SUMMARY
Regardless of genetic makeup, a female honey bee becomes a queen or worker depending on the food she receives as a larva. For decades, it has been known that nutrition and juvenile hormone (JH) signaling determine the caste fate of the individual bee. However, it is still largely unclear how these factors are connected. To address this question, we suppressed nutrient sensing by RNA interference (RNAi)-mediated gene knockdown of IRS (insulin receptor substrate) and TOR (target of rapamycin) in larvae reared on queen diet. The treatments affected several layers of organismal organization that could play a role in the response to differential nutrition between castes. These include transcript profiles, proteomic patterns, lipid levels, DNA methylation response and morphological features. Most importantly, gene knockdown abolished a JH peak that signals queen development and resulted in a worker phenotype. Application of JH rescued the queen phenotype in either knockdown, which demonstrates that the larval response to JH remains intact and can drive normal developmental plasticity even when IRS or TOR transcript levels are reduced. We discuss our results in the context of other recent findings on honey bee caste and development and propose that IRS is an alternative substrate for the Egfr (epidermal growth factor receptor) in honey bees. Overall, our study describes how the interplay of nutritional and hormonal signals affects many levels of organismal organization to build different phenotypes from identical genotypes.
KEY WORDS: diphenic development, queen bee, worker bee, insulin/insulin-like signaling, target of rapamycin, epidermal growth factor receptor
INTRODUCTION
One of the key characteristics of eusocial insect societies (termites, ants, some bees and wasps) is the reproductive division of labor within the colony (Wilson, 1971). In honey bees, Apis mellifera L., this is achieved via a diphenism between females: highly fecund queens that perform the reproductive function of the colony, and essentially sterile workers, which carry out the vast majority of other tasks. Depending upon differential nurturing by workers, fertilized (female) honey bee eggs can develop into either queens or workers. Bee larvae that consistently receive the nutrient-rich royal jelly throughout the larval stages during development attain queen characteristics. These individuals have enlarged bodies and well-developed ovaries, enabling them to specialize in egg laying. In contrast, larvae fed a restricted diet from the 3rd instar onward become small-bodied workers with greatly reduced ovarian development (Winston, 1987). As a result of this nutritional dichotomy, differences occur in larval titers of juvenile hormone (JH), a major systemic lipophilic hormone and transcriptional regulator in insects (Hartfelder and Engels, 1998). Honey bee larvae show a predictable pattern of JH signaling, with queen-destined larvae expressing higher titers during the 4th to 5th instars (Rachinsky et al., 1990; Rachinsky and Hartfelder, 1990). Experiments have revealed that consumption of a diet rich in royal jelly increases larval JH titer (Rembold, 1987; Rachinsky et al., 1990; Rachinsky and Hartfelder, 1990) and that application of synthetic JH to larvae reared on a restricted diet is sufficient to cause the development of queen-like traits (Rembold et al., 1974; Goewie, 1977; Dietz et al., 1979). These findings have led to the conclusion that the influence of food intake on JH is the key determinant of female caste in honey bees. Because of this link, recent investigations have focused on understanding the molecular relationship between nutritional stimuli and the hormonal modulation that results in caste differences. Initially, studies pointed towards two highly conserved eukaryotic nutrient-sensing pathways, IIS (insulin/insulin-like signalling) and TOR (target of rapamycin) as key regulators in honey bee caste determination (Patel et al., 2007; Wolschin et al., 2011). Both pathways are known integrators of genetic and environmental stimuli, regulating cell growth, metabolism, reproduction and lifespan across taxa (Oldham and Hafen, 2003; Wullschleger et al., 2006). During a critical period of larval honey bee phenotypic differentiation, transcripts encoding ILP-1 (insulin-like peptide-1), IRS (insulin receptor substrate) (Wheeler et al., 2006) and InR (insulin receptor) (de Azevedo and Hartfelder, 2008) show reduced expression in worker-destined larvae compared with queen-destined larvae. Furthermore, RNA interference (RNAi)-mediated knockdown of TOR and IRS expression in 3rd instar larvae is sufficient to cause queen-destined individuals to develop worker traits (Patel et al., 2007; Wolschin et al., 2011). Also, methylation of CpG (cytosine–phosphate–guanosine) dinucleotides is associated with an alteration in gene expression patterns across taxa (Jaenisch and Bird, 2003) and queen diet results in lower (CpG) DNA methylation levels in larvae compared to worker-diet reared larvae (Kucharski et al., 2008). Therefore, somatic imprinting established by nutrient sensing may contribute to caste-specific gene expression patterns. In support of this hypothesis, larval knockdown of DNA methyltransferase 3 (dnmt3), an enzyme that mediates de novo CpG methylation, is sufficient to cause worker-destined larvae to develop queen-like traits (Kucharski et al., 2008). Reports of InR and chico (IRS) influencing JH levels in insects (Tatar et al., 2001; Tu et al., 2005) lend further support to a role for IIS in caste development as JH levels influence queen–worker dimorphism. However, the relationship between TOR and JH in insects is less clear (Shiao et al., 2008; Kapahi et al., 2004).
Recent work by Kamakura has greatly advanced the understanding of honey bee caste regulation (Kamakura, 2011). A key protein component of royal jelly, royalactin, was identified as a major activator of the queen developmental pathway. It was subsequently revealed that the reaction to royalactin is mediated by epidermal growth factor receptor (Egfr), leading to changes in JH titer and caste differentiation. In the course of the study, it was also concluded that IIS, as assessed by knockdown of one InR, is not involved in caste differentiation, and that TOR has only a downstream supporting role. Part of this conclusion could potentially be in conflict with the conclusions of a previous study, which showed that caste development is perturbed by knockdown of IRS (Wolschin et al., 2011), and thus presumably by impaired IIS.
