SecDF is a multi-path membrane protein required for efficient protein translocation and integration via the Sec translocon. Crystals of the first periplasmic domain of SecDF, an essential element for SecDF function, diffracted X-rays to 2.3 Å resolution.
Keywords: Sec translocon, SecDF, protein translocation
Abstract
A membrane-integrated Sec component, SecDF, associates with the SecYEG protein-conducting channel and facilitates protein secretion and membrane–protein integration. SecDF contains 12 transmembrane helices and two periplasmic domains. The first periplasmic domain (P1) plays an important role in protein translocation. Here, the overexpression, purification and crystallization of the P1 domain of Thermus thermophilus SecDF are reported. The crystals diffracted X-rays to 2.3 Å resolution and belonged to space group C2, with unit-cell parameters a = 161.1, b = 35.8, c = 181.6 Å, suggesting that they contain four molecules per asymmetric unit. The initial phases were determined by the multiple-wavelength anomalous dispersion method using selenomethionine-labelled crystals.
1. Introduction
In bacteria, protein translocation across the cytoplasmic membrane is driven by dynamic interplay between the protein-conducting SecYEG channel (Sec translocon) and the SecA ATPase (Rapoport, 2007 ▶; du Plessis et al., 2011 ▶). Efficient protein translocation requires a membrane-integrated Sec component: the SecDF complex. The importance of SecDF in protein translocation has been demonstrated by the findings that strains genetically lacking SecDF are associated with severe growth retardation and protein-export defects in vivo (Pogliano & Beckwith, 1994 ▶). Previous studies have suggested that SecDF is involved in different steps of the translocation pathway: (i) facilitation of preprotein release to the periplasmic side in the late stage of translocation (Matsuyama et al., 1993 ▶), (ii) stabilization of the protein-translocation intermediate by preventing backward movement (Duong & Wickner, 1997 ▶) and (iii) stabilization of the membrane-inserted state of SecA (Economou & Wickner, 1994 ▶). It has also been proposed that SecDF facilitates the membrane–protein integration process by bridging SecYEG and YidC (Nouwen & Driessen, 2002 ▶; Chen et al., 2005 ▶; Xie et al., 2006 ▶), a membrane protein involved in the insertion/maturation of integral membrane proteins. Nevertheless, the exact role played by SecDF in the protein-translocation process remains unclear.
We have established experimental systems to understand the Sec machinery from Thermus thermophilus HB8 (Mori et al., 2003 ▶) and have determined the structures of T. thermophilus SecA and SecYE (Vassylyev et al., 2006 ▶; Tsukazaki et al., 2008 ▶). For further analysis of the Sec machinery, it was crucial to determine the structures of not only the core translocon complex but also of the auxiliary factors. TtSecDF, the counterpart of SecD and SecFs in T. thermophilus, which exists as a single polypeptide chain of 735 amino-acid residues, contains 12 transmembrane (TM) segments and six periplasmic regions, of which the first and fourth periplasmic domains (P1 and P4 of SecD and SecF, respectively) are characteristically large. The preliminary X-ray diffraction of full-length TtSecDF at 3.74 Å resolution has been achieved (Tsukazaki et al., 2006 ▶). Furthermore, structure determination of the periplasmic domains, which are functionally important (Nouwen et al., 2005 ▶), at high resolution would provide plenty of new insights into its function. Here, we report the purification, crystallization and X-ray diffraction analysis of the TtSecDF P1 domain in detail, including previously undescribed critical information for the crystallization of P1. The resulting crystal structure of the P1 domain (PDB entry 3aqo) enabled us to refine the model of the full-length TtSecDF structure. Its structural features and subsequent functional analyses suggested that SecDF functions as a membrane-integrated chaperone that is powered by the proton motive force to achieve ATP-independent protein translocation (Tsukazaki et al., 2011 ▶).
