the squash bug (Heteroptera: Coreidae) egg parasitoid Gryon pennsylvanicum (Hymenoptera: Scelionidae). Environ. Entomol. 24:454-458. [Google Scholar]
- 41.Potera, C. 1999. Microbiology—forging a link between biofilms and disease. Science 283:1838-1839. [DOI] [PubMed] [Google Scholar]
- 42.Pratt, L. A., and R. Kolter. 1999. Genetic analysis of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 43.Roos, I. M. M., and M. J. Hattingh. 1983. Scanning electron microscopy of Pseudomonas syringae pv. morsprunorum on sweet cherry leaves. Phytopathol. Z. 180:18-25. [Google Scholar]
- 44.Rouse, D. I., E. V. Nordheim, S. S. Hirano, and C. D. Upper. 1985. A model relating the probability of foliar disease incidence to the population frequencies of bacterial plant pathogens. Phytopathology 75:505-509. [Google Scholar]
- 45.Sabaratnam, S., and G. A. Beattie. 2003. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl. Environ. Microbiol. 69:1220-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timmer, L. W., J. J. Marois, and D. Achor. 1987. Growth and survival of xanthomonads under conditions nonconducive to disease development. Phytopathology 77:1341-1345. [Google Scholar]
- 47.Von Bodman, S. B., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, M., S. S. Hirano, and S. E. Lindow. 1999. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl. Environ. Microbiol. 65:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]