A modification of previously published protocols for the crystallization of homotetrameric R67 DHFR was used to crystallize a mixed wild-type and variant tandem dimer of R67 DHFR. Surprisingly, a fully wild-type crystal structure was obtained, apparently as a consequence of selective proteolytic degradation of the variant.
Keywords: dihydrofolate reductase, antibiotic resistance, in situ proteolysis, protein degradation, type II DHFR
Abstract
Trimethoprim is an antibiotic that targets bacterial dihydrofolate reductase (DHFR). A plasmid-encoded DHFR known as R67 DHFR provides resistance to trimethoprim in bacteria. To better understand the mechanism of this homotetrameric enzyme, a tandem dimer construct was created that linked two monomeric R67 DHFR subunits together and mutated the sequence of residues 66–69 of the first subunit from VQIY to INSF. Using a modified crystallization protocol for this enzyme that included in situ proteolysis using chymotrypsin, the tandem dimer was crystallized and the structure was solved at 1.4 Å resolution. Surprisingly, only wild-type protomers were incorporated into the crystal. Further experiments demonstrated that the variant protomer was selectively degraded by chymotrypsin, although no canonical chymotrypsin cleavage site had been introduced by these mutations.
1. Introduction
Dihydrofolate reductase (DHFR) is a highly conserved enzyme that catalyses the reduction of dihydrofolate (DHF) to tetrahydrofolate (THF) using NADPH as the hydride-donating cofactor. This process is critical in the synthesis of key metabolites, including purines and thymidylate. The central importance of DHFR in these biosynthetic processes has led to its use as a drug target for the treatment of a variety of conditions, including cancer and bacterial and protozoal infections (Volpato & Pelletier, 2009 ▶). Trimethoprim (TMP) has been shown to be an effective competitive inhibitor of the chromosomally encoded bacterial DHFRs. Despite the extensive similarity between bacterial and mammalian DHFRs (Margosiak et al., 1993 ▶), TMP displays a much lower affinity for mammalian DHFRs, making it an effective antibiotic. In response to the broad use of this compound as an antibiotic in humans and livestock, some bacteria have acquired a plasmid-encoded DHFR known as R67 DHFR (dfrB gene family), which is structurally unrelated to the chromosomally encoded DHFRs (dfrA gene family) (Pattishall et al., 1977 ▶). This lack of similarity, combined with the loose structural resemblance between TMP and the substrate DHF, allows R67 DHFR to evade inhibition by TMP, thus providing resistance to this antibiotic.
1.1. Structural features of R67 DHFR
The structure of wild-type R67 DHFR has been solved numerous times (Divya et al., 2007 ▶; Krahn et al., 2007 ▶; Matthews et al., 1986 ▶; Narayana, 2006 ▶; Narayana et al., 1995 ▶). The enzyme is a homodimer of homodimers, with four identical protomers forming a toroidal structure possessing a central pore. The fold of each protomer resembles an SH3-like domain, the first 16 N-terminal residues of which are presumably unstructured (Narayana et al., 1995 ▶). For structural studies, these residues are removed by limited proteolysis to enable crystallization (Divya et al., 2007 ▶; Krahn et al., 2007 ▶; Narayana, 2006 ▶; Narayana et al., 1995 ▶). Importantly, the removal of these residues does not alter the activity or stability of the purified enzyme (Reece et al., 1991 ▶). With respect to the quaternary structure, the first dimerization interface is mediated by residues 26 and 45–47 and effectively expands a β-sheet to span the two protomers. The ‘dimer-of-dimers’ interface is mediated by loop regions that link the β-strands, primarily through residues 60–65 (Narayana et al., 1995 ▶). It should also be noted that only minimal contact exists between protomers that are ‘diagonal’ to each other in the complete tetramer, which includes van der Waals interactions between residues 36 and 50.
The central pore has been identified as the active site, with the four protomers together being able to bind two ligands: two DHFs, two NADPHs or a combination of one DHF and one NADPH, with this last configuration representing the productive ternary complex (Bradrick et al., 1996 ▶). The active-site residues are for the most part distinct from those residues that are important for stabilization of the tetramer. The residues involved in ligand binding and catalysis include residues 32, 35–36, 50, 64–70 and 72–73. Of these, only residues 64–65 and 67 play any role in stabilizing the quaternary structure (Narayana et al., 1995 ▶).
1.2. Mutational analysis of key active-site residues
The key ligand-binding portion of the pore is defined by residues 66–69 (sequence VQIY), which are responsible for binding both the pteroyl moiety of DHF and the nicotinamide ribose of NADPH (Schmitzer et al., 2004 ▶; Strader et al., 2001 ▶; Krahn et al., 2007 ▶). As the binding of a single molecule of the substrate and the cofactor breaks the symmetry of the pore, this implies that each of these four residues can in fact play up to four different roles, depending on where they sit relative to the substrate and cofactor at any given time (Schmitzer et al., 2004 ▶). This also implies that the mutation of any of these residues would cause more convoluted perturbations in the binding and catalytic properties of the enzyme compared with typical structure–function studies.