2. Materials and methods
2.1. Plasmid preparation
The gene encoding N-terminally Met-His6-Leu-Glu-Val-Leu-Phe-Gln-Gly-tagged TtSecDF31–274 (residues 31–274, corresponding to the P1 domain) was PCR-amplified from pTT206 (Tsukazaki et al., 2006 ▶) using the primers 5′-CTGTGGCCATATGCACCATCACCATCACCATCTGGAAGTTCTGTTCCAGGGGCCCAAGGTCCGCCTGG-3′ and 5′-CTGGGTACCGTCGACTACCCCGCCTGGATGGCGTC-3′ and cloned into the NdeI and SalI sites of the pET-26b vector (Novagen). The resulting plasmid was used as a template for QuikChange mutagenesis (Stratagene) to construct plasmids encoding the C-terminally shortened or extended derivatives TtSecDF31–249, TtSecDF31–261, TtSecDF31–263, TtSecDF31–267, TtSecDF31–270 and TtSecDF31–281.
2.2. Purification
The purification of TtSecDF35–263 is described below. The other P1 mutants were purified using the same procedure. The resulting recombinant plasmid was introduced into Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene). The cells were cultivated at 310 K in LB medium supplemented with 0.4% glucose, 50 µg ml−1 kanamycin and 20 µg ml−1 chloramphenicol to a cell density (OD600) of approximately 0.6 and were then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 4 h. The cells were harvested, washed with 10 mM Tris–HCl pH 8.0 and resuspended in 50 mM Tris–HCl pH 8.0, 50 mM NaCl and 1 mM phenylmethanesulfonyl fluoride. After disruption of the plasma membrane using a French pressure cell (Thermo Fisher Scientific), the supernatant after centrifugation (9600g) for 30 min and ultracentrifugation (150 000g) for 30 min was applied onto an Ni–NTA agarose column (Qiagen) equilibrated with buffer A (50 mM Tris–HCl pH 8.0, 300 mM NaCl) containing 10 mM imidazole, which was then washed with buffer A containing 30 mM imidazole and further eluted with a 50–300 mM imidazole gradient in buffer A. The eluate was dialyzed against 50 mM Tris–HCl pH 8.0, 300 mM NaCl. The N-terminal tag and the subsequent four residues of P1 were cleaved off using trypsin (0.825 mg per gram of TtSecDF P1) for 2 h at 277 K. The solution containing the cleaved sample (TtSecDF35–263) was applied onto a Benzamidine column (GE Healthcare) equilibrated with buffer A to remove the trypsin; the flowthrough sample passed through an Ni–NTA agarose column equilibrated with buffer A containing 10 mM imidazole was subsequently collected. The sample was dialyzed against 30 mM Tris–HCl pH 8.5 and then separated using a Mono Q 10/100 GL column (GE Healthcare) and eluted with a 0–0.3 M linear NaCl gradient in 30 mM Tris–HCl pH 8.5. For further purification, the TtSecDF35–263 fractions were concentrated using an Amicon Ultra 10K filter (Millipore), subjected to HiLoad 16/60 Superdex 200 (GE Healthcare) size-exclusion column chromatography and eluted with 30 mM Tris–HCl pH 8.5, 150 mM NaCl. Typically, we obtained approximately 5 mg purified TtSecDF35–263 from a 1 l culture. Protein concentration was estimated by assuming an A 280 of 0.06 for a 1 mg ml−1 solution.
In order to solve the three-dimensional structure of TtSecDF35–263, which contains no Met residues, using the multiple-wavelength anomalous dispersion method, we prepared a selenomethionine-labelled TtSecDF35–263(L59M, S96M, L113M, L204M) mutant. The mutations were introduced by QuikChange (Stratagene) mutagenesis. The plasmid encoding the SecDF P1 domain mutant was introduced into the methionine-auxotrophic E. coli strain B834 (DE3) (Novagen) which additionally harboured the CodonPlus (DE3)-RIL plasmid (Stratagene). Cells were grown in Core medium (Wako) supplemented with 0.4% glucose, 250 µg ml−1 MgSO4·7H2O, 4.2 µg ml−1 FeSO4·7H2O, 2 µg ml−1 thiamine, 50 µg ml−1 kanamycin, 20 µg ml−1 chloramphenicol and 25 µg ml−1 l-selenomethionine (Nakalai Tesque). The selenomethionine-labelled TtSecDF35–263(L59M, S96M, L113M, L204M) was purified by the same procedures as used for purification of the native protein.
