A complex of the human DNA-repair proteins XRCC4 and XLF was crystallized under high-salt and extreme dehydration conditions to produce diffraction to 3.9 Å resolution. Initial phasing information was obtained from molecular replacement with single-wavelength anomalous diffraction using tantalum bromide clusters.
Keywords: XRCC4–XLF, protein complexes, low resolution, dehydration, tantalum bromide
Abstract
XRCC4 and XLF are key proteins in the repair of DNA double-strand breaks through nonhomologous end-joining. Together, they form a complex that stimulates the ligation of double-strand breaks. Owing to the suggested filamentous nature of this complex, structural studies via X-ray crystallography have proven difficult. Multiple truncations of the XLF and XRCC4 proteins were cocrystallized, but yielded low-resolution diffraction (∼20 Å). However, a combination of microseeding, dehydration and heavy metals improved the diffraction of XRCC4Δ157–XLFΔ224 crystals to 3.9 Å resolution. Although molecular replacement alone was unable to produce a solution, when combined with the anomalous signal from tantalum bromide clusters initial phasing was successfully obtained.
1. Introduction
DNA double-strand breaks (DSBs) are a serious threat to chromosomal stability and when left unrepaired cause genomic rearrangements or cell death. Mammals have two distinct pathways for repair of DNA DSBs: homologous recombination and nonhomologous end-joining (NHEJ). Of these, NHEJ is the primary repair method owing to its unrestricted use throughout the cell cycle. NHEJ requires a core set of seven proteins for binding, processing and ligating broken DNA ends (reviewed in Lieber et al., 2010 ▶).
Two of these proteins, XRCC4 and XLF, have no known enzymatic function yet are essential for repair, as shown by XLF−/− and XRCC4−/− mammalian cells, which display severe defects in DSB repair (Giaccia et al., 1990 ▶; Zha et al., 2007 ▶). XLF and XRCC4 have been shown to directly interact with one another and are responsible for stimulating ligase IV to repair DNA ends (Ahnesorg et al., 2006 ▶; Tsai et al., 2007 ▶). However, the mechanism by which this occurs is unknown. XRCC4 and XLF are structural homologues, with each existing as a homodimer. Both proteins contain an N-terminal head domain and an extended C-terminal tail domain. Each head domain is comprised of a seven-stranded antiparallel β-sheet interrupted by a helix–turn–helix motif between strands 4 and 5. The C-terminal α-helical tail extends away from the base of the head domain and constitutes the primary dimerization interface. In XRCC4 this tail region remains fully extended, while in XLF it wraps back up and around towards the head domain (Junop et al., 2000 ▶; Andres et al., 2007 ▶; Li et al., 2008 ▶).
The head domains of XLF and XRCC4 are required for association with one another. Initial mutational studies that identified interacting surfaces between these proteins suggested that the oligomeric state of the XRCC4–XLF complex may consist of an extended filament (Andres et al., 2007 ▶; Malivert et al., 2010 ▶). This idea was further supported by recent data from small-angle X-ray scattering (Hammel et al., 2010 ▶). Given the flexible and filamentous nature of the XRCC4–XLF complex, structural studies via X-ray crystallography have proven to be challenging. Here, we report the successful crystallization and initial diffraction of XRCC4–XLF crystals to 3.9 Å resolution.
2. Materials and methods
2.1. Molecular cloning and protein expression
Full-length XRCC4, XRCC4Δ265, XRCC4Δ202, XRCC4Δ202,A60E-BRCT, full-length XLF and XLFΔ224 were cloned and expressed as described previously (Table 1 ▶; Modesti et al., 1999 ▶; Junop et al., 2000 ▶; Andres et al., 2007 ▶; Wu et al., 2009 ▶). XRCC4Δ157 was generated using the Gateway Cloning system (Invitrogen, Canada) and expressed identically to full-length XRCC4. XRCC4Δ136 was created by inserting a stop codon after residue 136 in XRCC4Δ157 using QuikChange mutagenesis (Stratagene, USA) and was expressed using the M9 SeMet growth-medium kit (Medicilon Inc., People’s Republic of China).
