Abstract
We have proposed the novel concept that the macrophage ubiquitin-proteasome (UP) pathway functions as a key regulator of LPS-induced inflammation signaling. Our findings suggest that proteasome-associated protease subunits X, Y, and Z, are replaced by LMP subunits after LPS treatment of RAW 264.7 cells. Our objective here was to determine the contribution of selective LMP proteasomal subunits to LPS-induced nitric oxide (NO) and TNF-α production in primary murine macrophages. Accordingly, thioglycollate-elicited macrophages from LMP7, LMP2, LMP10 (MECL-1), and LMP7/MECL-1 double knockout mice were stimulated in vitro with LPS, and were found to generate markedly reduced NO levels compared to wild-type (WT) mice, whereas TNF-α levels responses were essentially unaltered relative to wild-type responses. Our recent studies suggest that the TRIF/TRAM pathway is defective in LMP knockouts which may explain why iNOS/NO are not robustly induced in LPS-treated macrophages from knockouts. Treating these macrophages with IFN-γ and LPS, however, reverses this defect, leading to robust NO induction. TNF-α is induced by LPS in the LMP knockout macrophages because IκB and IRAK are degraded normally via the MyD88 pathway. Collectively, these findings strongly support the concept that LMP7/MECL-1 proteasomes subunits actively function to regulate LPS-induced NO production by affecting the TRIF/TRAM pathway.
Keywords: inflammation, endotoxic shock, macrophages, cytokines, nitric oxide
INTRODUCTION
Lipopolysaccharide (LPS) is the prototype activator of macrophages via Toll-like receptors, leading to the generation of several pro and anti-inflammatory mediators through the MyD88-dependent or MyD88-independent TRIF/TRAM pathways. When produced in excess, these cause inflammation that can lead to development of septic shock. Results of our recent studies indicate that the macrophage proteasome is a key regulator of macrophage-dependent inflammation and plays a pivotal role in LPS-induced signaling and gene expression (1-4). The 20S proteasome contains three distinct subunits (X, Y, and Z), with well-characterized protease sites: subunits X (β5), Y (β1), and Z (β2) display chymotrypsin-like, post-acidic (post-glutamyl), and trypsin-like activities, respectively (5). Treatment of macrophages with IFN-γ, well recognized for its capacity to amplify the response of these cells to LPS (6), has been shown to alter the subunit composition of proteasomes (7, 8). Following stimulation with IFN-γ, subunits X, Y, and Z are replaced by “immunoproteasome” subunits LMP7, LMP2, and MECL-1, respectively (7, 8) that, in turn, have traditionally been reported to enhance antigen processing in the development of specific acquired immune response. The contribution of these IFN-γ-induced proteasome subunit substitutions to IFN-γ-induced amplification of inflammatory responses however, has not been extensively explored.
Results of our recent studies have provided strong evidence that, in addition to IFN-γ LPS also induces expression of immunoproteasomes (9, preceding manuscript). Based upon these findings, we have hypothesized a model in which LPS interaction with RAW 267.4 cells, that express X, Y, and Z subunits predominantly, leads to activation of the basal proteolytic activities associated with the X, Y, and Z subunits, while inducing the LMP immunoproteasome subunits. Thus, activation and induction of immunoproteasome subunits, in turn, thereby modifies the immune response by altering specific proteolytic cleavage profiles of the activated immunoproteasome. The development of immunoproteasomes also effects changes in the expression of specific signaling molecules and transcription factors. This affects levels of gene expression, as well as potential degradation of mediator signaling proteins and late stage mediators such as NO. However, the relative contribution of each of the 3 individual subunits of immunoproteasomal proteases in innate immunity is not known. Whether the proteasomes play an active role in cytokine activation, NO formation, and signaling pathways in response to LPS has not been defined in detail.