Here, we assessed the roles of IRS and TOR in queen/worker development by parallel and concurrent downregulation of these pathways in combination with analyses of several layers of molecular detail culminating in effects on JH signaling. To achieve this, we used RNAi to knock down (i) IRS, (ii) TOR and (iii) IRS and TOR simultaneously, in bee larvae reared on queen-inducing diet. Knockdown and control larvae were tested for expression of genes in the IRS, TOR and Egfr pathways, proteomic patterns, JH content and DNA methylation, while morphological traits were scored in adult individuals. We also examined abdominal lipid stores, as this trait is characteristically affected (increased) by IRS silencing in fruit flies (Bohni et al., 1999). Individual gene knockdowns resulted in an adult worker phenotype with effects on gene expression, protein abundance, lipid levels, DNA methylation patterns, and reduced larval JH levels. We further tested whether queen development could be rescued by restoration of JH signaling after gene knockdown. Our study provides new evidence that places IRS and TOR upstream of JH during honey bee caste differentiation. We propose that this is not mutually exclusive with other findings if IRS also provides an alternative substrate for Egfr.
MATERIALS AND METHODS
Bees
We used wild-type honey bees (unselected commercial stocks), maintained in the apiary of Arizona State University. Queens (N=2) were caged on a wax comb for 24 h for egg laying, and after 3 days, newly hatched (12–18 h old) larvae were grafted into the VS diet as previously described (Patel et al., 2007). Larvae from both queens were assigned randomly to all treatments throughout the study.
In vitro feeding regime
The queen-inducing feeding regime was as described previously (Patel et al., 2007), except that the amount of double-stranded RNA (dsRNA) directed toward IRS was adjusted to 250 μg ml–1 concentration in the diet. For IRS–/TOR–, 150 μg ml–1 of TOR and 250 μg ml–1 of IRS were mixed and fed to the larvae. The quantities used were chosen as the lowest concentration resulting in robust gene knockdown (Wolschin et al., 2011). Control larvae were fed with 400 μg ml–1 of dsGFP (green fluorescent protein) RNA. In total, 150 larvae per treatment group were used and each experimental set up was replicated twice. Details of the feeding regime are shown in supplementary material Fig. S1. All experiments were performed in an incubation chamber at 33°C and 80% relative humidity. Daily mortality during larval development was at 5% in all treatment groups except double knockdown where ∼15–20% of the larvae died. During pupation, mortality of about 50% was observed in all groups, with only two eclosed as adults in the double knockdown IRS–/TOR– group. For the JH treatments, 100 larvae per treatment group were used.
RNA isolation, first strand cDNA synthesis and RT-PCR
Total RNA was isolated from larvae using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the procedure provided by the manufacturer. For RT-PCR, total RNA was treated with DNaseI (Ambion, Austin, TX, USA) following standard instructions. Following RNA isolation, 5.0 μg was used as a template in cDNA synthesis using SuperScriptR III First-Strand Synthesis System for RT-PCR (Invitrogen) with oligoDT primers. PCR was done using primers listed in supplementary material Table S3; primers for some genes (actin, TOR, ILP-1 and ILP-2) were adapted from previous studies (Wheeler et al., 2006; Patel et al., 2007). Subsequently, 1.0 μl of first strand cDNA was used as a template for PCR, for which we used primers at 0.3 μmol l–1 concentration and 15.0 μl PCR master mix (Promega, Madison, WI, USA) in a final volume of 30.0 μl. Primer sequences and GenBank accession numbers of the genes analyzed by RT-PCR are listed in supplementary material Table S3. Briefly, PCR conditions were 94°C for 5 min, followed by 28 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. PCR products (20 μl) were examined on 2% agarose gels.
Protein extraction and proteomic analysis
These steps (see Appendix for details) were carried out as previously described (Wolschin et al., 2011), except larvae were subjected to extensive washing with nuclease-free water and rolled over Kimwipes in order to minimize potentially confounding effects of food-derived proteins on their cuticle. Liquid chromatography coupled to tandem mass spectrometry (LC–MS2) was used for proteomics analysis. Five individuals were assayed per group.
Lipid quantification
Fifth instar larvae were collected directly into methanol:chloroform buffer and subjected to lipid analysis (see Toth and Robinson, 2005). Absorbance was read at 525 nm on a spectrophotometer (Ultraspec 2100 pro, Amersham Biosciences, GE Healthcare, Piscataway, NJ, USA). A standard curve using known amounts of pure cholesterol was used to calculate lipid amounts. Each sample was run in duplicate and averages of these paired sample values were used for analysis.
JH analysis
We collected 5th instar larvae of the IRS–, TOR–, IRS–/TOR– and control groups. Each larva was gently rolled on a Kimwipe to remove diet, weighed and then sampled into 1 ml of 50% acetonitrile/50% water solution and stored at –80°C until purification. The larvae were ground using a glass rod, centrifuged and the supernatant collected. Samples were processed using the GC–MS method as reported elsewhere [(Dolezal et al., 2009) as modified from Shu et al. (Shu et al., 1997)]. Purified and derivatized JH was analyzed using a HP 6890 Series GC equipped with a 30 m×0.25 mm Carbowax Econo-Cap GC column (Alltech, Nicholasville, KY, USA) coupled to a HP 5973N inert MSD/DS running in SIM mode. Helium was used as a carrier gas. JH was quantified and normalized as pg μg–1 wet mass against a calibration curve established with JH III standards (Sigma-Aldrich, St Louis, MO, USA).
JH treatment
Bee larvae were grafted from experimental colonies and provisioned with one of the in vitro diets (see above). At the beginning of day 4, and after the completion of the described dsRNA feeding schedule, the topical application of either 1.0 μl JH (10 μg JH/1 μl acetone) or 1.0 μl of sham control (acetone) was performed twice at 24 h intervals. The IRS and TOR knockdown bees were split between JH-treated groups and acetone (sham)-treated groups, and all control (dsGFP RNA)-fed larvae were treated with acetone. The in vitro diet continued until the larvae began defecating. Defecating larvae were removed from the diet prior to pupation and cleaned to remove excess diet, then transferred into Petri plates with filter paper (Whatman) at the bottom and allowed to pupate in an incubator at 33°C. The time from larval defecation to adult eclosion was recorded. On the day of adult emergence, bee age and body mass were recorded, caste-associated morphologies were identified, and ovariole number was assessed (see Appendix for details).