2.3. Trypsin digestion
The TtSecDF31–274 sample (0.1 mg ml−1) after Ni–NTA column chromatography was incubated in the presence of trypsin (0, 0.8 and 4 µg ml−1) for 5 min at room temperature and then mixed with double the volume of 2× SDS–PAGE sample buffer and incubated for 5 min at 364 K. After SDS–PAGE, trypsin-digested proteins were stained with Coomassie Brilliant Blue.
2.4. Protein sequence and mass spectrometry
After separation by SDS–PAGE and electro-blotting onto PVDF membrane, a major Coomassie-stained protein band of the tryptic digest was excised and subjected to Edman degradation using a Procise 494 HT protein-sequencing system (Applied Biosystems). The tryptic digest was also subjected to matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI–TOF MS) on a Ultraflex (Bruker Daltonics) in linear mode using sinapinic acid as a matrix.
2.5. Crystallization
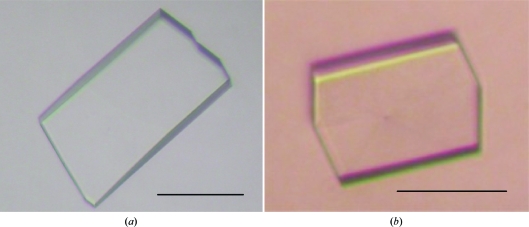
The purified P1 was concentrated to approximately 8 mg ml−1 using an Amicon Ultra 10K filter and dialyzed against 20 mM Tris–HCl pH 8.0. Initially, crystallization conditions were searched for using Crystal Screen, Crystal Screen 2, PEG/Ion, Index, Natrix and SaltRX kits from Hampton Research, MemSys/MemStart kits from Molecular Dimensions and JBScreen Classic 1, 2, 4 and 5 kits from Jena Bioscience. A Hydra II Plus One crystallization robot (Matrix Technologies) was used for screening using the sitting-drop vapour-diffusion method, mixing equal volumes (0.2 µl) of protein solution (8 mg ml−1 in Tris–HCl pH 8.0) and reservoir solution. The drops were equilibrated at 293 K against 100 µl reservoir solution. Small crystals of TtSecDF35–263 appeared in a few days using a reservoir solution consisting of 20% PEG 3350 and 0.2 M sodium thiocyanate. After further optimization of the reservoir conditions using Additive Screen (Hampton Research), the crystal size was improved by the addition of 3% trehalose. The best crystals of TtSecDF35–263 appeared in 1 d and grew to dimensions of approximately 100 × 200 × 30 µm within 6 d (Fig. 1 ▶ a) with a reservoir solution consisting of 18% PEG 3350, 0.6 M sodium thiocyanate and 3% trehalose. Crystals of selenomethionine-labelled TtSecDF35–263(L59M, S96M, L113M, L204M) were obtained under the same crystallization conditions as used for the native crystals (Fig. 1 ▶ b).
Figure 1.
Crystals of the SecDF P1 domain. Native (a) and selenomethionine-labelled (b) crystals were obtained by sitting-drop vapour diffusion with reservoir solution consisting of 20% PEG 3350, 0.2 M sodium thiocyanate and 3% trehalose. The scale bars represent 100 µm.