Mice with targeted mutations in specific proteasome subunit genes (i.e., those encoding LMP2, LMP7, and MECL-1) have been recently developed and characterized with respect to T cell activation and antigen presentation (10). In this respect, proteasomes are known to play an important role in the processing of antigenic peptides for the class I MHC pathway (8), as well as in the initiation of T cell proliferation, cell cycle progression, signal transduction pathways, and activation of NF-κB by degrading IκB (11,12). Caudill et al. (10) provided evidence that T cells lacking both MECL-1 and LMP7 proteasomal subunits proliferate in response to polyclonal T-cell mitogens, and that this mitogenic response is independent of class I MHC antigen processing. In addition, these investigators have shown that mice lacking the inducible proteasomal subunits have diminished influenza epitope-induced IFN-γ production in CD8+ T cells (13). Recently, a role for LMP2 has been described in antiviral humoral and innate immune responses in lymphocytes have been associated with altered NF-κB activity (14). In addition, immunoproteasomes have been involved in increasing the peptide supply for antigen presentation (15). Collectively, these results strongly support the conclusion that specific proteolytic subunits of the proteasome are likely to play a far greater role in regulating immune responses than simply contributing to antigen presentation or processing. However, the mechanisms by which immunoproteasomes regulate LPS-induced signal transduction, and whether or not these play an active role in innate immunity are not well understood.
In the current study, we demonstrate that macrophages derived from mice with targeted mutations in LMP genes exhibit highly dysregulated cytokine induction in response to LPS in vitro. Specifically, NO levels in supernatants of LPS-stimulated macrophages from LMP−/− mice are markedly reduced compared to macrophages from wild-type mice, whereas TNF-α levels are essentially unaltered or upregulated in LMP-deficient macrophages. In addition, we have shown that the LPS-induced MyD88 pathway was functional, while the TRIF/TRAM and IRF-3 pathways were defective, in LMP knockout macrophages. These studies reveal a novel active function of the proteasomal subunits, LMP2, LMP7, and MECL-1 in regulating LPS-induced pathways.
MATERIALS AND METHODS
Reagents
Highly purified, deep rough chemotype LPS (Re LPS) from E. coli D31m4 was prepared as described by Qureshi et al. (16). For tissue culture studies, Dulbecco’s Modified Eagle Medium (DMEM), heat-inactivated low-endotoxin fetal bovine serum (FBS), and gentamicin were all purchased from Cambrex (Walkersville, MD). Thioglycollate was purchased from Sigma, Aldrich (St. Louis, MO) and RNeasy mini kit from QIAGEN sciences (Germantown, MD).
Animals and cell culture
LMP7, LMP2, MECL-1, and LMP7/MECL-1double-null mice were generated and bred as described (10). Age-matched control mice (C57BL/6, Wild type [WT]) were purchased from Charles Rivers Laboratory (Wilmington, MA). Primary, thioglycollate-elicited peritoneal macrophages were prepared from C57BL/6, LMP7−/−, LMP2−/−, MECL-1−/−, and LMP7−/−/MECL-1−/− on a C57BL/6 background as described previously (3). All studies were conducted following institutional approval of animal protocols.
Detection of cell viability
Viability of peritoneal macrophages treated with LPS was determined by a quantitative colorimetric assay with 3-(4, 5)-dimethylthiozol-2, 5-diphenyltetrazolium bromide (MTT) as described previously (16).
Measurement of TNF-α and NO
The levels of TNF-α in cell culture supernatants were determined by Quantikine M ELISA kit (R&D System, Minneapolis, MN) according to the manufacturer’s instructions. The lower limit of detection for TNF-α using this method is approximately, 5.0 pg /ml. The presence of NO in the supernatants was assayed using the Griess reagent kit (Sigma-Aldrich, St. Louis, MO).
Western blot analysis
Thioglycollate-elicited macrophages from control C57BL/6 mice and LMP7−/−/MECL-1−/− knockouts were treated as described in the legends to the figures. After stimulation, macrophages were washed with PBS, and cytoplasmic extracts were prepared from cells using cell extraction buffer (Biosource, Camarillo, CA.) supplemented with a protease inhibitor cocktail, containing phenylmethylsulfonyl fluoride and phosphatase inhibitors, according to the manufacturer’s directions as described previously (3). Protein concentrations were measured with BCA protein assay kits (source). Western blots were used to measure the relative proportions of the proteins of control C57BL/6 macrophages and macrophages from LMP7−/−/MECL-1−/− knockouts (a Western blot analysis showed the presence of X, Y, Z and LMP2 subunits in the thioglycollate-elicited macrophages from the double knockouts). Each well of the gel was loaded with 20-40 μg of protein and samples were electrophoresed at a constant 150 V in 1 × Tris glycine buffer for 50 min. Proteins in the gels were transferred onto the Immobilon Transfer Membranes (IPVH 15150; Millipore, Bedford, Mass) using the semidry transfer cell and, after the appropriate antibody treatments, bands were visualized with an enhanced chemiluminescence detection kit (Pierce) as described previously (4).
