Crystal structures of the putative ATP pyrophosphatase PF0828 reveal a new domain (EGT domain), and a large conformational change upon ATP binding.
Keywords: ATP pyrophosphatases, PF0828, ATP binding, Pyrococcus furiosus
Abstract
ATP pyrophosphatases (ATP PPases) are widely distributed in archaea and eukaryotes. They share an HUP domain at the N-terminus with a conserved PP-motif that interacts with the phosphates of ATP. The PF0828 protein from Pyrococcus furiosus is a member of the ATP PPase superfamily and it also has a 100-residue C-terminal extension that contains a strictly conserved EGG(E/D)xE(T/S) motif, which has been named the EGT-motif. Here, crystal structures of PF0828 alone and in complex with ATP or AMP are reported. The HUP domain contains a central five-stranded β-sheet that is surrounded by four helices, as in other related structures. The C-terminal extension forms a separate domain, named the EGT domain, which makes tight interactions with the HUP domain, bringing the EGT-motif near to the PP-motif and defining the putative active site of PF0828. Both motifs interact with the phosphate groups of ATP. A loop in the HUP domain undergoes a large conformational change to recognize the adenine base of ATP. In solution and in the crystal PF0828 is a dimer formed by the side-by-side arrangement of the HUP domains of the two monomers. The putative active site is located far from the dimer interface.
1. Introduction
ATP pyrophosphatases (ATP PPases) belong to the adenine nucleotide α-hydrolase superfamily. These enzymes are widely distributed in nature and include class I aminoacyl-tRNA synthetases (aaRSs), universal stress protein (UspA), GMP synthetases, ATP sulfurylases and others. The nucleotide-binding catalytic domains of these enzymes, with approximately 120 residues, share a conserved PP-motif near the N-terminus which binds the phosphate moieties of ATP (analogous to the P-loop; Bork & Koonin, 1994 ▶). This domain is also known as the HUP domain, after the HIGH signature motif (at the nucleotide-binding site of aaRSs), UspA and ATP PPases (Aravind et al., 2002 ▶).
The structure of the HUP domain is a variant of the Rossmann fold, consisting of a five-stranded parallel β-sheet with 3-2-1-4-5 strand order that is surrounded by two α-helices on each face (Dessailly et al., 2010 ▶). The PP-motif, like the P-loop, is located in a loop between the first strand and the first helix. Insertions in this fold (embellishments) play important roles in defining the exact functions of the individual proteins (Dessailly et al., 2010 ▶).
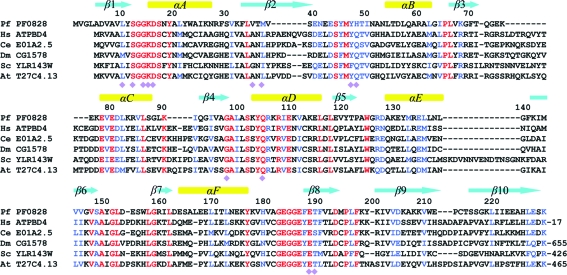
Based on sequence analysis, the Pyrococcus furiosus protein PF0828 is a member of the ATP PPase superfamily. It contains a PP-motif, 12-(S/T)GGKDS-17, that is strictly conserved among its homologs in archaea and eukaryotes (Fig. 1 ▶). However, PF0828 (with 229 residues) is significantly larger than the other members of this family, primarily owing to a 100-residue C-terminal extension to the HUP domain. This extension contains a strictly conserved 183-EGG(E/D)xE(T/S)-189 motif, which we have named the EGT-motif after the conserved residues in it (Fig. 1 ▶). This feature is unique to PF0828 and its homologs, suggesting that they form a distinct subfamily of the ATP PPases. The PF0828 homologs in some eukaryotes (yeast, plants and Drosophila) also contain an additional C-terminal extension (Fig. 1 ▶), two segments of which share weak sequence homology with L-PSP endoribonucleases (Morishita et al., 1999 ▶), which may be homotrimers (Sinha et al., 1999 ▶). The human homolog of PF0828, with 38% identity, is known as ATP-binding domain-containing protein 4 (ATPBD4; Fig. 1 ▶), although its function is unknown. The yeast homolog, YLR143W, is not essential for growth.
