The recombinant RPB5 subunit of human RNA polymerase II has been crystallized by vapour diffusion in hanging drops.
Keywords: RNA polymerase II, RPB5 subunit
Abstract
RPB5 is an essential subunit of eukaryotic RNA polymerase II. It has been proposed to interact with DNA and several key transcription factors during transcription. These interactions are crucial for transcription and its regulation. Here, prior to obtaining complex structures of human RPB5 and its binding partners, recombinant human RPB5 was crystallized alone by vapour diffusion in hanging drops. A complete data set was collected from a single frozen crystal employing an in-house X-ray source. The crystal diffracted to 2.8 Å resolution and belonged to space group P43212. The likely Matthews coefficient and solvent content of 2.67 Å3 Da−1 and 53.92%, respectively, suggested the presence of two protein subunits in the asymmetric unit. The structure was solved using molecular replacement.
1. Introduction
RNA polymerase II (RNAPII) is the central enzyme responsible for the transcription of all pre-mRNAs in eukaryotes. Direct protein interactions between RNAPII subunits and transcription factors are essential for the regulation of transcription during the processes that encompass initiation, elongation and termination. Extensive studies have been performed on the structure and function of Saccharomyces cerevisiae RNAP II and several general transcription factors (Wang et al., 2009 ▶; Fuda et al., 2009 ▶; Selth et al., 2010 ▶). However, very limited structural knowledge is available on mammalian RNAPII and its interactions with cofactors, mainly owing to the difficulty of protein purification and the likely existence of RNAPII isoforms (Hu et al., 2006 ▶).
Prior to structure determination of mammalian RNAPII, obtaining structures of individual RNAPII subunits and their complexes with cofactors should be a practical way of understanding how mammalian RNAPII interacts with other components of the transcriptional machinery. To date, structures of hRPB6, hRPB8 and hRPB4–hRPB7 subunits have been reported (del Río-Portilla et al., 1999 ▶; Kang et al., 2006 ▶; Meka et al., 2005 ▶).
In eukaryotes there are three RNA polymerases; RPB5 is a subunit that is common to all three eukaryotic RNAPs, while RPB5 homologues are not found in either prokaryotes or archaebacteria (Eloranta et al., 1998 ▶; Thiru et al., 1999 ▶). It is highly conserved throughout evolution and plays an essential role in eukaryotic cells (Edwards et al., 1991 ▶; McKune et al., 1995 ▶; Shpakovski et al., 1995 ▶). According to structures of RNAPII from S. cerevisiae, RPB5 consists of an N-terminal domain located at the far end of the DNA channel of RNAPII (Kim et al., 1997 ▶; Cramer et al., 2001 ▶) and a strongly conserved C-terminal module which is tightly connected to the largest subunit RPB1 (Cramer et al., 2001 ▶; Langer et al., 1995 ▶). RPB5 is thought to thought to grab the incoming DNA template, since RPB5 makes up part of the RNAPII lower jaw which surrounds the central cleft that houses the incoming DNA. RPB5 is largely exposed to interactions with general transcription factors or possibly with specific gene regulators such as TFIIF and Taf15 (Wei et al., 2001 ▶; Le et al., 2005 ▶).
Previous studies have suggested that the interaction of HBx, a multi-functional protein encoded by the X gene of human hepatitis B virus, with human RPB5 (hRPB5) can facilitate HBx transactivation (Le et al., 2005 ▶; Lin et al., 1997 ▶; Murakami et al., 2005 ▶; Wei et al., 2001 ▶). Experiments have demonstrated that the HBx transactivation domain and the central region of hRPB5 are necessary for their specific binding. Considering the key role of HBx in the transactivation of viral and host genes through a variety of cis-acting enhancer elements present in RNAPII promoters, interruption of HBx–hRPB5 binding may inhibit human hepatitis B virus proliferation.
We are interested in investigating the structural basis of the interaction of cofactors and human RNAPII and especially the potential for utilizing this information for drug development. The detailed molecular architecture of the HBx–hRPB5 complex is essential for the design of peptide or small-molecule inhibitors and this paper presents the first step towards this aim, i.e. the structure determination of hRPB5.