Real time RT-PCR
Total RNA was extracted from cells using RNeasy mini kit (Qiagen Sciences, Germantown, MD) according to manufacturer’s instruction. First-strand cDNA was prepared by TaqMan reverse transcriptase reagents (Applied Biosystem, Foster City, CA) using Peltier thermal cycler from MJ Research (Waltham, MA). The reverse transcribed cDNA was amplified and quantified using power SYBR green PCR master mix by Step One real time PCR system (Applied Biosystem) with specific primers according to manufacturer’s protocol. The primer sequences were used as follows: TNF-α, 5-CATCTTCTCAAAATTCGAGTGACAA-3 (forward primer), 5-TGGGAGTAGACAAGGTACAACCC-3 (reverse primer), IL-1β, 5-CCATGGCACATTCTGTTCAAA-3 (forward primer), 5-GCCCATCAGAGGCAAGGA-3 (reverse primer), IL-6, 5-GAGGATACCACTCCCAACAGACC-3 (forward primer), 5-AAGTGCATCATCGTTGTTCATACA-3 (reverse primer), iNOS, 5-GGATCTTCCCAGGCAACCA-3 (forward primer), 5- TCCACAACTCGCTCCAAGATT (reverse primer), IP-10, 5-CCTCATCCTGCTGGGTCTG-3 (Forward primer), 5-CTCAACACGTGGGCAGGA-3 (Reverse Primer), GAPDH, 5-TGTAGACCATGTAGTTGAGGTCA-3 (Forward primer), 5-AGGTCGGTGTGAACGGATTTG-3 (reverse primer). Following amplification, melting curve analysis was performed at temperature between 60°C to 95°C. Target genes mRNA copies were normalized to housekeeping gene GAPDH mRNA levels.
RESULTS
LPS-induced NO, but not TNF-α, is markedly reduced in macrophages from LMP knockout mice
RAW 264.7 macrophages which contain predominantly X, Y and Z subunits in their proteasomes (preceding manuscript). In contrast, thioglycollate-elicited macrophages from the C57BL/6 macrophages have mixed proteasomes that contain X, Y, Z, LMP7, LMP2, and MECL-1 subunits (data not presented).
Our previous experimental results demonstrated that LPS stimulation of WT C3H/FeJ thioglycollate-elicited macrophages induced an increase in the expression of LMP proteasome subunits (3). We also showed in the RAW 264.7 cell line that changes in the proteasomal subunits can be correlated with increases in the ratio of chymotrypsin-like activity/post-acidic activity and LPS-triggered NO production in these macrophages (preceding manuscript). Therefore, we hypothesized that absence of LMP proteasome subunits would adversely affect the ability of macrophages to produce NO in response to LPS stimulation. To test this hypothesis, we evaluated responses of macrophages derived from mice with targeted mutations in genes that encode several of the proteasome subunits, relative to macrophages from WT mice.
For these studies, we used macrophages from C57BL/6 mice as WT controls because the LMP7−/−, LMP2−/−, MECL-1−/− and LMP7/MECL-1−/− knockout mice (12) were backcrossed onto a C57BL/6 background. We compared LPS-induced levels of TNF-α and NO in supernatants from in vitro cultures of thioglycollate-elicited peritoneal macrophages obtained from both WT and knockout mice (Figure 1). TNF-α levels in culture supernatants of LPS-stimulated macrophages from knockout mice were not significantly reduced in LMP7−/−, LMP2−/−, MECL-1−/− and LMP7−/−/MECL-1−/− when compared to the controls (Figure 1). Conversely, our results demonstrate markedly reduced LPS-induced secretion of NO in culture supernatants of LPS-stimulated macrophages from all LMP knockout mice compared to control mice (Figure 1). Collectively, these findings support the concept that inducible LMP proteasome subunits are potentially key regulatory components of the activation pathway for LPS-induced NO release, but do not appear to play an equivalent role in LPS-induced TNF-α production.