In order to better examine the roles of individual active-site residues, we prepared a construct of R67 DHFR that reduced the symmetry in the active site. This variant of the enzyme consisted of two protomers connected by a short linker and allowed mutational manipulation of only two, instead of all four, of the protomers within the homotetramer. It is conceptually similar to the tandem dimer of Zhuang et al. (1993 ▶), but carries one mutated monomer. Specifically, for this study residues 66–69 in the N-terminal protomer were mutated from VQIY to INSF (V66I/Q67N/I68S/Y69F). This quadruply mutated construct is of interest since the two first substitutions (V66I/Q67N) are included in a previously characterized monomeric triple variant V66I/Q67N/I68R, which had a catalytic efficiency (k cat/K m DHF) threefold greater than the wild type (Schmitzer et al., 2004 ▶), while the point substituent Y69F had a catalytic efficiency fivefold lower than the wild type (Stinnett et al., 2004 ▶). In the following, we will refer to this construct as the INSF tandem variant; as it resulted from a functional selection, it is necessarily catalytically active (Schmitzer et al., 2004 ▶). We therefore characterized its kinetic properties and, to complement enzymatic studies, we pursued structure determination of this tandem variant. For this, we modified the most common crystallization method for R67 DHFR (Divya et al., 2007 ▶; Narayana, 2006 ▶; Narayana et al., 1995 ▶; Krahn et al., 2007 ▶) by incorporating in situ proteolysis to simplify the crystallization process. Surprisingly, the resulting crystallized tetramer does not include the mutated protomers and instead appears to have reassembled in an all-wild-type form in the crystal structure.
2. Materials and methods
2.1. Creation of the R67 DHFR INSF tandem variant construct and protein purification
The INSF tandem dimer was functionally selected from a library of tandem R67 DHFR variants. The detailed procedure for library creation will be described elsewhere. Briefly, the DNA sequence encoding the first protomer was combinatorially mutated at active-site residues 66–69 (Schmitzer et al., 2004 ▶) while keeping the second protomer native. The two protomers were linked by a Glu-Leu dipeptide, similar to a previous report (Zhuang et al., 1993 ▶). The tandem dimer was preceded by an N-terminal hexahistidine tag in a pQE-32-derived vector (Qiagen). DNA sequencing was used to confirm the successful generation of the INSF tandem dimer construct (DNA-sequencing platform at IRIC, Montreal). Clones that were active and TMP-resistant were selected from the resulting library, thus identifying the INSF tandem variant. The plasmid containing the INSF tandem variant (Fig. 1 ▶) was transformed into Escherichia coli BL21 (pREP4) and subsequently used for protein expression using standard procedures. Purification of the protein employed a nickel-affinity column followed by size-exclusion chromatography in 100 mM Tris buffer pH 8.0. SDS–PAGE was used to assess the purity of the protein samples. The protein was concentrated to 20 mg ml−1 for crystallization. The molecular weight of the purified protein was confirmed to be 18 639 Da by mass spectroscopy, which agreed well with the predicted molecular weight of 18 638.6 Da.
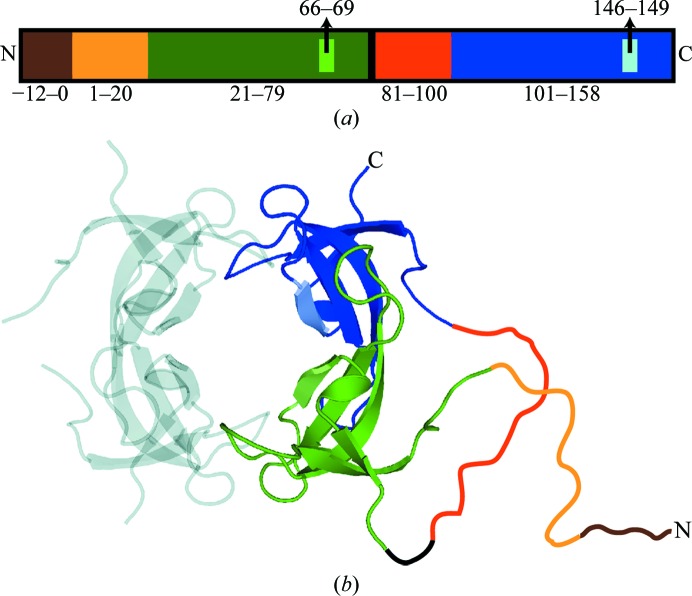
Figure 1.
Organization of the INSF tandem dimer gene. (a) The different segments of the primary structure of the INSF tandem dimer are shown to scale. The colours represent the various segments of the gene. Brown (residues −12 to 0) represents the His tag, light orange (residues 1–20) and orange (residues 81–100) represent the N-terminal unstructured regions in the variant and wild-type protomers, respectively, green (residues 21–78) and blue (residues 101–158) represent the structured regions in the variant and wild-type protomers, respectively, and black (residues 79–80) represents the interprotomer linker. Lighter green (residues 66–69) and blue (residues 146–149) represent the mutation sites in the variant (INSF) and wild-type (VQIY) protomers, respectively. (b) The same colour scheme is used to show the proposed structure of the tandem dimer in the context of the assembled tetramer. The positions of the unstructured regions and linker (drawn in) are speculated.