Statistics
We used the non-parametric Kruskal–Wallis test, and thereafter Mann–Whitney U-tests for post hoc comparisons of pupal length, lipid levels and JH levels (four treatment groups). χ2 goodness of fit test was used for comparison of the DNA methylation data. For comparison of protein levels between control and knockdown groups, statistical analysis was conducted as described previously (Wolschin et al., 2011). Non-parametric Kruskal–Wallis ANOVA was used to test overall differences in protein levels between all treatment groups followed by Mann–Whitney U-tests for post hoc comparisons. Type 1 error inflation was controlled by bootstrap correction of the Kruskal–Wallis alpha level: 1000 bootstrap iterations were run for each protein. All analyses except the proteomics analysis (conducted in R 2.10.1; www.r-project.org) were performed with Statistica 6.0.
RESULTS
IRS and TOR affect gene expression, lipid levels, protein abundance and DNA methylation levels
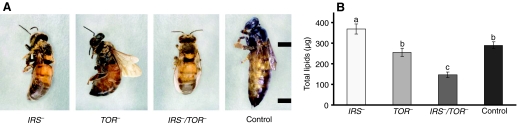
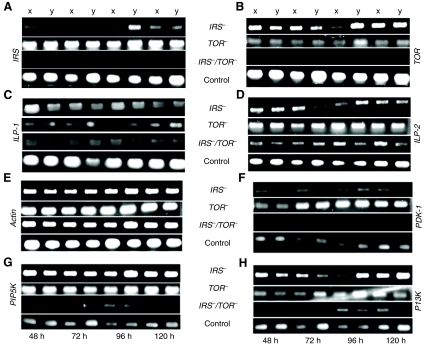
In order to gain broader insight into how nutrient sensing by IIS, Egfr and TOR pathways acts as an integrator of honey bee caste differentiation, we generated IRS– and TOR– single knockdown phenotypes as well as a double knockdown (IRS–/TOR–) phenotype (Fig. 1; supplementary material Fig. S2). All treatment groups were scored for caste-specific morphological characteristics: presence or absence of corbicula (pollen basket on the hindleg) and mandibular notch, ovariole count per ovary, pupation time and adult wet mass (see Materials and methods for details). Knockdowns developed worker traits while controls attained the characteristics of queens (see Fig. 1). We verified the downregulation of IRS, TOR and other essential pathway components, including those also involved in Egfr signaling, such as PDK-1 and PI3K (Navolanic et al., 2003), in response to RNAi by using semiquantitative PCR (Fig. 2). IRS–/TOR– double knockdowns showed cumulative effects on transcript levels with respect to controls (Fig. 2A,B). Downregulation of IRS expression further resulted in a decrease of TOR mRNA levels (Fig. 2B), but TOR RNAi did not affect IRS expression (Fig. 2A). Knockdowns showed a decrease in ILP-1 expression compared with ILP-2 (insulin-like peptide-2), which was expressed at higher levels in IRS– and TOR– knockdowns but not in IRS–/TOR– double knockdowns (Fig. 2C,D). Prior studies have shown that ILP-1 is naturally low in worker larvae, while ILP-2 is elevated (Wheeler et al., 2006; de Azevedo and Hartfelder, 2008; Ament et al., 2008). These findings are in accordance with our results from the IRS– and TOR– phenotypes (Fig. 2C,D), in which ILP-1 expression is diminished but ILP-2 is upregulated (or probably little affected) in TOR– knockdowns, and this supports the argument for different roles for these peptides in honey bee caste development. As expected from previous work on flies, total abdominal lipid levels increased significantly in IRS– larvae (Bohni et al., 1999), while this was not the case in TOR– larvae compared with controls, and IRS–/TOR– larvae had the least abdominal lipids (Fig. 1B).
Fig. 1.
IRS (insulin receptor substrate) and TOR (target of rapamycin) expression affect female caste fate in honey bees. (A) Final adult phenotypes in IRS–, TOR– and IRS–/TOR– gene knockdowns and control groups. Distance between tick marks, 1cm. (B) Total lipid levels were elevated in IRS– compared with TOR–, IRS–/TOR– and control 5th instar larvae. There was no significant difference between the TOR– and control larvae (N=10). Bars represent means ± s.e. and different letters (a–c) refer to groups showing differences according to the post hoc Mann–Whitney U-test (P<0.05).
Fig. 2.
Semi-quantitative RT-PCR analysis. mRNA expression profile from two larvae (x and y) representative of knockdown and control groups collected at different time points (48, 72, 96 and 120 h) after first dsRNA feeding. (A) Verification of downregulation of IRS (GenBank accession no. XM_391985) expression in IRS– and IRS–/TOR– but not in TOR– larvae relative to control larvae. (B) TOR (XM_625127) expression was downregulated in TOR–, IRS– and IRS–/TOR– relative to control larvae. (C) ILP-1 (insulin-like peptide-1; GB_17332) expression was reduced in IRS–, TOR– and IRS–/TOR– larvae when compared with ILP-2. (D) ILP-2 (GB_10174) was expressed at higher levels in IRS– and TOR– larvae compared with ILP-1. (E) Actin (XM_623378) was used as a control gene. (F) PDK-1 (XM_396869) was up-regulated in TOR– larvae compared with the other groups. (G) PIP5K (XM_001120893) is up-regulated in IRS– and TOR– larvae when compared with controls. (H) PI3K (XM_394208) is initially down-regulated but later up-regulated after 96–120 h of dsRNA feeding in IRS– and TOR– larvae compared with controls. Increased transcription of PIP5K and PI3K may lead to greater production of diacylglycerol that promotes lipid storage (Zemel et al., 2000; Watt et al., 2003), a syndrome observed in IRS– larvae. General down-regulation of gene expression was observed in IRS–/TOR– larvae (B–F).