Figure 1. NO and TNF-α responses to LPS in thioglycollate-elicited macrophages derived from C57BL/6 WT mice (C); LMP7/MECL-1−/−(DK), LMP7−/−, LMP2−/−, and MECL-1−/− knockouts.
A. Thioglycollate-elicited peritoneal macrophages were cultured in vitro and treated with medium or ReLPS at 0 (M), and 10 (L10) ng/ml for 8 h, and then supernatants were assayed for TNF-α by ELISA. Thioglycollate-elicited peritoneal macrophages were also cultured in vitro and treated with medium, or ReLPS at 100 (L100) ng/ml, for 48 h, and supernatants were assayed for NO using the Greiss reagent. This experiment was repeated three times. Data is plotted as % over WT controls.
We and others have shown previously IFN-γ synergizes with LPS to induce NO. We therefore tested the effect of IFN-γ on the LPS-induced NO in murine macrophages. The addition of IFN-γ to the cultures reversed this effect, showing significant increases in the LPS-induced NO in macrophages from all LMP single knockout mice; however, the levels of NO were still reduced compared to the responses of WT macrophages (Figure 2) at 48 h. In contrast, the levels of NO in macrophage culture supernatants from double knockout mice approached the control values. These data suggested that the major defect in the LMP7−/−/MeCL-1−/− macrophages is the absence of an IFN-mediated signal. In support of this interpretation, when IFN-γ is added exogenously, NO is again induced in response to LPS.
Figure 2. NO responses to LPS, and LPS plus IFN-γ in thioglycolate-elicited macrophages derived from C57BL/6 WT mice (C); LMP7/MECL-1−/− (DK), LMP7−/−, LMP2−/−, and MECL-1−/− double knockouts.
Thioglycollate-elicited macrophages were cultured in vitro and treated with medium, or ReLPS at 100 (L100) ng/ml, and IFN-γ (50 units) for 48 h, and supernatants were assayed for NO using the Greiss reagent. This experiment was repeated three times. Data is plotted as % over WT controls.
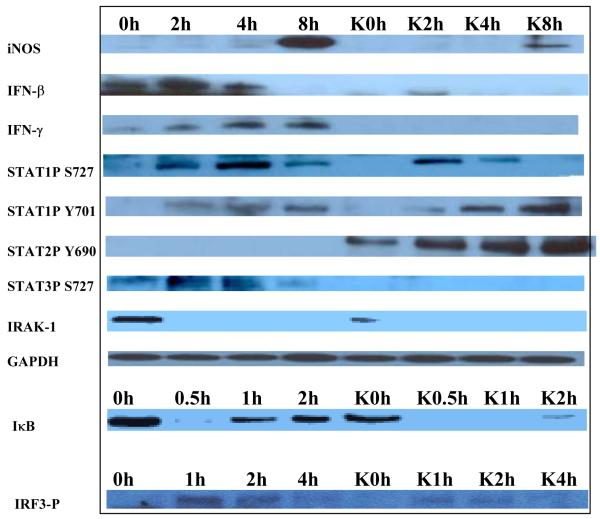
Since the induction of NO requires the MyD88-independent, TRIF/TRAM pathway, we hypothesized that this pathway, leading to the phosphorylation of IRF3, may not be functional in the macrophages of both single and double LMP knockouts. However, the effect was more pronounced in the double knockouts. To test this hypothesis we treated primary macrophages from wild-type control and LMP7−/−/MECL-1−/− double knockout mice without or with LPS for 0, 0.5, 1, 2, 4 and 8 h, and measured the relative levels of iNOS, IFN-γ, and signaling proteins in the cell lysates using Western Blotting (Figure 3). Upon stimulation with LPS, we observed a time-dependent change in the levels of iNOS, IFN-β, IFN-γ, and key signaling proteins such as P-STAT1 S727. P-STAT3 S727 and P-IRF3 were either undetectable, or were not as robustly induced in the LPS-treated LMP knockout macrophages as compared with the WT controls. In contrast, STAT2P Y690 was more robustly induced in the macrophages from knockouts, as compared to the macrophages from control mice. Degradation of ubiquitinated IκB by the proteasome for NF-κB activation occurred in macrophages from both controls and LMP knockouts suggesting that the defect is upstream of NF-κB. Collectively, these results suggested that the TRIF/TRAM pathway involving P-IRF3, STAT1 and IFN induction is not functional in the macrophages from the double knockouts.