2.2. Crystallization and data collection
Immediately prior to setting up crystal trays, chymotrypsin was added to the protein sample to a final ratio of 1:100 chymotrypsin:R67 DHFR by mass. In addition, the protein sample was diluted to 3/4 using MPD, thus reducing the protein concentration to 15 mg ml−1 and resulting in a final MPD concentration of 25%. Crystals were obtained using the hanging-drop vapour-diffusion method in a Greiner 24-well hanging-drop crystallization plate. Reservoirs were prepared using 1 ml 100 mM sodium phosphate pH 7.2 and 55% MPD. On a siliconized glass cover slip (Hampton Research), 2.5 µl protein sample was mixed with 1.5 µl reservoir solution. The plate was incubated at 277 K and crystals were obtained within a week (Table 1 ▶). Data were collected (Table 2 ▶) under standard cryogenic conditions using a Rigaku RU-H3R generator equipped with an R-AXIS IV++ detector and Confocal Blue optics. The data were processed using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶).
Table 1. Sample information.
| Macromolecule details | |
| Database code(s) | PDB code 3sfm; UNP code dyr21 ecolx |
| Component molecules | Dihydrofolate reductase type 2 (modification; EC 1.5.1.3) |
| Macromolecular assembly | R67 DHFR is a homotetramer. The other three parts of the biological assembly are generated by the following symmetry operations: (y, x, z), (x, y, z) and (y, x, z). |
| Mass of macromolecular assembly (Da) | 26912 (method: mass spectrometry) |
| Source organism | E. coli |
| Crystallization and crystal data | |
| Crystallization method | Vapor diffusion, hanging drop |
| Temperature (K) | 277 |
| Crystallization solutions | |
| Macromolecule | 2.5l 15mgml1 R67 DHFR tandem dimer, 75mM Tris pH 8, 25%(v/v) 2-methyl-2,4-pentanediol, 0.15mgml1 chymotrypsin |
| Precipitant | 1.5l 100mM sodium phosphate pH 7.2, 55%(v/v) 2-methyl-2,4-pentanediol |
| Reservoir | 1 ml 100mM sodium phosphate pH 7.2, 55%(v/v) 2-methyl-2,4-pentanediol |
| Crystal data | |
| Matthews coefficient V M (3Da1) | 2.35 |
| Solvent content (%) | 47.70 |
| Unit-cell data | |
| Crystal system, space group | Tetragonal, I4122 |
| Unit-cell parameters (, ) | a = b = 67.69, c = 51.76, = = = 90 |
| No. of molecules in unit cell Z | 16 |
Table 2. Data-collection and structure-solution statistics.
Values in parentheses are for the outer shell.
| Diffraction source | Rigaku RU-H3R rotating anode |
| Diffraction protocol | Single wavelength |
| Monochromator | Rotating copper anode |
| Wavelength () | 1.5418 |
| Detector | Rigaku R-AXIS IV++ image plate |
| Temperature (K) | 93 |
| Resolution range () | 501.40 (1.431.40) |
| No. of unique reflections | 12169 (802) |
| No. of observed reflections | 12169 |
| Completeness (%) | 100 (100) |
| Multiplicity | 23.8 (13.9) |
| I/(I) | 18.5 |
| R merge | 0.061 (0.371) |
| Data-processing software | HKL-2000 |
| Phasing method | Molecular replacement |
| Starting model data set | PDB entry 2rh2 |
| Solution software | REFMAC |
2.3. Structure solution and refinement
The integrated and scaled data were imported into the CCP4 suite of programs (Winn et al., 2011 ▶). The isomorphic crystal structure of wild-type R67 DHFR (PDB entry 2rh2; Krahn et al., 2007 ▶), which has one protomer in the asymmetric unit, was used to obtain phases for the new crystal structure. As the INSF tandem variant R67 DHFR crystallized in the same space group and with the same unit-cell parameters as the wild-type model, care was taken to ensure that the same R free test set was selected as was used in the 2rh2 structure. Refinement of the structure was performed using the program REFMAC (Murshudov et al., 2011 ▶; Table 3 ▶). TLS refinement was introduced part way through the refinement and was ultimately switched to anisotropic B-factor refinement in the late stages of refinement. Manual model building was employed periodically using Coot (Emsley & Cowtan, 2004 ▶). Residues and solvent molecules that were revealed to have multiple positions in the electron-density maps were modelled accordingly. The finalized coordinates and structure factors have been deposited in the PDB (PDB entry 3sfm). All figures of protein structures were prepared using PyMOL (DeLano, 2002 ▶).
Table 3. Structure refinement and model validation.