Figure 1.
Sequence alignment of PF0828 homologs. Alignment of the sequences of P. furiosus (Pf) PF0828, Homo sapiens (Hs) ATPBD4, Caenorhabditis elegans (Ce) E01A2.5, Drosophila melanogaster (Dm) CG1578, Saccharomyces cerevisiae (Sc) YLR143W and Arabidopsis thaliana (At) T27C4.13. The secondary-structure elements are labeled. Residues involved in ATP binding are indicated by purple diamonds. The number of additional residues at the C-termini of ATPBD4, CG1578, YLR143W and T27C4.13 is indicated. The PP-motif is located in the β1–αA loop and the EGT-motif in the αF–β8 loop.
Here, we present crystal structures of PF0828 alone and in complex with ATP and with AMP at 2.3, 2.3 and 2.4 Å resolution, respectively. The C-terminal extension forms a separate domain, which we have named the EGT domain. It makes extensive interactions with the HUP domain, bringing the EGT-motif near the PP-motif and defining the putative active site of PF0828. Both motifs interact with the phosphate groups of ATP. A large conformational change in the β2–αB loop in the HUP domain is required to recognize the adenine base of ATP and possibly also for catalysis. The movement and stabilization of this loop may also be coupled to a change in the organization of the dimer of PF0828.
2. Materials and methods
The production of the PF0828 protein from P. furiosus was carried out as part of the high-throughput protein-production process at the Northeast Structural Genomics consortium (NESG; Acton et al., 2005 ▶). The PF0828 protein corresponds to NESG target PfR23. The full-length PF0828 gene from P. furiosus (strain DSM 3638) was cloned into a pET21d (Novagen) derivative, generating plasmid pPfR23-21. The recombinant protein contains eight non-native residues (LEHHHHHH) at the C-terminus. The construct was verified by standard DNA-sequence analysis. Escherichia coli BL21 (DE3) pMGK cells, a rare-codon-enhanced strain, were transformed with pPfR23-21. A single isolate was cultured in MJ9 minimal medium (Jansson et al., 1996 ▶) supplemented with selenomethionine, lysine, phenylalanine, threonine, isoleucine, leucine and valine for the production of selenomethionine-labeled PF0828 (Doublié et al., 1996 ▶). Initial growth was carried out at 310 K until the OD600 of the culture reached 0.6–0.8. The incubation temperature was then decreased to 290 K and protein expression was induced by the addition of IPTG (isopropyl β-d-1-thiogalactopyranoside) to a final concentration of 1 mM. Following overnight incubation, the cells were harvested by centrifugation. This expression-vector plasmid for PF0828 (NESG PfR23-21.3) has been deposited in the PSI Materials Repository (http://psimr.asu.edu/).
Selenomethionyl PF0828 was purified by standard methods. Cell pellets were resuspended in lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole and 5 mM β-mercaptoethanol) and disrupted by sonication. The resulting lysate was clarified by centrifugation at 26 000g for 45 min at 277 K. The supernatant was loaded onto an Ni–NTA column (Qiagen) and eluted in lysis buffer containing 250 mM imidazole. Fractions containing partially purified PF0828 were pooled and optimized buffer conditions providing monodisperse samples were determined by analytical gel filtration monitored by static light-scattering experiments, as described elsewhere (Acton et al., 2005 ▶). Preparative gel filtration (Superdex 75, GE Healthcare) was then performed using a buffer consisting of 10 mM Tris pH 7.5, 100 mM NaCl, 5 mM DTT and 0.02% NaN3. The purified PF0828 protein was concentrated to 12 mg ml−1, flash-cooled in aliquots and used for crystallization screening. Sample purity (>95%) and molecular weight were verified by SDS–PAGE and MALDI–TOF mass spectrometry, respectively. The yield of purified PF0828 protein was approximately 30 mg per litre.