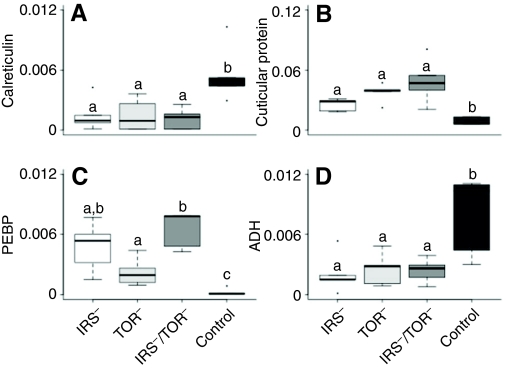
A proteomic analysis further validated the biochemical differences between knockdowns and controls (Fig. 3; supplementary material Fig. S3 and Table S1). For example, calreticulin and acetaldehyde dehydrogenase were reduced in the knockdown groups compared with controls (Fig. 3A,D), while levels of cuticular protein 4 and PEPB (phosphatidylethanolamine binding protein) were higher in knockdowns (Fig. 3B,C). The findings were in general agreement with an earlier study on IRS– (Wolschin et al., 2011). Additional results are provided in supplementary material Fig. S3 and Table S1.
Fig. 3.
Abundance levels of selected proteins in knockdown vs control 5th instar larvae as determined by tandem mass spectrometry (LC–MS2). Protein abundance was measured in normalized spectrum count and displayed in box plots with interquartile ranges from 25% to 75% (N= 5 per group). (A) Calreticulin (GenBank accession no. XP_392689) is a multifunction protein involved in signal transduction; (B) cuticular protein 4 (XP_001122907) may be influenced by JH (Roberts and Willis, 1980); (C) phosphatidylethanolamine binding protein (PEBP; XP_623194) is a potential inhibitor of the MAP kinase pathway (Vallee et al., 2003); and (D) aldehyde dehydrogenase CG3752-PA isoform 1 (ADH; XP_623084) and insulin levels are positively correlated in mice (Demozay et al., 2004).
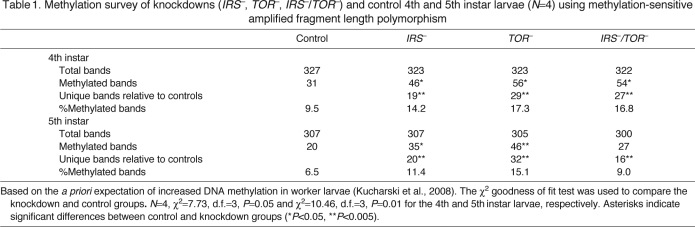
Finally, we tested relative de novo DNA methylation in response to downregulation of IRS–, TOR– and IRS–/TOR– larvae using a methylation-sensitive amplified fragment length polymorphism (AFLP) technique (supplementary material Fig. S4). Single and double gene knockdown individuals showed significant addition of methylated loci overall (Table 1; post hoc comparisons between all the treatments are shown in supplementary material Table S2). Our data are in agreement with the finding that worker-destined larvae show higher levels of DNA methylation than queen-destined bees (Kucharski et al., 2008) and with the observation that about 11% of AFLP loci are methylated during worker bee development (Kronforst et al., 2008). The effect on DNA methylation was most pronounced in TOR– 4th and 5th larvae when compared with controls.
Table 1.
Methylation survey of knockdowns (IRS–, TOR–, IRS–/TOR–) and control 4th and 5th instar larvae (N=4) using methylation-sensitive amplified fragment length polymorphism
Knockdown of IRS and TOR reduces JH content in larvae
In this experiment, we sampled 5th instar larvae corresponding to the time point when the JH level shows the largest difference between queen- and worker-destined bees (Rachinsky et al., 1990). We found that JH content was significantly lower in IRS–, TOR– and IRS–/TOR– larvae compared with controls (Fig. 4A). This result was consistent with natural differences in JH titers observed in worker- and queen-destined larvae (Rachinsky et al., 1990). It also illustrates that, although the IRS–, TOR– and IRS–/TOR– larvae are characterized by some complementary effects at the level of gene expression (Fig. 2B–H) and protein abundance (Fig. 3; supplementary material Fig. S3 and Table S1), the gene knockdowns shared a common influence on JH during the 5th instar.
Fig. 4.
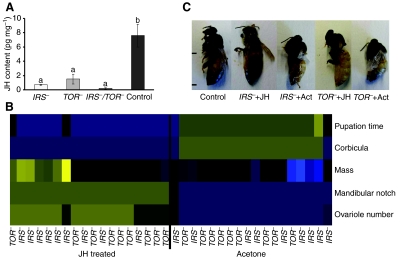
IRS and TOR expression affect endocrine signaling during honey bee development. (A) Juvenile hormone (JH) content of early 5th instar larvae (S1 stage) of IRS– (N=10), TOR– (N=9), IRS–/TOR– (N=9) and control (N=7). Bars represent means ± s.e and different letters (a or b) refer to groups showing differences according to the post hoc Mann–Whitney U-tests (P<0.05). (B) The heatmap is based on unsupervised hierarchical clustering and displays morphological differences between individuals treated with JH and controls. Characteristics that were investigated included the presence (queen character) or absence (worker character) of a mandibular notch, the presence (worker character) or absence (queen character) of a structure on the hindleg (corbicula), mass in grams (queens weigh more than workers), time spent as a pupa (in this case, total time from larval defecation until adult eclosion), and ovariole number (a higher value is indicative of a queen phenotype). All values were z-transformed before generating the heatmap. Values increase from blue (low value) over black (medium value) to yellow (high value). (C) JH treatment rescues the queen phenotype in IRS– and TOR– larvae. Final adult phenotypes in the control, IRS–+JH, IRS–+Ac (acetone), TOR–+JH and TOR–+Ac treatment groups. Distance between tick marks, 1 cm.