Values in parentheses are for the outer shell.
| Refinement software | REFMAC v.5.5.0109 |
| Refinement on | F |
| Resolution range () | 23.971.40 (1.4351.40) |
| No. of reflections used in refinement | 11533 (826) |
| No. of reflections above cutoff in final cycle | 11533 (826) |
| Final overall R factor | 0.146 |
| Atomic displacement model | Anisotropic |
| Overall average B factor (2) | 17.8 |
| No. of protein atoms | 443 |
| No. of ligand atoms | 24 |
| No. of solvent atoms | 59 |
| Total No. of atoms | 526 |
| Bulk-solvent model | Mask |
| Final R work | 0.144 (0.525) |
| No. of reflections for R free | 626 (42) |
| Final R free | 0.164 (0.565) |
| Ramachandran plot analysis | |
| Most favoured regions (%) | 100 |
| Additionally allowed regions (%) | 0 |
| Generously allowed regions (%) | 0 |
| Disallowed regions (%) | 0 |
2.4. Analysis of the mutation sites
A priori, given the INSF tandem variant construct and the presence of a single protomer in the asymmetric unit, the density for residues 66–69 should reflect 50% occupancy for the wild-type sequence VQIY and 50% occupancy for the INSF sequence. In order to assess whether this was the case, a series of 11 ‘hybrid’ models were generated. These models included both the wild-type (VQIY) and variant (INSF) residues modelled as alternate conformers for residues 66–69. In the first model, the wild-type occupancies were set to 0 and those of the variant residues were set to 1. In each subsequent model the wild-type occupancies were increased by 0.1, while the variant occupancies were decreased by the same amount. For the variant residue Ser68, two alternate conformations for serine at half the occupancy of the other variant residues were used. An additional ten cycles of refinement in REFMAC were performed and the R factors and the sizes of key peaks in F o − F c maps were noted.
2.5. Analysis of protein samples before and after digestion
In order to better understand the results from the crystal structure, three analyses were performed: SDS–PAGE, an activity assay and mass-spectral analysis. A protein sample was prepared in an identical manner as for crystallization and the above three analyses were performed prior to the addition of chymotrypsin and then repeated on the same sample the day after addition of chymotrypsin. One week later, the SDS–PAGE and activity-assay analyses were repeated.
Enzyme activity was monitored in 50 mM phosphate buffer pH 7 in the presence of 100 µM each of NADPH and DHF. Activity was measured with a Cary100 Bio UV–Vis spectrophotometer (Varian Canada, Montréal, Quebec, Canada) by monitoring NADPH and DHF depletion (Δ∊340 = 12 800 M −1 cm−1). In those cases where the protein had been digested with chymotrypsin prior to the activity assay, the protein was diluted in buffer and then reconcentrated to remove small peptide fragments.
Mass-spectral analysis was performed using an HPLC-MS system composed of an Agilent 1100 HPLC coupled with a TOF instrument equipped with an electrospray source in positive mode. The chromatographic column was a Poroshell 300SB-C8 (2.1 × 75 mm, 5 µm particle size; Agilent Technologies). The eluents used were 0.1% formic acid in H2O and 0.1% formic acid in acetonitrile.
3. Results and discussion
3.1. Protein purification
The INSF tandem dimer was purified as described and determined to be pure by SDS–PAGE. The oligomerization state of the protein under native conditions was determined to be the expected dimer of tandem dimers (i.e. a tetramer of the 8.4 kDa natural protomer) by gel filtration. The activity of the tandem dimer was determined to be 20% of the wild-type activity under conditions that are saturating for the wild type (0.078 ± 0.008 U mg−1 for mutant INSF versus 0.45 ± 0.05 U mg−1 for the wild type). This strongly suggests that the INSF tandem dimer is correctly folded and assembles as a tetramer. We cannot preclude, however, that the quadruple mutation introduces some minor perturbations to the native fold of the protein.
3.2. Crystallization and structure determination
The conditions used to crystallize R67 DHFR represent a modification (and simplification) of the crystallization conditions previously used to crystallize R67 DHFR (Divya et al., 2007 ▶; Krahn et al., 2007 ▶; Narayana, 2006 ▶; Narayana et al., 1995 ▶). Previously, purification of this enzyme for use in crystallization had incorporated a limited proteolysis step using chymotrypsin-linked beads. After removal of the beads, the enzyme was further purified to remove the proteolysed fragments. In this case, chymotrypsin was simply added to the crystallization experiment and allowed to function in situ, a technique that is increasingly being adopted by structural genomics groups (Dong et al., 2007 ▶; Wernimont & Edwards, 2009 ▶). Crystals were produced easily using this simplified technique and the crystal quality did not appear to suffer, as demonstrated by the high resolution of the crystals obtained by this method. It should be noted that the composition of both the protein crystallization buffer and the reservoir conditions were modified to make the protocol more amenable to commonly used automated crystallization protocols.