PF0828 was first crystallized in apo and ATP-bound forms by the vapor-diffusion method using hanging-drop and sitting-drop trays at 277 and 291 K, respectively. 2 µl protein solution containing PF0828 (12 mg ml−1 in 10 mM Tris pH 7.5, 100 mM NaCl, 5 mM DTT and 0.02% NaN3) was mixed with 2 µl precipitant solution consisting of 380 mM potassium/sodium tartrate for crystallization of the apo crystals and 100 mM Tris pH 8.5, 300 mM (NH4)H2PO4, 1%(w/v) PEG 8000, 5 mM DTT and 5 mM ATP for crystallization of the ATP complex. Crystals were subsequently cryoprotected by supplementing the precipitant solutions with 25%(v/v) glycerol and 25%(v/v) ethylene glycol, respectively, and then flash-frozen in liquid propane for data collection at 100 K. Attempts to grow diffraction-quality crystals of PF0828 in the presence of both ATP and Mg2+ were unsuccessful.
We subsequently screened many ATP analogs and obtained crystals in the presence of dephospho-coenzyme A using the microbatch method. 2 µl protein solution containing 10 mM dephospho-coenzyme A was mixed with 2 µl precipitant solution consisting of 20%(w/v) PEG 3350 and 200 mM magnesium formate. The crystals were cryoprotected by supplementing the precipitant solution with 20% ethylene glycol for data collection at 100 K. Crystallographic analysis revealed the binding of only AMP.
Crystals of PF0828 belonged to space group P41212 for the apo and AMP-bound enzymes and P212121 for the ATP-bound enzyme. There is one molecule of PF0828 in the crystallographic asymmetric unit of the apo and AMP-bound structures and two molecules in the ATP-bound structure. For apo PF0828, a single-wavelength anomalous diffraction (SAD) data set to 2.3 Å resolution was collected at the peak absorption wavelength (0.9793 Å) of selenium on the X4A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the HKL-2000 package (Otwinowski & Minor, 1997 ▶). An anomalous signal of approximately 1% was observed in the diffraction data and the selenium sites were located with the program SnB (Weeks & Miller, 1999 ▶). SOLVE/RESOLVE (Terwilliger, 2003 ▶) was used for phasing the reflections and automated model building, which correctly placed 45% of the residues with side chains. The majority of the model was built manually with the program XtalView (McRee, 1999 ▶). Structure refinement was performed with CNS (Brünger et al., 1998 ▶). Noncrystallographic symmetry restraints were applied in most stages of the refinement of the structure.
The model of the apo structure was used to determine the structure of the ATP and AMP complexes using the molecular-replacement method with the program COMO (Jogl et al., 2001 ▶). The data-processing and refinement statistics are summarized in Table 1 ▶.
Table 1. Summary of crystallographic information.
Values in parentheses are for the highest resolution shell.
| Ligand | None | ATP | AMP |
|---|---|---|---|
| Space group | P41212 | P212121 | P41212 |
| Unit-cell parameters (Å) | a = b = 85.0, c = 74.5 | a = 59.0, b = 64.1, c = 140.1 | a = b = 85.2, c = 74.2 |
| Maximum resolution (Å) | 2.3 | 2.3 | 2.4 |
| No. of observations | 147793 | 165951 | 305025 |
| Rmerge (%) | 8.2 (24.7) | 8.5 (33.1) | 7.0 (36.2) |
| 〈I/σ(I)〉 | 26.9 (6.1) | 15.2 (2.9) | 46.8 (8.2) |
| Resolution range used for refinement (Å) | 19.3–2.3 (2.4–2.3) | 19.9–2.3 (2.4–2.3) | 19.9–2.4 (2.5–2.4) |
| No. of reflections† | 21129 | 37778 | 18724 |
| Completeness (%) | 91.3 (75.8) | 83 (58) | 91.8 (78.9) |
| R factor (%) | 23.9 (22.9) | 20.3 (23.3) | 22.9 (25.7) |
| Free R factor (%) | 27.3 (29.7) | 25.3 (27.0) | 27.8 (34.1) |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.008 | 0.007 |
| Bond angles (°) | 1.1 | 1.3 | 1.0 |
| PDB code | 3rjz | 3rk1 | 3rk0 |
The number for the apo enzyme (selenomethionyl protein) includes both Friedel pairs.