Figure 3. Signaling proteins present in LPS-treated thioglycollate-elicited macrophages derived from C57BL/6 WT and LMP7−/−/MECL-1−/− double knockout mice.
Macrophages were cultured in vitro and treated with LPS (200 ng/ml) for 0, 0.5, 1, 2, 4 and 8 h. Cell lysates were prepared and the cytoplasmic signaling proteins were analyzed by Western Blotting. This experiment was repeated three times.
Levels of LPS-induced gene expression of IL-1, IL-6, and iNOS, but not TNF-α, are markedly reduced in macrophages from LMP knockout mice
We have shown that the proteasomal LMP subunits contribute to the LPS-induced secretion of NO but not TNF-α. We hypothesized that this may be due to decreased gene expression of iNOS, but not TNF-α. To test this hypothesis, we treated primary macrophages from both control and LMP7−/−/MECL-1−/− double knockout mice with or without LPS for 0, 1, 4 and 8 h and monitored levels of gene expression using real-time RT-PCR (Figure 4). Our results demonstrate that transcription of IL-1, IL-6, and iNOS, in response to LPS are dependent on LMP7 and MECL-1 subunits. In contrast, levels of transcription of TNF-α in response to LPS are independent of these subunits. An IFN-inducible IP-10 gene was also partially affected in LPS-treated macrophages from knockouts, as its kinetics changed and this gene was expressed later as compared to control macrophages. Overall, the results demonstrate that the subunits X, Y, Z in part, may be responsible for the transcription of TNF-α as shown in the preceding manuscript, while the LMP7 and MECL-1 subunits are likely responsible for the transcription of iNOS, IL-1β, and IL-6.
Figure 4. Gene-expression of cytokine genes in LPS-treated thioglycollate-elicited macrophages derived from C57BL/6 WT and LMP7−/−/MECL-1−/− double knockouts.
Thioglycollate-elicited peritoneal macrophages from wild type (WT) and LMP7/MECL-1−/− (DKO) mice were cultured in vitro and treated with medium or LPS at 10 ng/ml concentration for 0, 1, 4 and 8 hours. Total RNA was extracted and real time RT-PCR was carried out as described in materials and methods for the following genes; A. IL-1β, B. TNF-α, C. IL-6, D. iNOS and E. IP-10.
DISCUSSION
In this study, we have used LMP knockout mice to confirm and extend our previous studies in RAW 264.7 cells and in primary murine macrophages. Collectively, the results of this investigation provide strong evidence, for the first time, IFN-γ, IFN-β and LPS-inducible proteasome protease sites are central to the expression of LPS-induced NO, IL-1β and IL-6. In stark contrast to these results, LPS-induced TNF-α production was not detectably decreased in macrophages derived from LMP2−/−, LMP7−/−, MECL-1−/−, or MECL-1−/−/LMP7−/− knockout mice relative to macrophages from WT mice. Consequently, the results of these studies support and extend our previous observations that TNF-α and NO are induced in macrophages in response to LPS through distinct proteasome-regulated pathways (1-5). The specific protease functionalities defined by specific subunits in the proteasome play a crucial active role in agonist-induced signal transduction.