The crystals obtained diffracted to a resolution of 1.4 Å in space group I4122 with one protomer per asymmetric unit (crystallization, data-collection and refinement statistics are reported in Tables 1 ▶, 2 ▶ and 3 ▶). Upon examination of symmetry-related molecules, the characteristic toroidal structure of R67 DHFR could be observed. Residues 21–78 (numbering using equivalent wild-type residue numbers) could be modelled into the density, while residues 1–20 were not visible. Based on MS results, a portion of these residues were removed by proteolysis by chymotrypsin, while the rest are likely to be disordered in the crystal structure. The N-terminal His tag and linker which connected the two protomers in the modified gene were also not visible. As would be expected in a crystal structure at this resolution, six residues were observed in multiple conformations, as were some water molecules. Several larger peaks in the solvent were modelled as MPD.
As the crystals that were obtained were isomorphous to those used to obtain previously published structures (Divya et al., 2007 ▶; Krahn et al., 2007 ▶; Matthews et al., 1986 ▶; Narayana, 2006 ▶; Narayana et al., 1995 ▶), it is expected that the structure reported here would be very similar to these. The highest resolution structure of R67 DHFR that has been solved to date is an apo structure with a resolution of 0.96 Å (PDB entry 2rh2; Krahn et al., 2007 ▶). As such, this structure was used as the basis for all further comparisons. Of the six residues that are present in dual conformations (Arg31, Trp45, Cys47, Asn49, Leu50 and Ser65), all six were also present in multiple conformations in 2rh2. However, there were seven residues (Lys32, Trp38, Gln41, Pro52, Pro63, Gln67 and Glu75) that were only modelled as having a single conformation even though two conformations were modelled in 2rh2. This variation is likely to be a consequence of the higher resolution of 2rh2 and does not represent a significant difference. A comparison of both proteins in their entirety revealed that the main chain essentially did not vary between the two structures and that most side-chain atoms occupied effectively identical positions in both structures. The only residues to vary significantly from one structure to the other were the first and last residues in the chain, as well as some of the residues that were modelled in dual conformations. The r.m.s.d. for all main-chain atoms, excluding the first and last residues, is 0.08 Å and that for all atoms is 0.38 Å; however, if multiple conformations are omitted from the comparison the all-atom value drops to 0.08 Å. From this analysis, it is clear that the two structures are essentially identical to each other, with the side chains of those residues known to be somewhat flexible being the only sites to show some differences between the two structures.
3.3. Examination of the mutation sites in the crystal structure
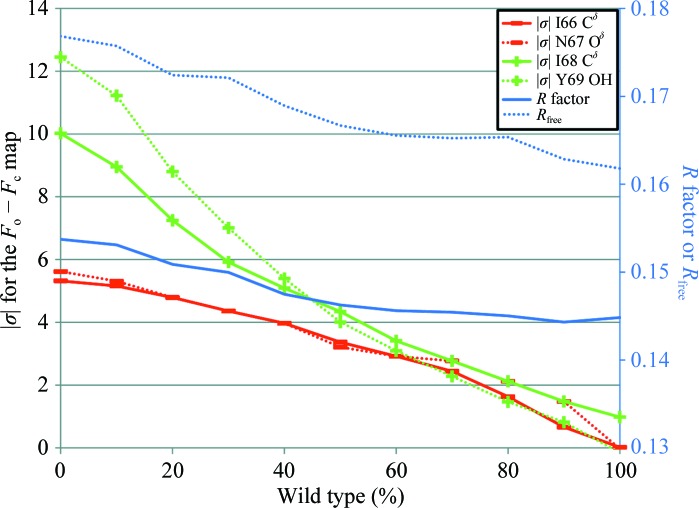
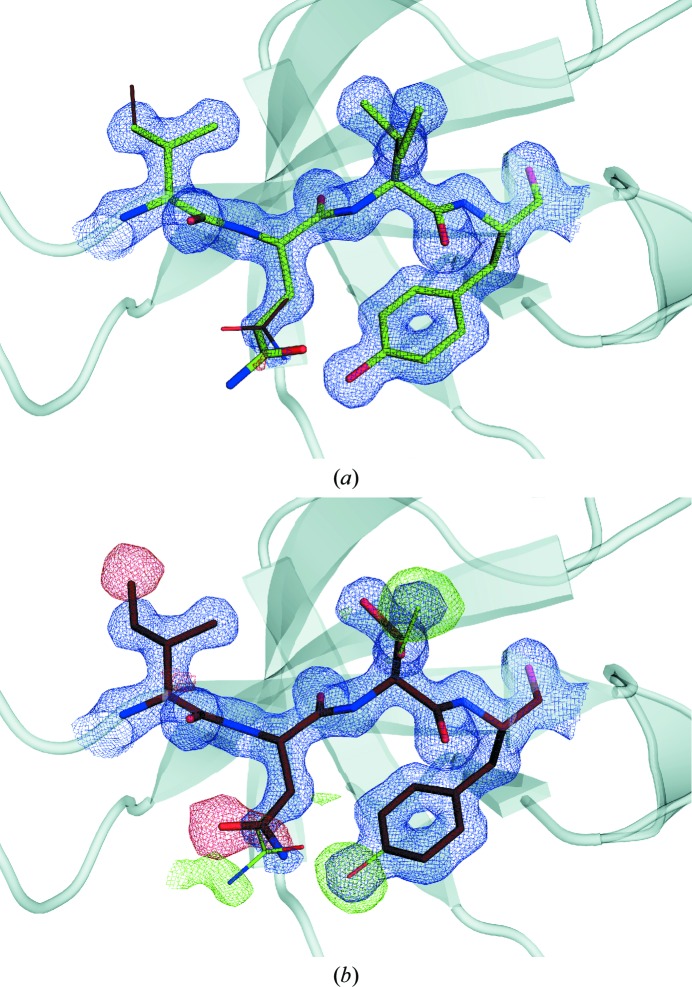
The mutated residues are located in a well ordered part of the structure that is critical for binding both of the ligands. The crystallization of the protein in the presence of chymotrypsin led us to expect that the linked dimers would be cleaved apart. This would be likely to abolish the ordered architecture of the homotetramer over the timescale of crystallization. Irrespective of whether the architecture is affected, since the crystallization condition contained an exact 1:1 stoichiometric ratio of wild-type to variant protomers, there should be an equal probability of each type being incorporated into the growing crystal. The two sequences are sufficiently different to be easily distinguishable in the crystal structure at this resolution. It was therefore quite surprising that the series of models with varying occupancies of the wild-type and variant forms revealed that the best agreement with the diffraction data was achieved when the wild-type sequence was assigned an occupancy of 0.9–1.0 (Fig. 2 ▶). This was confirmed both in reciprocal space using R factors and in real space by assessing the σ level in F o − F c maps for distinguishing atoms in each of the four mutated residues (Fig. 3 ▶). This implies that the variant protomer is selectively excluded from the crystal and that it is likely that not even one variant protomer is incorporated into the majority of functional R67 DHFR tetramers in the crystal structure.