3. Results and discussion
The crystal structure of P. furiosus PF0828 apoenzyme was determined using the selenomethionyl single-wavelength anomalous diffraction (SAD) method (Hendrickson, 1991 ▶) and refined at 2.3 Å resolution (Table 1 ▶). Crystals of the ATP complex were prepared by cocrystallization with ATP in the absence of Mg2+ and the structure of this complex was determined and refined at 2.3 Å resolution. Crystals were also obtained in the presence of 10 mM 3′-dephospho-CoA and 200 mM Mg2+. However, the subsequent crystallographic analysis at 2.4 Å resolution (Table 1 ▶) showed the presence of only AMP, suggesting that PF0828 may have catalyzed the hydrolysis of the α–β phosphodiester bond of dephospho-CoA, consistent with its ATP PPase activity. This activity appears to be dependent on divalent cations, as we observed binding of ATP in the absence of Mg2+.
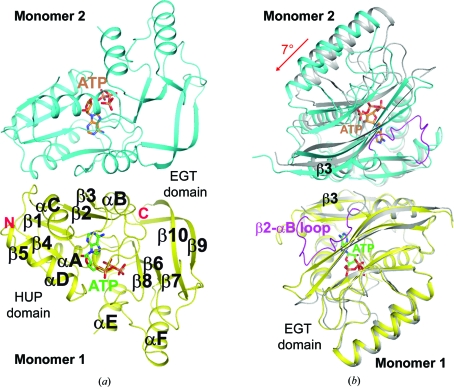
As expected, the HUP domain of PF0828, covering residues 1–123, contains a five-stranded parallel β-sheet (β1–β5) with four helices (αA–αD; Fig. 2 ▶ a). The C-terminal 100-residue extension forms a separate domain, which we have named the EGT domain. It contains five β-strands (β6–β10) and two helices (αE–αF) (Fig. 2 ▶ a) and makes extensive interactions with the HUP domain. In particular, the strictly conserved 183-EGG(E/D)xE(T/S)-189 motif (the EGT-motif) is located near the PP-motif and contributes to the putative active site of PF0828 (see below). The structure of the EGT domain was not similar to any other structures in the Protein Data Bank based on an analysis by DaliLite (Holm et al., 2008 ▶), except for the structure of the PF0828 homolog from P. horikoshii (PH1257; PDB entry 2d13; N. K. Lokanath & N. Kunishima, unpublished work). The two proteins share ∼72% sequence identity and the overall structures of the free enzyme monomer and dimer of PH1257 are generally similar to those of PF0828.
Figure 2.
Crystal structure of the P. furiosus PF0828 homodimer. (a) Schematic drawing of the structure of the PF0828 homodimer in complex with ATP. Monomer 1 is colored yellow and monomer 2 is colored cyan; their bound ATP molecules are shown with green and brown C atoms, respectively. The twofold axis of the dimer is in the horizontal direction. (b) Structural overlay of the PF0828 dimer in complex with ATP (in color) and the free enzyme (gray) viewed down the twofold axis of the dimer. This view is related to that of (a) by a 90° rotation around the vertical axis. Only monomer 1 (in yellow) is included in the overlay and a 7° rotation is observed in the other monomer compared with the apo enzyme. The β2–αB loop is shown in magenta in the ATP complex, while it is mostly disordered in the free enzyme. All structure figures were created with PyMOL (http://www.pymol.org).