JH application rescues queen development in IRS and TOR knockdowns
Topical application of either 1.0 μl (10 pg μl–1) JH III in acetone or 1.0 μl of sham (vehicle) control (acetone) was performed twice at 24 h intervals to 5th instar larvae (supplementary material Fig. S1). As before, all treatment groups were scored for caste-specific morphological characteristics: presence or absence of corbicula (pollen basket on the hindleg), ovariole count per ovary, and mandibular notch; pupation time and adult wet mass were also recorded (Fig. 4B,C). Double knockdowns (IRS–/TOR–) were not treated because of the high mortality rate (see Materials and methods for details).
JH treatment resulted in bees that eclosed with queen characteristics (Fig. 4C) and exhibited a queen-like adult phenotype with enlarged body and more developed ovaries, while the majority of sham controls emerged with worker traits (Fig. 4C and supplementary material Movie 1). Adult phenotypes are shown in Fig. 4B. We conclude that JH can direct honey bee development independent of disruptions of major nutrient-sensing genes.
DISCUSSION
In eusocial insects, one genotype provides molecular roadmaps to multiple morphologically and behaviorally distinct phenotypes that are induced and shaped by environmental variables such as nutrition, temperature and humidity (Winston, 1987; Page and Amdam, 2007). Honey bee caste differentiation is frequently used as a model for understanding the molecular basis and regulation of these polyphenisms. However, different results do not always come together to form consistent explanations. Here, we generated RNAi knockdowns of IRS and TOR, and a corresponding double knockdown (IRS–/TOR–) to address one such challenge.
As shown previously (Patel et al., 2007; Wolschin et al., 2011), modulation of either IRS or TOR was sufficient to alter the developmental fate of a queen-destined larva into a worker phenotype. Although perturbation of IRS and TOR both lead to a worker phenotype, their effects on the transcriptome (Fig. 2), proteome (Fig. 3; supplementary material Fig. S3) and total lipid level (Fig. 1B) are different. These results suggest that, despite some common downstream effects, which culminate in the JH level (Fig. 4A), IRS and TOR can have separate functions during honey bee development. This conclusion may be extended to Egfr-mediated pathways that also regulate changes in JH signaling to affect queen fate (Kamakura, 2011). IRS, TOR and Egfr could, thereby, illustrate how multiple molecular sensors converge onto one central signal integrator (JH) to ensure robust decision making during a crucial developmental stage.
An in vitro experiment using cultured corpora allata (the glands that produce JH) from Blatella germanica showed that downregulation of TOR leads to reduced JH production (Maestro et al., 2009). Our results take this insight a step forward to establish the interactions between IRS, TOR and JH at an organismal level. Our data suggest that these interactions participate in the orchestration of honey bee caste development, such that perturbations in either pathway are sufficient to alter caste fate from queen to worker. However, even after downregulation of IRS and TOR, queen development could be rescued by established protocols for artificial reinitiation of JH signaling (Rembold, 1974; Goewie and Beetsma, 1976; Dietz et al., 1979), showing that developmental plasticity is maintained. This evidence also clearly places the JH signal downstream of nutrient-sensing genes in the route of caste development.
Our results may seem to conflict with conclusions of Kamakura (Kamakura, 2011). We propose two possible explanations that resolve this inconsistency. A first possibility is that our knockdown of IRS effectively downregulated any and all InR-dependent signals, while Kamakura perturbed one predicted InR gene of honey bees (InR-2), thus perhaps accommodating redundant or compensatory signaling via an alternative gene (InR-1). However, the expression of both predicted honey bee InR genes is naturally suppressed during caste differentiation (de Azevedo and Hartfelder, 2008). This finding suggests that no redundant or compensatory signaling takes place.
It follows that there is a second possibility: IRS is not only mediating InR signals during the critical window of queen–worker development. While not characterized in the honey bee, it has been shown in mammals that IRS-1 and IRS-2 are key mediators of Egfr activation (Fujioka and Ui, 2001; Fujioka et al., 2001). In addition, research on human cell lines has shown that Egfr is a main recruiter of the IRS protein, sometimes dominant even over IGF-IR (insulin-like growth factor type 1 receptor) (Knowlden et al., 2008). Thus, it is quite possible that IIS, as mediated by the InR proteins, is less important than Egfr signaling during queen–worker differentiation. However, it can be equally correct that IRS is central to this phenotypic divergence: as a mediator of Egfr.
It was previously argued that Egfr is a ‘stiff’ component of its pathway; that is, where small perturbations result in large phenotypic effects. Such stiff components can be contrasted with ‘soft’ components that withstand more variation with less significant consequences for the phenotype (Brown et al., 2004). These system properties are widespread in biology and physics; they are formalized mathematically (Brown and Sethna, 2003) and result in circuits that produce very reliable outcomes (Brown and Sethna, 2003; Brown et al., 2004). Considering the selective advantage of reproductive division of labor in eusocial insects, and the costs of colonies producing too many, or inopportune, queens, it follows that reliability is highly desirable during caste differentiation (Nijhout, 2003). Variable phenotypic impact is thus expected for components of pathways that influence caste fate, and this variation may be exemplified by Egfr, IRS, TOR and S6-kinase (Patel et al., 2007; Wolschin et al., 2011; Kamakura, 2011). Therefore, rather than focusing on single explanations, it may be important to ask how molecular pathways act together during insect caste development to produce reliable outcomes in the face of environmental and genetic noise.