Table 1. Details of the constructs used in crystallization trials.
| Protein | Mutation | Plasmid backbone | Affinity tags | Expression cell line |
|---|---|---|---|---|
| XRCC4 | Full length | pACYC184 | C-terminal His6 | BL21 (DE3) |
| XRCC4 | Δ265 | pET-28a | C-terminal His6 | BL21 (DE3) |
| XRCC4 | Δ202 | pACYC184 | None | BL21 (DE3) |
| XRCC4 | Δ157 | pDEST14 | C-terminal His6 | BL21 (DE3) |
| XRCC4 | Δ136 | pDEST14 | None | BL21 (DE3) |
| XRCC4 | Δ202, A60E | pACYC184 | None | BL21 (DE3) |
| XLF | Full length | pET-Duet1 | C-terminal His6 | Rosetta pLysS (DE3) |
| XLF | Δ224 | pDEST14 | C-terminal His6 | Rosetta (DE3) |
| Ligase IV tandem BRCTs | 654–911 | pPRO-EXb | N-terminal His6 | BL21 (DE3) |
2.2. Purification
All constructs of XLF and XRCC4 were purified as described previously. His-tag fusions were not removed from any of the expressed proteins (Andres et al., 2007 ▶; Junop et al., 2000 ▶). XRCC4Δ136 purification differed from that of wild-type XRCC4 as follows: after nickel-affinity purification, XRCC4Δ136 was loaded onto a 5 ml HiTrap Q HP followed by a 5 ml HiTrap SP HP (GE Healthcare, USA), both of which were equilibrated with 20 mM Tris pH 8, 10 mM DTT, 10% glycerol, 1 mM EDTA and 150 mM KCl. XRCC4Δ136 did not bind to either column and was collected in the unbound fraction. Following cation exchange, XRCC4Δ136 was further purified via gel filtration (HiLoad 16/60 Superdex 200; GE Healthcare, USA) using cation-exchange buffer at 200 mM KCl.
2.3. General crystallization
XRCC4 and XLF were mixed in varying ratios: 25:50, 50:100 or 50:50 µM XLF:XRCC4. Crystallization was performed using hanging-drop vapour diffusion, combining 1 µl each of protein and crystallization solutions. Crystallization solutions from commercially available kits were used (Classics I and II, Ammonium Sulfate, PEGs, pH Clear I, Nucleix and JCSG I, II, III and IV Suites from Qiagen, Canada; Index Screen from Hampton Research, USA; Extension, Cryo, Membrane and Low Ionic Screens from Sigma–Aldrich, Canada; Original Screen from Biogenova, USA). Crystallization trials were initially performed with well solutions consisting of 2.5 M ammonium sulfate (800 µl) and incubated at 277, 293 or 303 K. Other well solutions tested included 2.5–4 M ammonium sulfate, 1.5–4 M sodium chloride and 20% PEG 3350. Crystals were further optimized by systematically varying each component of the primary crystallization condition. Additives were included during the optimization of initial crystallization conditions (Opti-Salts screen from Qiagen, USA; Silver Bullets and Additive Screen from Hampton Research, USA).