A dimer of PF0828 is observed in all three crystals, consistent with our gel-filtration analysis (data not shown). The dimer is formed by the side-by-side arrangement of the HUP domains of the two monomers such that their β3 strands are hydrogen bonded to each other, extending the central β-sheet to ten strands (Fig. 2 ▶ a). In addition, residues in helices αB and αC also contribute to the formation of the dimer interface, which buries approximately 1300 Å2 of the surface area of each monomer in the apo enzyme and the AMP complex. The buried surface area in the ATP complex is smaller, 900 Å2 per monomer, owing to a large conformational change in the β2–αB loop (Fig. 2 ▶ b) which is crucial for ATP binding (see below). The overall organization of the dimers is similar in the apo enzyme and the AMP complex, but there is a 7° rotation of the two monomers relative to each other in the ATP complex (Fig. 2 ▶ b).
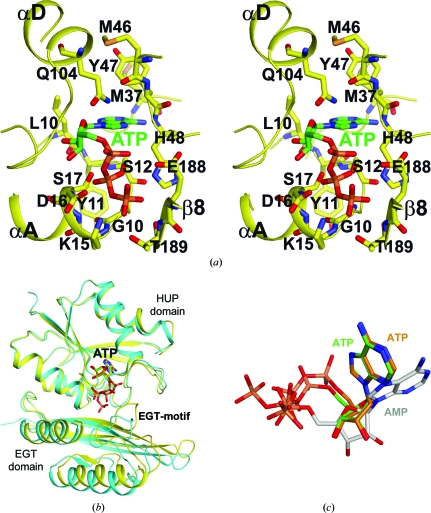
The binding site for ATP, and therefore the likely active site of PF0828, is located at the C-terminal end of the central β-sheet of the HUP domain, away from the dimer interface (Fig. 2 ▶ a). The N1, N6 and N7 atoms of adenine are recognized by direct hydrogen bonding to the main-chain amide and carbonyl groups of residues Met37 and His48 (Fig. 3 ▶ a) in the β2–αB loop, suggesting that PF0828 may be specific for adenine nucleotides. One face of adenine is placed against the peptide bond between residues 11 and 12, while the other face makes van der Waals interactions with the side chains of Met46 and Tyr47, both of which are well conserved among PF0828 homologs (Fig. 1 ▶).
Figure 3.
The active site of PF0828 in complex with ATP. (a) Stereo drawing of the interactions between ATP and PF0828. ATP is shown with green C atoms. (b) Superposition of the two ATP-bound monomers of PF0828. A large difference in the position of the EGT domain, and particularly the EGT-motif, is visible between the two monomers. (c) Superposition of the bound conformations of the two ATP molecules (in green and brown) and AMP (in gray).
The β2–αB loop is mostly disordered in the structures of the apo enzyme and the AMP complex (Fig. 2 ▶ b). The few residues in this loop that are ordered are located near the dimer interface (Fig. 2 ▶ b) and are not positioned correctly for binding ATP. In the ATP complex, the loop becomes completely ordered and recognizes the adenine base and it also helps to cover the putative active site. The 7° rotation of the two monomers relative to each other in the ATP complex appears to be coupled with the movement of this loop (Fig. 2 ▶ b). These observations suggest that a large conformational change may be required for ATP binding and possibly for catalysis by PF0828.
Two distinct conformations of the triphosphate groups of ATP were observed in the crystal, one in each active site of the dimer (Fig. 3 ▶ b). This difference in the binding mode of the triphosphates is observed together with structural changes in the EGT domain of PF0828, especially the EGT-motif, as well as the β2–αB loop (Fig. 3 ▶ b).
In the first monomer, the triphosphate groups interact with the PP-motif as well as the EGT-motif, which is located in the αF–β8 loop (Fig. 3 ▶ a). The PP-motif interacts with the β- and γ-phosphates of ATP. The side-chain hydroxyls of Ser12 and Ser17 are located close to each other and interact with the β-phosphate. The γ-phosphate interacts with the side-chain ammonium ion of Lys15 and the side chain of Thr189. The β- and γ-phosphates are also placed at the N-terminus of helix αA, making favorable interactions with the dipole of this helix. The α-phosphate is located near Glu188 of the EGT-motif. The conserved Asp16 residue in the PP-motif may also be important for catalysis.