APPENDIX
Cloning of IRS and preparation of dsRNA
These procedures were conducted as described previously (Wang et al., 2010). Briefly, after dsRNA synthesis (see Patel et al., 2007), the PCR products were excised from low melting temperature 1% agarose gels and purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). The dsRNA was then made using an AmpliScribe T7 transcription kit (Epicentre Biotechnologies, Madison, WI, USA) following the manufacturer's protocol. dsRNA was purified using phenol:choloform extraction and run on a 1% agarose gel for verification of size and purity (Mutti et al., 2008).
Scoring morphological characteristics that distinguish queens and workers
Queens were identified as having >100 ovarioles per ovary, notched mandibles, a smooth stinger and no corbicula (pollen basket) (Snodgrass, 1956). Workers were identified as having 2–30 ovarioles per ovary and a barbed stinger, and by the presence of a corbicula. For worker ovariole scoring, ovaries were dissected and placed in a drop of water and ovarioles were counted under a microscope. For queen ovariole scoring, ovarioles were dissected and fixed in 4% paraformaldehyde and embedded into a 3% agarose in PBS (137 mmol l–1 NaCl, 2.7 mmol l–1 KCl, 10 mmol l–1 Na2HPO4, 1.8 mmol l–1 KH2PO4; pH 7.4). Following embedding, the agarose was cut into thin sections using a microtome and ovarioles were counted under a microscope at 40× magnification. For queens, ovariole number was estimated within an accuracy of 10 ovarioles per ovary. Detailed data on morphological characteristics and their occurrence were in agreement with our previously reported results (Patel et al., 2007; Wolschin et al., 2011).
Protein extraction from larval tissue
Larvae were placed on a Kimwipe and gently rinsed with water to minimize contamination from royal jelly (Wolschin et al., 2011). Then, larvae were ground for 1 min in 100 μl of extraction buffer (50 mmol l–1 Tris pH 8.5, 2% SDS, 100 mmol l–1 DTT, 10% glycerol). After vortexing, samples were heated for 5 min at 95°C and vortexed again. Following centrifugation for 5 min at 12,000 g at room temperature, the supernatant was transferred to a new tube and proteins were precipitated using methanol:chloroform (Luong et al., 2006). Pellets were redissolved in 40 μl of buffer [50 mmol l–1 Tris pH 8.5, 6 mol l–1 urea, 2 mol l–1 thiourea, 2% acetonitrile (MeCN), 1 mmol l–1 CaCl2] using two incubation periods of 5 min and vigorous vortexing. Then, 160 μl of dilution buffer (50 mmol l–1 Tris pH 8.5, 2% MeCN, 1 mmol l–1 CaCl2) was added and the samples were vortexed for 5 min. The resulting samples were centrifuged for 10 min at 10,000 g and the supernatant was transferred to a new tube. Protein concentrations were determined with the Bradford assay using albumin as a standard (Bradford, 1976). For protein digestion, 1 μg of trypsin was added to 40 μg of protein and proteins were incubated overnight at 30°C. Peptides were desalted according to Rappsilber and colleagues (Rappsilber et al., 2003).
Peptide analysis
Briefly, peptides (10 μg per sample) were separated using a monolithic column and eluted into a linear ion trap mass spectrometer (Thermo Electron, San Diego, CA, USA). In order to minimize potential carry-over effects between runs, samples were separated by a blank and were run in a semi-randomized fashion that ensured an equal distribution of all sample types over the time of the analysis. Settings for the HPLC and the mass spectrometer were essentially as described previously (Wolschin and Amdam, 2007a; Wolschin and Amdam, 2007b; Wolschin et al., 2011), but MS2 were triggered for the five most abundant peaks of every MS recording. OMSSA 2.0.0 (Greer et al., 2004) was used for searching the A. mellifera database with the following tolerance windows: 0.8 Da fragment tolerance, 0.8 Da precursor tolerance, maximum of two missed cleavages, only tryptic sequences allowed, 10 hits per peptide reported (after analysis the output was restricted to only the best matches), variable modifications: methionine oxidation, deamidation of glutamine and asparagine. For quantification purposes, only proteins with a spectrum count ≥4 in at least four out of the six replicates were considered. Statistical analysis was conducted as described before (Wolschin and Amdam, 2007a). For each protein, the individual spectral count was divided by the total spectral count of the sample in order to achieve normalization.
Methylation-sensitive AFLP
DNA was extracted from individual IRS–, TOR–, IRS–/TOR– and control 4th and 5th instar larvae as described previously (Wang et al., 2006). Two replicates per treatment per instar were used. For methylation-sensitive AFLP DNA display, 500 ng of DNA was digested with EcoRI/MspI and EcoRI/HpaII restriction endonucleases, and respective oligo primers (adaptors) were ligated to the ends of the DNA fragments to generate template DNA for amplification. The PCR reactions were performed in a total volume of 20 μl containing 20 ng of the DNA template, 5 μmol l–1 of each dNTP, 0.4 μl of Taq polymerase, 5 μmol l–1 of the MspI/HpaII primer, 20 μmol l–1 of EcoRI primer, 1.5 mmol l–1 MgCl2, 2 μl of 10× PCR reaction buffer and 0.2 μl of 35S dATP isotope. The ligated products were PCR amplified in the presence of 35S isotope using primer combinations with one and three restrictive bases at the 3′ end of the primers. The generic primer used for EcoRI was: 5′-GACTGCGTACCAATTN-3′; for MspI/HpaII: 5′-GATGAGTCTAGAACGGAN-3′. The primer combinations used were adapted from Kronfrost et al. (Kronfrost et al., 2008) (supplementary material Table S4). The PCR conditions for single and three selective nucleotides were performed as described before (Vos et al., 1995). After amplification, the PCR products were size separated on a 0.4 mm thick denaturing 8% polyacrylamide/8 mol l–1 urea gel, following a standard sequencing gel protocol. Only 1/5th (5 μl) of the amplification product was loaded on the gel. The gels were pre-run at 33 mA constant current for 30 min and then at 70 W constant power for 4 h. An X-ray film was placed on the dried gel and exposed for 3–5 days. Two independent AFLP libraries per sample were made and each PCR was repeated twice for accuracy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Baker for assistance with general bee keeping and members of the Amdam lab for helpful discussions. We are grateful to T. Flatt, C. Smith, Ø. Halskau, G. Reeck, J. Reese, M.-S. Chen, K. Traynor, S. Pratt, J. Hunt, M. Coulombe, D. Moore and O. Rueppell for helpful comments on the manuscript, and A. Smith and Z. Shaffer for assistance with the photography.