2.6. X-ray data collection and processing
The crystals of TtSecDF35–263 were transferred into a cryoprotectant solution consisting of 20% PEG 400, 20% PEG 3350, 0.6 M sodium thiocyanate, 3% trehalose and 20 mM Tris–HCl pH 8.0. The treated crystals were then flash-cooled in a nitrogen-gas stream at 100 K. X-ray diffraction data sets for TtSecDF35–263 were collected at 100 K in a nitrogen-gas stream using an ADSC Quantum 315 detector on beamline BL41XU at SPring-8 (Sayo, Japan), whereas those for selenomethionine-labelled TtSecDF35–263(L59M, S96M, L113M, L204M) were collected using an ADSC Quantum 210 detector on beamline NW12 at the Photon Factory (Tsukuba, Japan). The diffraction data sets were processed using DENZO/SCALEPACK (Otwinowski & Minor, 1997 ▶). The data-collection statistics are summarized in Table 1 ▶ and the refinement statistics for the model of the P1 structure have been described previously (Tsukazaki et al., 2011 ▶)
Table 1. Data-collection statistics.
| Selenomethionine-labelled | |||||
|---|---|---|---|---|---|
| Native | Peak | Inflection | High remote | Low remote | |
| X-ray source | SPring-8 BL41XU | Photon Factory AR-NW12 | |||
| Crystal dimensions (mm) | 0.20 × 0.10 × 0.30 | 0.15 × 0.10 × 0.30 | |||
| Wavelength (Å) | 0.97942 | 0.97923 | 0.97931 | 0.98319 | 0.96408 |
| Crystal-to-detector distance (mm) | 300 | 263.5 | |||
| Temperature (K) | 100 | 100 | 100 | 100 | 100 |
| Total oscillation range (°) | 360 | 720 | 360 | 180 | 180 |
| Oscillation range (°) | 1 | 1 | 1 | 1 | 1 |
| Exposure time (s) | 1 | 1 | 1 | 1 | 1 |
| Detector | ADSC Q315 | ADSC Q210 | |||
| Space group | C2 | C2 | |||
| Unit-cell parameters (Å, °) | a = 161.1, b = 35.8, c = 181.6, β = 113.7 | a = 160.6, b = 35.8, c = 180.9, β = 113.5 | |||
| Resolution (Å) | 50–2.34 (2.38–2.34) | 50–2.70 (2.75–2.70) | 50–2.70 (2.75–2.70) | 50–2.70 (2.75–2.70) | 50–2.70 (2.75–2.70) |
| Observed reflections | 260026 | 248311 | 120784 | 61211 | 61659 |
| Unique reflections | 40237 | 26074 | 25400 | 23886 | 23914 |
| Multiplicity | 6.5 (4.0) | 9.5 (7.9) | 4.9 (4.0) | 2.6 (2.1) | 2.6 (2.2) |
| Completeness (%) | 97.5 (86.2) | 97.2 (96.4) | 94.4 (92.4) | 89.0 (83.6) | 88.4 (83.1) |
| 〈I/σ(I)〉 | 20.0 (1.9) | 50.5 (17.1) | 35.6 (11.1) | 26.2 (8.0) | 26.0 (7.9) |
| Rmerge | 0.110 (0.541) | 0.066 (0.106) | 0.064 (0.109) | 0.051 (0.095) | 0.054 (0.102) |
3. Results and discussion
3.1. Construct for crystallization
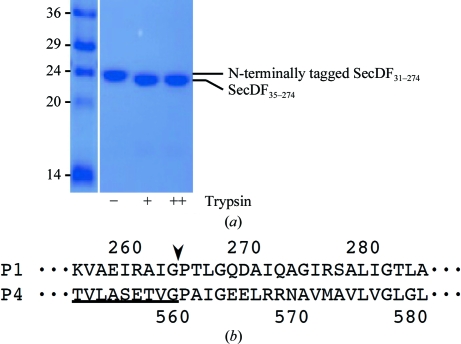
We initially planned to remove the N-terminal His6 tag using PreScission protease, which specifically cleaves between the Gln and Gly residues of the N-terminal tag recognition sequence Leu-Glu-Val-Leu-Phe-Gln-Gly. Nevertheless, we found that the purified P1 domain of TtSecDF, except for the N-terminus, was trypsin-resistant (Fig. 2 ▶ a). The slightly downshifted band in Fig. 2 ▶(a) was shown to be composed of the N-terminally truncated P1 domain, TtSecDF35–274, from its N-terminal sequence (LGLDLKGGLRIVLEADVEN) and by mass spectrometry. The trypsin-digestion procedure enables us to effectively remove the N-terminal tag and the subsequent four residues of the P1 domain, which were supposed to be located in the flexible N-terminal region. We thus decided to