2. Materials and methods
2.1. Cloning, expression and purification
POLR2E cDNA (NM_002695) encoding hRPB5 was purchased from GeneCopoeia China and subcloned into the pET15b expression vector between NdeI and BamHI sites, with 20 additional amino acids (MGSSHHHHHHSSGLVPRGSH–), including a hexahistidine tag, at the N-terminus. Escherichia coli strain BL21-CodonPlus (Stratagene) was used for protein expression. The transformant bacteria were grown to an OD600 of 1.5 or higher in LB medium containing 100 µg ml−1 ampicillin at 310 K and were induced with 0.2 mM IPTG at 289 K for 20 h. According to the pre-treatment protocol proposed by Magnusdottir et al. (2009 ▶), the cells were harvested by centrifugation and were washed three times with sucrose buffer (20% sucrose, 1 mM EDTA, 50 mM HEPES pH 7.9) and cold distilled water containing 5 mM MgCl2 alternately. The cells were resuspended in cold lysis buffer (300 mM NaCl, 20 mM imidazole, 10% glycerol and 5 mM β-mercaptoethanol, 1 mg ml−1 lysozyme, 20 mM Tris pH 8.0). The suspension was sonicated and the soluble proteins in the supernatant were recovered by centrifugation at 13 000g for 30 min at 277 K. The supernatant was loaded onto an Ni2+-chelating HisTrap FF column (GE Healthcare, USA) which had been pre-equilibrated with binding buffer (lysis buffer without lysozyme). The column was washed with binding buffer followed by washing buffer (50 mM NaCl, 40 mM imidazole, 30% glycerol, 5 mM β-mercaptoethanol, 20 mM Tris pH 8.0). The recombinant protein was eluted with elution buffer (50 mM NaCl, 250 mM imidazole, 10% glycerol, 5 mM β-mercaptoethanol, 20 mM Tris pH 8.0). The purified protein was ultrafiltered and concentrated using Amicon Ultra-10 (Millipore, USA) in crystallization buffer consisting of 50 mM NaCl, 5% glycerol, 5 mM Tris pH 8.0 prior to crystallization setup. Protein concentrations were determined using the method of Bradford (1976 ▶). All column-chromatography and enzyme-concentration steps were performed at 277 K. The purity of the protein was examined by 10% SDS–PAGE and was determined to be >98%.
2.2. Crystallization
Initial crystallization screening was performed manually using Crystal Screen and Crystal Screen 2 (Hampton Research) by the hanging-drop vapour-diffusion technique at 289 K. 1.5 µl reservoir solution and 1.5 µl hRPB5 solution at 15–24 mg ml−1 were mixed and equilibrated against 500 µl reservoir solution. Four conditions were found to give tiny crystals: Nos. 16, 33 and 39 of Crystal Screen and No. 14 of Crystal Screen 2, of which No. 39 from Crystal Screen [2%(v/v) PEG 400, 2.0 M ammonium sulfate, 0.1 M HEPES pH 7.5] gave the best-shaped crystals. The buffer was modified to permit more robust crystal growth, with the final crystallization condition consisting of 2%(v/v) PEG 400, 2.0 M ammonium sulfate, 0.1 M HEPES pH 6.0. Biconvex lens-shaped crystals were obtained in approximately two months at 289 K.