FOOTNOTES
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/214/23/3977/DC1
FUNDING
This work was supported by the Research Council of Norway (180504 and 185306, 191699), US National Science Foundation (0615502), National Institute on Aging (NIA P01 AG22500), PEW Foundation (G.V.A.) and the Alexander-von-Humboldt-Foundation (F.W.). Deposited in PMC for release after 12 months.
REFERENCES
- Ament S. A., Corona M., Pollock H. S., Robinson G. E. (2008). Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl. Acad. Sci. USA 105, 4226-4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., Andruss B. F., Beckingham K., Hafen E. (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97, 865-875 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248-254 [DOI] [PubMed] [Google Scholar]
- Brown K. S., Sethna J. P. (2003). Statistical mechanical approaches to models with many poorly known parameters. Phys. Rev. E 68, 021904 [DOI] [PubMed] [Google Scholar]
- Brown K. S., Hill C. C., Calero G. A., Myers C. R., Lee K. H., Sethna J. P., Cerione R. A. (2004). The statistical mechanics of complex signaling networks: nerve growth factor signaling. Phys. Biol. 1, 184-195 [DOI] [PubMed] [Google Scholar]
- de Azevedo S. V., Hartfelder K. (2008). The insulin signaling pathway in honey bee (Apis mellifera) caste development-differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 54, 1064-1071 [DOI] [PubMed] [Google Scholar]
- Demozay D., Rocchi S., Mas J. C., Grillo S., Pirola L., Chavey C., Van Obberghen E. (2004). Fatty aldehyde dehydrogenase – potential role in oxidative stress protection and regulation of its gene expression by insulin. J. Biol. Chem. 279, 6261-6270 [DOI] [PubMed] [Google Scholar]
- Dietz A., Hermann H. R., Blum M. S. (1979). Role of exogenous Jh-I, Jh-Iii and Anti-Jh (Precocene-Ii) on queen induction of 4.5-day-old worker honey bee larvae. J. Insect Physiol. 25, 503-512 [Google Scholar]
- Dolezal A. G., Brent C. S., Gadau J., Holldobler B., Amdam G. V. (2009). Endocrine physiology of the division of labour in Pogonomyrmex californicus founding queens. Anim. Behav. 77, 1005-1010 [Google Scholar]
- Fujioka T., Ui M. (2001). Involvement of insulin receptor substrates in epidermal growth factor induced activation of phosphatidylinositol 3-kinase in rat hepatocyte primary culture. Eur. J. Biochem. 268, 25-34 [DOI] [PubMed] [Google Scholar]
- Fujioka T., Kim J. H., Adachi H., Saito K., Tsujimoto M., Yokoyama S., Ui M. (2001). Further evidence for the involvement of insulin receptor substrates in epidermal growth factor-induced activation of phosphatidylinositol 3-kinase. Eur. J. Biochem. 268, 4158-4168 [DOI] [PubMed] [Google Scholar]
- Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X. Y., Shi W. Y., Bryant S. H. (2004). Open mass spectrometry search algorithm. J. Proteome Res. 3, 958-964 [DOI] [PubMed] [Google Scholar]
- Goewie E. A. (1977). Induction of caste differentiation in honey bee (Apis mellifera) after topical application of JH-III. Insectes Soc. 24, 265 [Google Scholar]
- Goewie E. A., Beetsma J. (1976). Induction of caste differentiation in honey bee (Apis mellifera) after topical application of JH-III. Proc. Kon. Ned. Akad. Wetenesch 79, 466-469 [Google Scholar]
- Hartfelder K., Engels W. (1998). Social insect polymorphism: hormonal regulation of plasticity in development and reproduction in the honey bee. Curr. Top. Dev. Biol. 40, 45-77 [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Bird A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Gen. 33, 245-254 [DOI] [PubMed] [Google Scholar]
- Kamakura M. (2011). Royalactin induces queen differentiation in honey bees. Nature 473, 478-483 [DOI] [PubMed] [Google Scholar]
- Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden J. M., Jones H. E., Barrow D., Gee J. M. W., Nicholson R. I., Hutcheson I. R. (2008). Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signalling: implication for Gefitinib (‘Iressa’) response and resistance. Breast Cancer Res. Treat. 111, 79-91 [DOI] [PubMed] [Google Scholar]
- Kronforst M. R., Gilley D. C., Strassmann J. E., Queller D. C. (2008). DNA methylation is widespread across social Hymenoptera. Curr. Biol. 18, R287-288 [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R. (2008). Nutritional control of reproductive status in honey bees via DNA methylation. Science 319, 1827-1830 [DOI] [PubMed] [Google Scholar]
- Luong N., Davies C. R., Wessells R. J., Graham S. M., King M. T., Veech R., Bodmer R., Oldham S. M. (2006). Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 4, 133-142 [DOI] [PubMed] [Google Scholar]
- Maestro J. L., Cobo J., Belles X. (2009). Target of rapamycin (TOR) mediates the transduction of nutritional signals into juvenile hormone production. J. Biol. Chem. 284, 5506-5513 [DOI] [PubMed] [Google Scholar]
- Mutti N. S., Louis J., Pappan L. K., Pappan K., Begum K., Chen M. S., Park Y., Dittmer N., Marshall J., Reese J. C., et al. (2008). A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA 105, 9965-9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navolanic P. M., Steelman L. S., McCubrey J. A. (2003). Egfr family signaling and its association with breast cancer development and resistance to chemotherapy (Review). Int. J. Oncol. 22, 237-252 [PubMed] [Google Scholar]