perform crystallization trials using the N-terminally trypsin-digested samples. However, the purified TtSecDF35–274 could not be crystallized using more than 1000 conditions. We then attempted to crystallize several C-terminally shortened or extended variations: TtSecDF35–249, TtSecDF35–261, TtSecDF31–267, TtSecDF31–270 and TtSecDF31–281. As a result, TtSecDF31–267 and TtSecDF31–270 were crystallized as microneedles, which implies that the length of the C-terminal region is critical for the crystallization ability of the P1 domain. In parallel to our trials to crystallize another construct of the P1 domain, the structure of the P4 domain, TtSecDF480–559, was determined by nuclear magnetic resonance (PDB entry 2rrn; Tsukazaki et al., 2011 ▶). Considering that the C-terminal tail of P4 is homologous to that of P1 but proved to be partially disordered (Fig. 2 ▶ b), we performed crystallization screening using TtSecDF35–263, the C-terminus of which was adjusted to the ordered C-terminal tail of P4. This enabled us to successfully obtain plate-like crystals (Fig. 1 ▶ a).
Figure 2.
Flexibility of the N-terminus and the C-terminus of the SecDF P1 domain. (a) Digestion of the P1 domain using trypsin (0, 0.8 and 4 µg ml−1). (b) Sequence alignment of the C-termini of the P1 and P4 domains. The residue numbers are shown. Underlined residues are ordered in the NMR structure of P4; the subsequent residues of P4 are disordered. The border between the ordered and disordered regions is indicated by an arrowhead.
3.2. Data processing
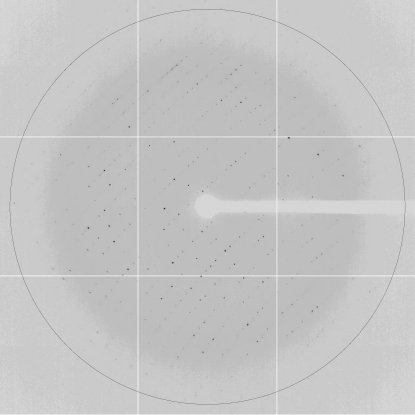
The crystals of native TtSecDF35–263 (molecular weight 24.7 kDa) diffracted X-rays to 2.3 Å resolution (Fig. 3 ▶) and belonged to space group C2, with unit-cell parameters a = 161.1, b = 35.8, c = 181.6 Å. The calculated Matthews coefficient (V M) of 2.43 Å3 Da−1 suggested the presence of four molecules in the asymmetric unit, with a solvent content of 49.3% (Matthews, 1968 ▶). The cystals of selenomethionine-labelled TtSecDF35–263(L59M, S96M, L113M, L204M) diffracted X-rays to 2.7 Å resolution and belonged to the same space group as the native crystals. We collected a four-wavelength multiwavelength anomalous diffraction (MAD) data set from selenomethionine-labelled TtSecDF35–263 at the absorption peak (λ = 0.97923 Å), the inflection point (λ = 0.97931 Å), a high-energy remote point (λ = 0.98319 Å) and a low-energy remote point (λ = 0.96408 Å) (Table 1 ▶). We identified the 12 Se sites using the programs SHELXC and SHELXD (Sheldrick, 2008 ▶) and obtained an initial electron-density map of TtSecDF35–263 using the program SHARP (de La Fortelle & Bricogne, 1997 ▶), which clearly showed a clear solvent–protein boundary. The subsequent procedures for determination of the P1 structure and the crystal structure have been described elsewhere (Tsukazaki et al., 2011 ▶). The head and base subdomains that are the major components of the P1 domain were separately fitted into the ambiguous density map of full-length TtSecDF determined by single-wavelength anomalous dispersion analysis at 3.7 Å resolution. We finally refined the full-length SecDF structure at 3.3 Å resolution and examined the function of SecDF (Tsukazaki et al., 2011 ▶).