Our present results indicate that LPS-induced NO is not detectable in culture supernatants of macrophages derived from LMP knockouts; therefore, we hypothesized that the TRIF/TRAM pathway responsible for the activation of IRF-3 and IFN-β production does not occur. We monitored the levels of key LPS-dependent signaling proteins induced in macrophages of controls and LMP7−/−/MECL-1−/− double knockout mice. We found that the LPS-induced P-IRF3 pathway was not operative in macrophages from the double knockout mice while I-κB-α was being degraded, albeit, at a somewhat lower level of IκB at 0 time as compared to control WT macrophages. We found that IRAK1 was degraded normally by the proteasomes of controls and double knockouts. These results suggest that the MyD88 pathway is functionally intact, but the TRIF/TRAM pathway has defects in the phosphorylation of IRF3 and phosphorylation of STAT1. Phosphorylation of STAT1 is known to be required for the LPS-induced iNOS. NO is not induced by LPS in the macrophages of LMP double knockouts and this is likely due to diminished induction of P-IRF3 and P-STAT1, leading to diminished levels of IFN-β, IFN-γ, and iNOS in macrophages from double knockout mice.
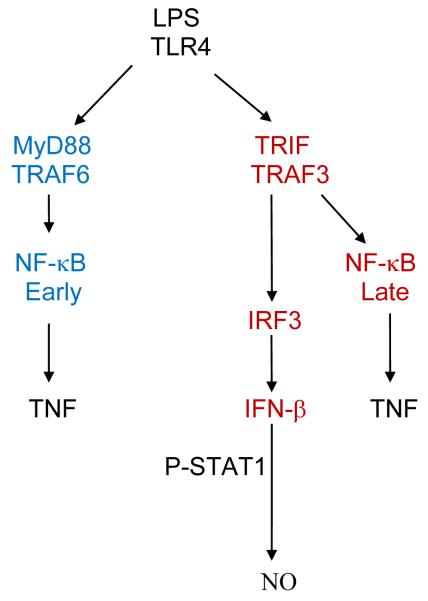
We had previously suggested a pivotal role for the proteasome in LPS-induced signaling and inflammation (1-4, 18). LPS treatment of cells triggers two distinct pathways via TLR4 (19-23), i.e., the MyD88 and the TRIF/TRAM pathways (Figure 5). The latter involves the degradation of two regulatory proteins, TRAF6 and TRAF3, both of which are K63 ubiquitinated (24). Based in part on present results, as well significant contributions from other investigators, we propose that LPS interacts at the cell surface with the TLR4 receptor signaling complex via the MyD88 pathway. After LPS is internalized, shown previously by us to occur within 30 seconds (25), signaling via the TRIF/TRAM pathway is initiated within the endosome (26). This leads to activation of the proteasome in its basal state (i.e., in certain cells like RAW 264.7 cells, there is a lower basal expression of LMP subunits compared to thioglycollate-elicited macrophages), followed by a time-dependent modification of the relative distribution and activation of the 3 LMP proteases that characterize the immunoproteasome. Following activation of the macrophage by LPS, the proteolytic activity of the proteasome changes and results in the activation of selective kinases and transcription factors, leading to induction of cytokines, such as TNF-α. As the inflammatory response continues, TRIF activation leads to P-IRF3 and IFN-β Both IFN-β and IFN-γ activate the STAT1, a key transcription factor for the induction of iNOS and NO (27). Interferons also promote production of proinflammatory cytokines such as IL-1β and IL-6.
Figure 5. Two pathways for the LPS-induced cytokines in macrophages.
LPS-induced cytokines are triggered by TLR4 and are dependent in part on the MyD88 pathway and/or the MyD88-independent TRIF pathway.
The results of the present study provide important confirmation of this hypothetical mechanism. Absence of the LMP subunits in macrophages affected the endosomal TRIF/TRAM signaling normally triggered by LPS, with the net results that IRF-3 was not robustly phosphorylated, IFN-γ was not induced, and P-STAT1 and P-STAT3 were diminished. This may have been due to absence of K63-ubiquitinated TRAF3 in the endosomes. With diminished activation of the TRIF/TRAM pathway, the MyD88/TRAF6 pathway becomes favored (24). The consequences of these events are that NO, IL-1β, and IL-6 are not induced in LPS-treated cells. However, the pathway to TNF-α induction remained intact because IκB or some other signaling proteins may be degraded by the X, Y, and Z subunits of the proteasome, as we have observed with RAW 264.7 cells (9). Further confirmation of the hypothetical mechanism can be found in our previously published results, which established that proteasomal subunits are involved in LPS-induced inflammation (1-4). A low dose of 5μM lactacystin, which binds to subunits X, LMP7 and to a lesser extent Z and MeCL-1, blocks NO, IL-1β, and IL-6 in response to LPS to a much greater extent than TNF-α(3). In summary, proteasomal protease sites present in cells selectively actively regulate signal transduction pathway by degrading the signaling proteins, and alterations in the proteases, as seen in the immunoproteasome, can alter the signaling pathways.