- Nijhout H. F. (2003). Development and evolution of adaptive polyphenisms. Evol. Dev. 5, 9-18 [DOI] [PubMed] [Google Scholar]
- Oldham S., Hafen E. (2003). Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell. Biol. 13, 79-85 [DOI] [PubMed] [Google Scholar]
- Page R. E., Jr, Amdam G. V. (2007). The making of a social insect: developmental architectures of social design. BioEssays 29, 334-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Fondrk M. K., Kaftanoglu O., Emore C., Hunt G., Amdam G. V. (2007). The making of a queen: TOR pathway governs diphenic caste development. PLoS ONE 6, e509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinsky A., Hartfelder K. (1990). Corpora allata activity, a prime regulating element for caste-specific juvenile-hormone titer in honey-bee larvae (Apis-mellifera-carnica). J. Insect Physiol. 36, 189-194 [Google Scholar]
- Rachinsky A., Strambi C., Strambi A., Hartfelder K. (1990). Caste and metamorphosis: hemolymph titers of juvenile hormone and ecdysteroids in last instar honey bee larvae. Gen. Comp. Endocrinol. 79, 31-38 [DOI] [PubMed] [Google Scholar]
- Rembold H. (1987). Caste specific modulation of juvenile-hormone titers in Apis mellifera. Insect Biochem. 17, 1003-1006 [Google Scholar]
- Rembold H., Czoppelt C., Rao P. J. (1974). Effect of juvenile hormone treatment on caste differentiation in the honey bee, Apis mellifera. J. Insect Physiol. 20, 1193-1202 [DOI] [PubMed] [Google Scholar]
- Roberts P. E., Willis J. H. (1980). Effects of juvenile hormone, ecdysterone, actinomycin D, and mitomycin C on the cuticular proteins of Tenebrio molitor. J. Embryol. Exp. Morphol. 56, 107-123 [PubMed] [Google Scholar]
- Shiao S. H., Hansen I. A., Zhu J., Sieglaff D. H., Raikhel A. S. (2008). Juvenile hormone connects larval nutrition with target of rapamycin signaling in the mosquito Aedes aegypti. J. Insect Physiol. 54, 231-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S. Q., Park Y. I., Ramaswamy S. B., Srinivasan A. (1997). Hemolymph juvenile hormone titers in pupal and adult stages of southwestern corn borer [Diatraea grandiosella (Pyralidae)] and relationship with egg development. J. Insect Physiol. 43, 719-726 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663-670 [DOI] [PubMed] [Google Scholar]
- Snodgrass R. E. (1956). Anatomy of the Honey Bee. New York: Comstock Publishing Associates; [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M. P., Yin C. M., Garofalo R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107-110 [DOI] [PubMed] [Google Scholar]
- Toth A. L., Robinson G. E. (2005). Worker nutrition and division of labour in honeybees. Anim. Behav. 69, 427-435 [Google Scholar]
- Tu M. P., Yin C. M., Tatar M. (2005). Insulin signal regulation of juvenile hormone synthesis in Drosophila melanogaster. Gen. Comp. Endocrinol. 142, 347-356 [DOI] [PubMed] [Google Scholar]
- Vallee B. S., Coadou G., Labbe H., Sy D., Vovelle F., Schoentgen F. (2003). Peptides corresponding to the N- and C-terminal parts of PEBP are well-structured in solution: new insights into their possible interaction with partners in vivo. J. Pept. Res. 61, 47-57 [DOI] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nuc. Acids Res. 23, 4407-4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jorda M., Jones P. L., Maleszka R., Ling X., Robertson H. M., Mizzen C. A., Peinado M. A., Robinson G. E. (2006). Functional CpG methylation system in a social insect. Science 314, 645-647 [DOI] [PubMed] [Google Scholar]
- Wang Y., Mutti N. S., Ihle K. E., Siegel A., Dolezal A. G., Kaftanoglu O., Amdam G. V. (2010). Down-regulation of honey bee IRS gene biases behavior toward food rich in protein. PLoS Genet. 6, e1000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt M. J., Steinberg G. R., Heigenhauser G. J., Spriet L. L., Dyck D. J. (2003). Hormone-sensitive lipase activity and triacylglycerol hydrolysis are decreased in rat soleus muscle by cyclopiazonic acid. Am. J. Physiol. Endocrinol. Metab. 285, E412-E419 [DOI] [PubMed] [Google Scholar]
- Wheeler D. E., Buck N., Evans J. D. (2006). Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect. Mol. Biol. 15, 597-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. O. (1971). The insect societies. Cambridge MA: Belknap Press of Harvard University Press; [Google Scholar]
- Winston M. L. (1987). The Biology of the Honey Bee. Cambridge, MA: Harvard University Press; [Google Scholar]
- Wolschin F., Amdam G. V. (2007a). Comparative proteomics reveal characteristics of life-history transitions in a social insect. Proteome Sci. 5, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschin F., Amdam G. V. (2007b). Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera). Anal. Bioanal. Chem. 389, 1095-1100 [DOI] [PubMed] [Google Scholar]
- Wolschin F., Mutti N. S., Amdam G. V. (2011). Insulin receptor substrate influences female caste development in honeybees. Biol. Lett. 7, 112-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 127, 5-19 [DOI] [PubMed] [Google Scholar]
- Zemel M. B., Shi H., Greer B., Dirienzo D., Zemel P. C. (2000). Regulation of adiposity by dietary calcium. FASEB J. 14, 1132-1138 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.