2.3. X-ray diffraction data collection
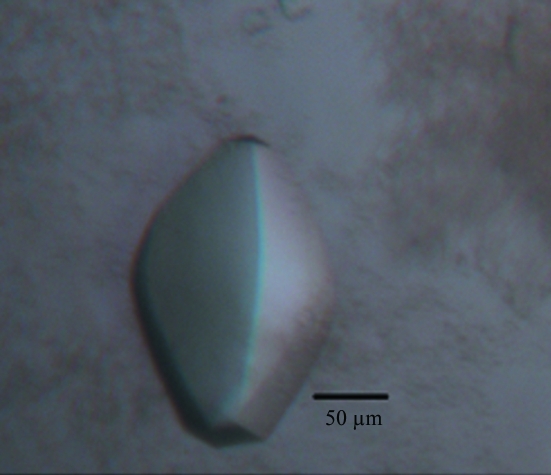
The crystals were collected from the crystallization drop using a nylon loop, flash-cooled in a dry nitrogen-gas stream at 100 K and tested for diffraction employing our in-house Oxford Diffraction Xcalibur Nova diffractometer operating at 50 kV and 0.8 mA with a wavelength of 1.5418 Å. We found that under the current crystallization conditions the diffraction quality of crystals from the same drop varied significantly from 10 to 3 Å resolution. A single crystal (Fig. 1 ▶) was used to collect a full data set. The crystal diffracted X-rays to 2.8 Å resolution. A total of 392 images were recorded using a 165 mm Onyx CCD detector. Data sets were processed and scaled using CrysAlisPro (v.1.171.33.49; Oxford Diffraction) and SCALA from the CCP4 suite (Winn et al., 2010 ▶) at a resolution cutoff of 2.8 Å. A typical diffraction image of the hRPB5 crystal is shown in Fig. 2 ▶. The crystal belonged to space group P43212, with unit-cell parameters a = b = 77.15, c = 179.06 Å, α = β = γ = 90.00°. The data-collection and processing statistics are summarized in Table 1 ▶.
Figure 1.
A biconvex lens-shaped crystal of hRBP5 with dimensions of 150 × 100 × 100 µm.
Figure 2.
A typical diffraction pattern of an hRPB5 crystal. The exposure time was 160 s, the crystal-to-detector distance was 120.0 mm and the oscillation range per frame was 0.5°. The diffraction image was collected on a 165 mm Onyx CCD detector.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.5418 |
| Temperature (K) | 100 |
| Resolution range (Å) | 24.85–2.80 (2.95–2.80) |
| Space group | P43212 |
| Unit-cell parameters (Å, °) | a = b = 77.15, c = 179.06, α = β = γ = 90.00 |
| Observed reflections | 140468 (13497) |
| Unique reflections | 14036 (1993) |
| Data completeness (%) | 99.9 (100.0) |
| Multiplicity | 10.0 (6.8) |
| 〈I/σ(I)〉 | 20.7 (2.7) |
| Rmerge (%) | 0.09 (0.60) |
| Matthews coefficient (Å3 Da−1) | 2.67 |
| Solvent content (%) | 53.92 |
| Average mosaicity (°) | 0.62 |
3. Results and discussion
The likely Matthews coefficient (Matthews, 1968 ▶) and solvent content of 2.67 Å3 Da−1 and 53.92%, respectively, suggested the presence of two molecules in the asymmetric unit. The hRPB5 structure has successfully been solved by molecular replacement using BALBES (Long et al., 2008 ▶) followed by autobuilding using ARP/wARP (Perrakis et al., 1999 ▶) on the YSBL web server. Several cycles of refinement with PHENIX (Adams et al., 2010 ▶) and manual rebuilding with Coot (Emsley & Cowtan, 2004 ▶) indicated a partially disordered C-terminal domain with residues absent from the loops, whereas in the crystal structure of S. cerevisiae RPB5 (PDB entry 1dzf) the C-terminal domain is nicely ordered (Todone et al., 2000 ▶). The model converged with an R factor of 0.27 and an R free of 0.35 at this stage. Since the C-terminus of hRPB5 obviously does not participate in the interaction of hRPB5 and cofactors, we propose that by carefully shortening the C-terminus we may significantly improve the quality of hRPB5 crystals and still be able to obtain structures of complexes of RPB5 with cofactors. Our efforts are now aimed at obtaining complex crystals of hRBP5 with HBx fragments and other cofactors.