In the second monomer, the β- and γ-phosphate groups are located near the α- and β-phosphates of the ATP in the first monomer, respectively (Fig. 3 ▶ c). The EGT-motif moves away by about 3 Å and no longer interacts with the triphosphates (Fig. 3 ▶ b). Overall, the interactions between ATP and this monomer appear to be weaker.
In the AMP complex, the α-phosphate is actually located within 0.4 Å of the β-phosphate of ATP in the first monomer. This is probably linked to a nearly 90° rotation in the positions of the ribose and the adenine base compared with those of ATP (Fig. 3 ▶ c). The N1, N6 and N7 atoms of adenine are not recognized by hydrogen-bonding interactions and the β2–αB loop is mostly disordered. Therefore, it is likely that the observed AMP-binding mode is not a productive one.
Overall, our studies have defined the binding mode of ATP to the likely active site of PF0828, a putative ATP pyrophosphatase. These three crystal structures revealed a large conformational change in the enzyme that is required for ATP binding and possibly catalysis. Most of the highly conserved polar and charged residues are located in the active-site region of the enzyme, although not all of them are involved in ATP binding. These include residues Ser44, Glu78, Ser101, Gln104, and Glu183 and Glu186 in the EGT-motif (Fig. 1 ▶). In addition, Trp124, which is located in the linker between the HUP domain and the C-terminal extension, is disordered in the structures of the ATP and AMP complexes. These conserved residues are likely to be involved in binding the substrate of this enzyme.
Supplementary Material
PDB reference: PF0828, 3rjz
PDB reference: AMP complex, 3rk0
PDB reference: ATP complex, 3rk1
Acknowledgments
We thank Angela Lauricella and George DeTitta for setting up initial crystal screenings, Randy Abramowitz and John Schwanof for setting up the X4A beamline and Jean Jakoncic for setting up the X6A beamline. This research was supported by the Protein Structure Initiative of the National Institutes of Health (U54 GM074958 and U54 GM094597).
References
- Acton, T. B. et al. (2005). Methods Enzymol. 394, 210–243. [DOI] [PubMed]
- Aravind, L., Anantharaman, V. & Koonin, E. V. (2002). Proteins, 48, 1–14. [DOI] [PubMed]
- Bork, P. & Koonin, E. V. (1994). Proteins, 20, 347–355. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Dessailly, B. H., Redfern, O. C., Cuff, A. L. & Orengo, C. A. (2010). Structure, 18, 1522–1535. [DOI] [PMC free article] [PubMed]
- Doublié, S., Kapp, U., Aberg, A., Brown, K., Strub, K. & Cusack, S. (1996). FEBS Lett. 384, 219–221. [DOI] [PubMed]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed]
- Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Jansson, M., Li, Y.-C., Jendeberg, L., Anderson, S., Montelione, B. T. & Nilsson, B. (1996). J. Biomol. NMR, 7, 131–141. [DOI] [PubMed]
- Jogl, G., Tao, X., Xu, Y. & Tong, L. (2001). Acta Cryst. D57, 1127–1134. [DOI] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol. 125, 156–165. [DOI] [PubMed]
- Morishita, R., Kawagoshi, A., Sawasaki, T., Madin, K., Ogasawara, T., Oka, T. & Endo, Y. (1999). J. Biol. Chem. 274, 20688–20692. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sinha, S., Rappu, P., Lange, S. C., Mäntsälä, P., Zalkin, H. & Smith, J. L. (1999). Proc. Natl Acad. Sci. USA, 96, 13074–13079. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003). Methods Enzymol. 374, 22–37. [DOI] [PubMed]
- Weeks, C. M. & Miller, R. (1999). J. Appl. Cryst. 32, 120–124.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PF0828, 3rjz
PDB reference: AMP complex, 3rk0
PDB reference: ATP complex, 3rk1