These results help to define an important active role for the proteasome in the development of innate immunity. The profile and magnitude of cytokine (such as TNF-α, IL-1, IL-6), and inflammatory mediator responses to LPS can be significantly modified by altering the protease subunit composition of proteasomes, or by using selective proteasome inhibitors (e.g., lactacystin). As a consequence, the differential profile of cytokine responses to inflammatory stimuli, as regulated by the proteasome, is likely to be important to host outcome in response to Gram negative infections, and can range from beneficial (e.g., eradication of infectious agents and repair of damaged tissue), to highly detrimental (e.g., shock, multiple organ failure, and death). Collectively, these findings support the proteasomal protease-sites as potentially valuable therapeutic targets in treatment of septic shock and other diseases in which inflammation may play a key role, such as cancer, asthma, and autoimmune diseases. With regard to cancer, IL-6 and STAT3 are critical tumor promoters during colitis-associated cancer and our results demonstrate that macrophages from the double knockout LMP7−/−/MeCL-1−/− mice induce diminished levels of IL-6 and no P-STAT3, in response to LPS (28), thus suggesting that composition of proteasomal subunits are likely to be critically important in defining the agonist-induced response.
Acknowledgments
These studies were supported in part by NIH grants GM-50870 (NQ), and AI-18797 (SNV). We thank Drs. Alfred L. Goldberg and Alexei Kisselev for helpful suggestions. Jing Shen is now with the WA Dental group, Bellevue, Washington.
Footnotes
Disclosures. The authors have no financial conflict of interest.
REFERENCES
- 1.Qureshi N, Perera P-Y, Splitter G, Morrison DC, Vogel SN. The Proteasome as a LPS-binding protein in macrophages. Toxic lipopolysaccharide activates the proteasome complex. J Immunol. 2003;171:1515–1525. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi N, Vogel SN, Van Way C, III, Papasian CJ, Qureshi AA, Morrison DC. The proteasome. A central regulator of Inflammation and macrophage function. Immuno Res. 2000;31:243–260. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 3.Shen J, Reis J, Morrison DC, Papasian C, Sreekumar R, Kolbert C, Qureshi AA, Vogel SN, Qureshi N. Key Inflammatory signaling pathways are regulated by the proteasome. Shock. 2006;25:472–484. doi: 10.1097/01.shk.0000209554.46704.64. [DOI] [PubMed] [Google Scholar]
- 4.Shen J, Gao JJ, Zhang G, Tan X, Morrison DC, Papasian C, Vogel SN, Qureshi N. Proteasome inhibitor, lactacystin blocks CpG DNA- and peptidoglycan induced inflammatory genes, cytokines and mitogen-activated protein kinases in macrophages. Shock. 2006;25:594–599. doi: 10.1097/01.shk.0000209555.46704.2d. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch C, Pleogh HL. Intracellular targeting of the proteasome. Trends Cell Biol. 2000;10:268–272. doi: 10.1016/s0962-8924(00)01768-2. [DOI] [PubMed] [Google Scholar]
- 6.Lorsbach RB, Murphy WJ, Lowenstein CJ, Snyder SH, Russell SW. Expression of the nitric oxide synthase gene in mouse macrophages activated for tumor cell killing: Molecular basis for the synergy between interferon-gamma and lipopolysaccharide. J Biol Chem. 1993;268:1908–1913. [PubMed] [Google Scholar]
- 7.Groettrup M, Khan S, Schwarz K, Schmidtke G. Interferon-γ inducible exchanges of 20S proteasome active subunits: Why? Biochimie. 2001;83:367–372. doi: 10.1016/s0300-9084(01)01251-2. [DOI] [PubMed] [Google Scholar]
- 8.Gaczynska M, Rock KL, Spies T, Goldberg AL. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. J Biol Chem. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis J, Guan X, Silverstein P, Kisselev A, Papasian CJ, Qureshi AA, Morrison DC, Van Way CW, III, Vogel SN, Qureshi N. LPS-induced formation of immunoproteasomes: TNF and NO regulated by altered composition of proteasomes-active sites. Cell Biophys. Biochem. doi: 10.1007/s12013-011-9182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caudill CM, Jayapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyperproliferate in response to polyclonal mitogens. J Immunol. 2006;176:4075–4082. doi: 10.4049/jimmunol.176.7.4075. [DOI] [PubMed] [Google Scholar]