Acknowledgments
The authors gratefully thank the Natural Science Foundation of China (30800169), the Research Fund for the Doctoral Program of Higher Education of China (200805581146) and the Fundamental Research Funds for the Central Universities (SYSU 11lgjc09).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Cramer, P., Bushnell, D. A. & Kornberg, R. D. (2001). Science, 292, 1863–1876. [DOI] [PubMed]
- Edwards, A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. (1991). J. Biol. Chem. 266, 71–75. [PubMed]
- Eloranta, J. J., Kato, A., Teng, M. S. & Weinzierl, R. O. (1998). Nucleic Acids Res. 26, 5562–5567. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fuda, N. J., Ardehali, M. B. & Lis, J. T. (2009). Nature (London), 461, 186–192. [DOI] [PMC free article] [PubMed]
- Hu, X., Malik, S., Negroiu, C. C., Hubbard, K., Velalar, C. N., Hampton, B., Grosu, D., Catalano, J., Roeder, R. G. & Gnatt, A. (2006). Proc. Natl Acad. Sci. USA, 103, 9506–9511. [DOI] [PMC free article] [PubMed]
- Kang, X., Hu, Y., Li, Y., Guo, X., Jiang, X., Lai, L., Xia, B. & Jin, C. (2006). J. Biol. Chem. 281, 18216–18226. [DOI] [PubMed]
- Kim, T.-K., Lagrange, T., Wang, Y.-H., Griffith, J. D., Reinberg, D. & Ebright, R. H. (1997). Proc. Natl Acad. Sci. USA, 94, 12268–12273. [DOI] [PMC free article] [PubMed]
- Langer, D., Hain, J., Thuriaux, P. & Zillig, W. (1995). Proc. Natl Acad. Sci. USA, 92, 5768–5772. [DOI] [PMC free article] [PubMed]
- Le, T. T., Zhang, S., Hayashi, N., Yasukawa, M., Delgermaa, L. & Murakami, S. (2005). J. Biochem. 138, 215–224. [DOI] [PubMed]
- Lin, Y., Nomura, T., Cheong, J., Dorjsuren, D., Iida, K. & Murakami, S. (1997). J. Biol. Chem. 272, 7132–7139. [DOI] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Magnusdottir, A., Johansson, I., Dahlgren, L.-G., Nordlund, P. & Berglund, H. (2009). Nature Methods, 6, 477–478. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McKune, K., Moore, P., Hull, M. & Woychik, N. (1995). Mol. Cell. Biol. 15, 6895–6900. [DOI] [PMC free article] [PubMed]
- Meka, H., Werner, F., Cordell, S. C., Onesti, S. & Brick, P. (2005). Nucleic Acids Res. 33, 6435–6444. [DOI] [PMC free article] [PubMed]
- Murakami, Y., Uemura, K., Sasaki, T., Hayashidani, Y., Sudo, T. & Sueda, T. (2005). Surgery, 138, 962–963. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Río-Portilla, F. del, Gaskell, A., Gilbert, D., Ladias, J. A. & Wagner, G. (1999). Nature Struct. Biol. 6, 1039–1042. [DOI] [PubMed]
- Selth, L. A., Sigurdsson, S. & Svejstrup, J. Q. (2010). Annu. Rev. Biochem. 79, 271–293. [DOI] [PubMed]
- Shpakovski, G. V., Acker, J., Wintzerith, M., Lacroix, J. F., Thuriaux, P. & Vigneron, M. (1995). Mol. Cell. Biol. 15, 4702–4710. [DOI] [PMC free article] [PubMed]
- Thiru, A., Hodach, M., Eloranta, J. J., Kostourou, V., Weinzierl, R. O. & Matthews, S. (1999). J. Mol. Biol. 287, 753–760. [DOI] [PubMed]
- Todone, F., Weinzierl, R. O., Brick, P. & Onesti, S. (2000). Proc. Natl Acad. Sci. USA, 97, 6306–6310. [DOI] [PMC free article] [PubMed]
- Wang, D., Bushnell, D. A., Huang, X., Westover, K. D., Levitt, M. & Kornberg, R. D. (2009). Science, 324, 1203–1206. [DOI] [PMC free article] [PubMed]
- Wei, W., Dorjsuren, D., Lin, Y., Qin, W., Nomura, T., Hayashi, N. & Murakami, S. (2001). J. Biol. Chem. 276, 12266–12273. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.