Figure 2.
Change in magnitude of map σ and R factor with increasing wild-type character. The map σ value is evaluated at the position of one atom per residue that is unique to either the wild-type or the variant protomers. The map σs (left axis) that are positive are shown in green and denoted with a + sign, while the map σs that are negative are shown in red with a − sign. R factors (right axis) are shown in blue.
Figure 3.
Electron-density map of the mutation sites. 2F o − F c (blue) and F o − F c (green and red) maps contoured at a map σ of 1.5 and 3.0, respectively, calculated using (a) the wild-type and (b) the variant models are shown. The wild-type model is shown in green and the variant model is shown in brown. The model used to calculate the map in each panel is shown using thicker lines.
3.4. Rationale for the variant protomers being excluded from the crystal
The observation of the variant protomers being excluded from the growing crystal may be the result of preferential degradation of the variant protomer and/or destabilization of the inter-protomer interactions in the variant relative to the wild-type protomer. We have demonstrated that the variant protomer is degraded by chymotrypsin prior to its possible incorporation into the crystal (see below), despite the fact that no canonical chymotrypsin cleavage site is introduced by mutating VQIY to INSF. We carefully considered the possibility that destabilization of the inter-protomer interactions is what caused the variant protomers to be excluded. This would most likely have to be mediated by loss of the interaction of Asn67 with Asn67 in the neighbouring protomer, as this is the only one of the four mutated residues that forms an inter-protomer interaction. However, the results shown below clearly indicate that a single species the size of the monomer is present after chymotrypsin digestion, and molecular-dynamics simulations failed to demonstrate any significant difference in interaction energy between the wild-type and variant forms.
When the INSF tandem dimer is exposed to chymotrypsin, as is required to remove the unstructured N-terminus for crystallization, the interprotomer linker is efficiently cleaved under crystallization conditions. This is clearly demonstrated by the elimination of at least 90% of the variant protomers from the crystal. In order to further investigate this, the protein sample was analysed using SDS–PAGE, activity assays and mass spectrometry before and after chymotrypsin digestion. Following 24 h of exposure to chymotrypsin at 277 K, resolution on tricine–SDS–PAGE showed no trace of the dimer (18.6 kDa) and the appearance of a 6 kDa band (Fig. 4 ▶). This corresponds roughly to the molecular weight expected for a single protomer having no additional residues (His tag, linker or termini). The 6 kDa band was estimated to contain approximately 25% of the protein initially digested, with some faint bands of lower molecular weight. This indicates full degradation of a fraction of the initial tandem dimer; a further fraction of at least one of the protomers remained intact, suggesting a fold that is sufficiently stable to prevent proteolysis. When these same samples were assayed for enzyme activity at 24 h, the chymotrypsin-digested tandem dimer revealed an unexpected increase in specific activity (in units per milligram of protein) from 20% of the wild-type specific activity for the undigested INSF tandem dimer to wild-type-like levels following chymotrypsin digestion. These data are consistent with the variant protomer being degraded more quickly than the wild-type protomer: as the variant protomers are degraded, the wild-type protomers are allowed to reassociate into a stable and fully wild-type tetramer. These analyses were repeated after one week and identical results were obtained, indicating that the sample was stable after 24 h.
Figure 4.

Comparison of the untreated and chymotrypsin-treated INSF tandem dimers on SDS–PAGE. The positions of the uncleaved (tandem dimer) and digested monomeric proteins are shown and the expected molecular weight as determined by mass spectroscopy is given.