- 11.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem J. 2000;345:1–15. [PMC free article] [PubMed] [Google Scholar]
- 12.Elenich LA, Nandi D, Kent AE, McCuskey TS, Cruz M, Lyer MN, Woodward EC, Conn CW, Ochoa AL, Ginsburg DB, Monaco JJ. The complete primary structure of mouse 20S proteasomes. Immunogenetics. 1999;49:835–842. doi: 10.1007/s002510050562. [DOI] [PubMed] [Google Scholar]
- 13.Pang KC, Sanders MT, Monaco JJ, Doherty PC, Turner SJ, Chen W. Immunoproteasome Subunit Deficiencies Impact Differentially on Two Immunodominant Influenza Virus-Specific CD8+ T Cell Responses. J Immunol. 2006;177:7680–7688. doi: 10.4049/jimmunol.177.11.7680. [DOI] [PubMed] [Google Scholar]
- 14.Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, Skon CN, Grebe KM, Griffin TA, Chen W, Bennink JR, Yewdell JW. J. Immunol. 2010;184:4115–4122. doi: 10.4049/jimmunol.0903003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voight A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel P-M, Kruger E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi N, Takayama K, Mascagni P, Honovich J, Wong R, Cotter RJ. Complete structural determination of lipopolysaccharides obtained from deep rough mutant of Escherichia coli: purification by high performance liquid chromatography and direct analysis by plasma desorption mass spectrometry. J Biol Chem. 1988;263:11971–11976. [PubMed] [Google Scholar]
- 17.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 18.Reis J, Xiaoyu T, Yang R, Rockwell CE, Papasian CJ, Vogel SN, Morrison DC, Qureshi AA, Qureshi N. A combination of proteasome inhibitors and antibiotics prevents lethality in a septic shock model. Innate Immun. 2008;14:319–329. doi: 10.1177/1753425908096855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill LA, Bryant JCE, Doyle SI. Therapeutic targeting of Toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacological reviews. 2009;61:177–197. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating SE, Bowie AG. Role of Non-degradative ubiquitination in interleukin and toll-like receptor signaling. J Biol Chem. 2009;284:8217–8221. doi: 10.1074/jbc.R800038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wertz IE, Dixit VM. Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perpect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poltorak AX, Smirnova HI, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freundenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 23.Vogel SN, Fitzgerald KA, Fenton MJ. TLRs: Differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Molecular Interventions. 2003;3:466–477. doi: 10.1124/mi.3.8.466. [DOI] [PubMed] [Google Scholar]
- 24.Tseng P-H, Matsuzawa A, Zhang W, Mino T, Vignali DAA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutuzova G, Albrecht R, Erickson C, Qureshi N. Diphosphoryl lipid A from Rhodobacter sphaeroides blocks the binding and internalization of toxic lipopolysaccharide in RAW 264.7 cells. J Immunol. 2001;167:482–489. doi: 10.4049/jimmunol.167.1.482. [DOI] [PubMed] [Google Scholar]
- 26.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. Tram couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao JJ, Filla MB, Fultz MJ, Vogel SN, Russell SW, Murphy WJ. Autocrine/Paracrine IFN-αβ mediates the lipopolysaccharide-induced activation of transcription factor Stat1α in mouse macrophages: Pivotal role of Stat1α in induction of the inducible nitric oxide synthase gene. J Immunol. 1998;161:4803–4810. [PubMed] [Google Scholar]
- 28.Grivennikov S, Karin E, Terzic J, Mucida D, Yu G-Y, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]