Figure 3.
Diffraction pattern from a native crystal of the SecDF P1 domain. The black circle is drawn at 2.3 Å resolution.
Acknowledgments
We thank Shuya Fukai, Hiroyuki Mori, Koreaki Ito and Hiroshi Nishimasu for useful suggestions, Rieko Yamasaki for secretarial assistance and the beamline staff at SPring-8 (Sayo, Japan) and the Photon Factory (Tsukuba, Japan) for technical help during data collection. This work was supported by grants from JSPS and MEXT KAKENHI.
References
- Chen, M., Xie, K., Yuan, J., Yi, L., Facey, S. J., Pradel, N., Wu, L.-F., Kuhn, A. & Dalbey, R. E. (2005). Biochemistry, 44, 10741–10749. [DOI] [PubMed]
- Duong, F. & Wickner, W. (1997). EMBO J. 16, 4871–4879. [DOI] [PMC free article] [PubMed]
- Economou, A. & Wickner, W. (1994). Cell, 78, 835–843. [DOI] [PubMed]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol. 276, 472–494. [DOI] [PubMed]
- Matsuyama, S., Fujita, Y. & Mizushima, S. (1993). EMBO J. 12, 265–270. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mori, H., Tsukazaki, T., Masui, R., Kuramitsu, S., Yokoyama, S., Johnson, A. E., Kimura, Y., Akiyama, Y. & Ito, K. (2003). J. Biol. Chem. 278, 14257–14264. [DOI] [PubMed]
- Nouwen, N. & Driessen, A. J. (2002). Mol. Microbiol. 44, 1397–1405. [DOI] [PubMed]
- Nouwen, N., Piwowarek, M., Berrelkamp, G. & Driessen, A. J. (2005). J. Bacteriol. 187, 5857–5860. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Plessis, D. J. du, Nouwen, N. & Driessen, A. J. (2011). Biochim. Biophys. Acta, 1808, 851–865. [DOI] [PubMed]
- Pogliano, J. A. & Beckwith, J. (1994). EMBO J. 13, 554–561. [DOI] [PMC free article] [PubMed]
- Rapoport, T. A. (2007). Nature (London), 450, 663–669. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tsukazaki, T., Mori, H., Echizen, Y., Ishitani, R., Fukai, S., Tanaka, T., Perederina, A., Vassylyev, D. G., Kohno, T., Maturana, A. D., Ito, K. & Nureki, O. (2011). Nature (London), 474, 235–238. [DOI] [PMC free article] [PubMed]
- Tsukazaki, T., Mori, H., Fukai, S., Ishitani, R., Mori, T., Dohmae, N., Perederina, A., Sugita, Y., Vassylyev, D. G., Ito, K. & Nureki, O. (2008). Nature (London), 455, 988–991. [DOI] [PMC free article] [PubMed]
- Tsukazaki, T., Mori, H., Fukai, S., Numata, T., Perederina, A., Adachi, H., Matsumura, H., Takano, K., Murakami, S., Inoue, T., Mori, Y., Sasaki, T., Vassylyev, D. G., Nureki, O. & Ito, K. (2006). Acta Cryst. F62, 376–380. [DOI] [PMC free article] [PubMed]
- Vassylyev, D. G., Mori, H., Vassylyeva, M. N., Tsukazaki, T., Kimura, Y., Tahirov, T. H. & Ito, K. (2006). J. Mol. Biol. 364, 248–258. [DOI] [PubMed]
- Xie, K., Kiefer, D., Nagler, G., Dalbey, R. E. & Kuhn, A. (2006). Biochemistry, 45, 13401–13408. [DOI] [PubMed]