Mass-spectral analysis of the same sample was performed 24 h following chymotrypsin treatment. For the digested INSF tandem dimer one predominant species at 6728 Da was observed (relative to 18 639 Da for the starting material), suggesting that one of the two protomers is the dominant species. Furthermore, this corresponds well to the theoretical molecular weight of 6727.5 Da for residues 17–78 of the wild-type protomer. As residue 16 is a phenylalanine, this would indicate cleavage C-terminal to a phenylalanine, which is typical of chymotrypsin activity. This is consistent with the digestion of almost the entire N-terminal region, as only the last four residues (17–20) of this unstructured region remain. In contrast, the same span of residues for the mutant protomer would have a theoretical molecular weight of 6685.4 Da. However, it is interesting to note that neither the wild-type VQIY sequence nor the variant INSF sequence contain a ‘canonical’ chymotrypsin cleavage site. Chymotrypsin typically cleaves C-terminal to Trp, Tyr or Phe, but not if the residue C-terminal to the aromatic residue is a Pro. The first residue after the Tyr69 or Phe69 in these sequences is Pro70, eliminating this site as a good chymotrypsin cleavage site. We are conducting more detailed analysis of the degradation in the context of this and other tandem dimers of R67 DHFR by modifying the cleavage conditions (time and the ratio of target to chymotrypsin) and analyzing the resulting fragments.
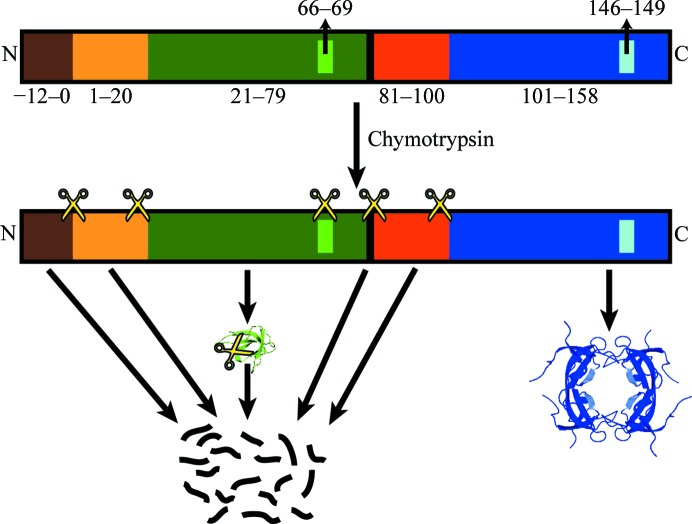
In addition to the apparent degradation of the variant protomer by chymotrypsin, the mutations may also destabilize the structure of the variant protomers themselves or impair the inter-protomer interactions. The latter could only be caused directly through the loss of the Gln67 interaction, although this seems unlikely given the lack of evidence from previous studies or from molecular-dynamics simulations. It is also possible that the mutations introduce disorder throughout the protein, as has recently been reported for a human DHFR variant (Volpato et al., 2009 ▶). This disordering could be sufficient to allow chymotrypsin to more easily degrade the variant protomers. We can also speculate that if the linker and N-terminus is cleaved prior to the degradation of the structured portion of the protein, then the mixture of wild-type and variant subunits in each complete tetramer would be allowed to reach an equilibrium. If the variant was excluded from the tetramer, either because of decreased stability or the loss of a direct interaction, it would follow that it would be less structured and more prone to proteolytic degradation. This could then lead to the rapid degradation of the monomeric INSF variant protomers prior to formation of the crystal (Fig. 5 ▶). This could explain the degradation of the variant even without a canonical chymotrypsin cleavage site.
Figure 5.
Proposed model for the degradation of the INSF tandem dimer by chymotrypsin. Proposed chymotrypsin cut sites are denoted using the scissors icon. The exact positions are speculative and there may be additional cut sites that are not shown. The colour scheme is the same as is used in Fig. 1 ▶.
3.5. Implications for structural and functional studies of wild-type and heterodimeric R67 DHFR variants
The crystallization protocol presented here provides a simplified route to rapidly and readily obtain high-quality crystals of R67 DHFR which will be useful for further studies of this enzyme aimed at addressing the issue of resistance to trimethoprim. However, the method does highlight the difficulties in obtaining the structure of a tandem variant of R67 DHFR. To begin with, the fact that in situ proteolysis appears to cause the interprotomer linker and the INSF variant protomer to be digested on a comparable timescale will be likely to make it impossible to use in situ proteolysis to crystallize the tandem variant. In addition, the requirement to have the N-terminal unstructured region within R67 DHFR removed in order to obtain crystals jeopardizes the structure elucidation of an ordered tetramer from a tandem variant without the use of proteolysis. It may be necessary to prepare a tandem variant that contains specific protease cleavage sites engineered in locations at the N- and C-termini of the SH3-like domain, eliminating the use of a nonspecific protease such as chymotrypsin. Alternatively, a tandem variant in which the first unstructured N-terminal sequence is made to be cleavable and the second has been eliminated in favour of a linker of appropriate length between the two SH3-like domains might prove effective. This would eliminate the need for nonspecific proteases and maintain both the N-terminal region that allows the stable expression of the protein in E. coli (Reece et al., 1991 ▶) and the linker that enforces the ordered tandem dimer in the crystal structure.
3.6. Implications for the use of in situ proteolysis as a crystallization method
With the rise of structural genomics consortia, in situ proteolysis is gaining popularity as a means to crystallize proteins that initially yield few or no crystals. The crystallization of R67 DHFR can be seen as one of the earliest examples of the use of this technique to obtain high-quality crystals. The present work demonstrates that while powerful, this technique can have many unforeseen consequences. While it was not surprising that the linkers were cleaved, it would have been very difficult to predict the destruction of the variant protomers of a protein that had previously been shown to be resistant to proteolytic degradation. This is especially true given that there is no canonical chymotrypsin cleavage site that is being introduced into the variant protomers, making a simple explanation of the degradation of the variant protomers difficult to make. While it is known that proteases can cleave proteins at noncanonical sequences, it is difficult to predict these additional cut sites prior to performing an in situ proteolysis experiment, especially when the susceptibility of the protein to proteolysis based on structural accessibility is taken into account. This demonstrates that our understanding of the effects of mutations on the global stability of protein structure is still lacking. Overall, we can see here that adapting a previously obtained in situ proteolysis protocol to a slightly different system, be it a mutant, a homologue or a ligand-bound structure, can be more difficult than one might initially expect.
Supplementary Material
PDB reference: R67 DHFR, 3sfm
Structure factors: contains datablock(s) r3sfmsf. DOI: 10.1107/S1744309111030417/gx51883sfmsup1.hkl
Acknowledgments
The authors would like to thank the past and present members of the Berghuis and Pelletier laboratories for their help, advice and useful suggestions. Mass-spectrometric data were collected at the Regional Center for Mass Spectrometry at Université de Montréal. This work was supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada awarded to AMB and JNP. AMB is the recipient of the Canada Research Chair in Structural Biology.
References
- Bradrick, T. D., Beechem, J. M. & Howell, E. E. (1996). Biochemistry, 35, 11414–11424. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Divya, N., Grifith, E. & Narayana, N. (2007). Protein Sci. 16, 1063–1068. [DOI] [PMC free article] [PubMed]
- Dong, A. et al. (2007). Nature Methods, 4, 1019–1021. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Krahn, J. M., Jackson, M. R., DeRose, E. F., Howell, E. E. & London, R. E. (2007). Biochemistry, 46, 14878–14888. [DOI] [PMC free article] [PubMed]
- Margosiak, S. A., Appleman, J. R., Santi, D. V. & Blakley, R. L. (1993). Arch. Biochem. Biophys. 305, 499–508. [DOI] [PubMed]
- Matthews, D. A., Smith, S. L., Baccanari, D. P., Burchall, J. J., Oatley, S. J. & Kraut, J. (1986). Biochemistry, 25, 4194–4204. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Narayana, N. (2006). Acta Cryst. D62, 695–706. [DOI] [PubMed]
- Narayana, N., Matthews, D. A., Howell, E. E. & Nguyen-huu, X. (1995). Nature Struct. Biol. 2, 1018–1025. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pattishall, K. H., Acar, J., Burchall, J. J., Goldstein, F. W. & Harvey, R. J. (1977). J. Biol. Chem. 252, 2319–2323. [PubMed]
- Reece, L. J., Nichols, R., Ogden, R. C. & Howell, E. E. (1991). Biochemistry, 30, 10895–10904. [DOI] [PubMed]
- Schmitzer, A. R., Lépine, F. & Pelletier, J. N. (2004). Protein Eng. Des. Sel. 17, 809–819. [DOI] [PubMed]
- Stinnett, L. G., Smiley, R. D., Hicks, S. N. & Howell, E. E. (2004). J. Biol. Chem. 279, 47003–47009. [DOI] [PubMed]
- Strader, M. B., Smiley, R. D., Stinnett, L. G., VerBerkmoes, N. C. & Howell, E. E. (2001). Biochemistry, 40, 11344–11352. [DOI] [PubMed]
- Volpato, J. P. & Pelletier, J. N. (2009). Drug Resist. Updat. 12, 28–41. [DOI] [PubMed]
- Volpato, J. P., Yachnin, B. J., Blanchet, J., Guerrero, V., Poulin, L., Fossati, E., Berghuis, A. M. & Pelletier, J. N. (2009). J. Biol. Chem. 284, 20079–20089. [DOI] [PMC free article] [PubMed]
- Wernimont, A. & Edwards, A. M. (2009). PLoS One, 4, e5094. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zhuang, P., Yin, M., Holland, J. C., Peterson, C. B. & Howell, E. E. (1993). J. Biol. Chem. 268, 22672–22679. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: R67 DHFR, 3sfm
Structure factors: contains datablock(s) r3sfmsf. DOI: 10.1107/S1744309111030417/gx51883sfmsup1.hkl