2.4. Crystallization and diffraction collection
Crystals of XRCC4Δ157–XLFΔ224 grew from a combination of 1 µl XRCC4Δ157 (100 µM) and XLFΔ224 (50 µM) in 20 mM Tris pH 8, 200 mM KCl, 1 mM EDTA, 10 mM DTT and 10% glycerol. The protein solution was combined with 0.8 µl 1.8 M triammonium citrate pH 8 containing varying dilutions of crushed XRCC4Δ157–XLFΔ224 crystals and 0.2 µl each of the additives 0.1 M barium chloride dihydrate and 2.0 M sodium thiocyanate. Hanging drops were initially dehydrated over 2.5 M ammonium sulfate pH 7 at 303 K. After 24 h crystal trays were moved to 293 K and after a further 24 h to 277 K. 4 d later, crystals were further dehydrated over 4 M ammonium sulfate. 5 d later, crystals were soaked in a combination of 1 µl 0.5 mM tantalum bromide and 0.5 µl 60% PEG 8000 for 3 h prior to flash-cooling in liquid nitrogen. Diffraction data were collected on NSLS beamline X25 (Brookhaven, New York, USA) to a resolution of 3 Å, using a wavelength of 1.2536 Å, in a nitrogen stream at 100 K. Data were collected in 30° wedges, with 0.5° oscillation and 2 s exposure per image. Initial scaling and space-group determination were performed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
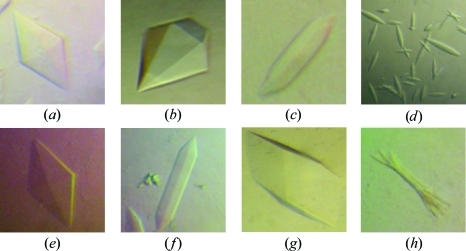
XRCC4–XLF complexes comprised of varying protein lengths produced crystals with similar hexagonal rod morphologies under many crystallization conditions (Table 2 ▶; Fig. 1 ▶). Additives were essential for reproducing the initial crystals. Diffraction was highly dependent on crystal size and was only observed from crystals of >0.4 mm in length. Excess XRCC4 promoted initial crystal growth, while micro-seeding of the crystallization solution and incubation at 303 K controlled the extent of nucleation and increased growth in all three dimensions. Extended incubation at 303 K (>24 h) produced larger crystals but with weaker diffraction (>20 Å). Therefore, the incubation temperature was slowly decreased from 303 to 277 K, greatly improving the resolution to 6–8 Å.
Table 2. Crystals of XRCC4–XLF complexes and associated diffraction limits.
| XRCC4 | XLF | Crystal of complex | Diffraction resolution (Å) |
|---|---|---|---|
| Full-length | Full length | Fig. 1 ▶(a) | >20 |
| Δ265 | Full length | Fig. 1 ▶(b) | ∼8 |
| Δ265 | Δ224 | Fig. 1 ▶(c) | >20 |
| Δ265, A60E + BRCT | Full length | Fig. 1 ▶(d) | >20 |
| Δ202 | Full length | Fig. 1 ▶(e) | >20 |
| Δ202 | Δ224 | Fig. 1 ▶(f) | >20 |
| Δ157 | Δ224 | Fig. 1 ▶(g) | ∼3 |
| Δ136 | Δ224 | Fig. 1 ▶(h) | ∼4 |
Figure 1.
Crystals of XRCC4–XLF complexes (see Table 2 ▶ for details).
Initial diffraction of XRCC4Δ157–XLFΔ224 crystals was limited to 6–8 Å resolution and was further hampered by the presence of ice rings owing to insufficient cryoprotectant. As observed with other protein crystals, dehydration of the XRCC4Δ157–XLFΔ224 crystals improved cryoprotection (Heras et al., 2003 ▶). The extent and duration of dehydration were optimized. Extreme dehydration was achieved by changing the well solution to 4 M ammonium sulfate and by soaking the crystals in 60% PEG 8000. This not only improved cryoprotection, but also increased the diffraction resolution to ∼3 Å. Recently, Hammel et al. (2011 ▶) published a similar XRCC4–XLF structure to 3.9 Å resolution, also using dehydration by PEG 3350 to achieve crystals, with a solvent content identical to that of the XRCC4Δ157–XLFΔ224 crystals discussed here (∼70%). Therefore, this effect may be more successful for crystals of high solvent content.
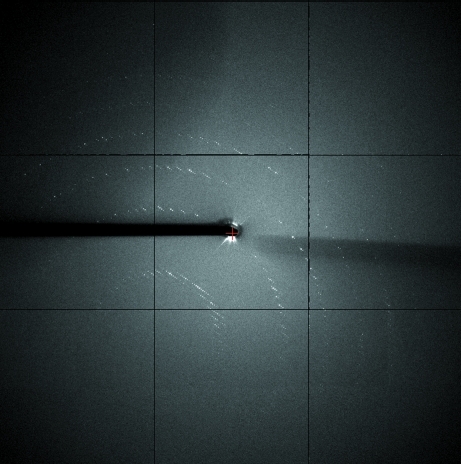
Data were collected on a microfocus beamline (X25 at NSLS), allowing multiple data sets to be collected from different regions of a single crystal. The crystals diffracted to 3 Å resolution; however, owing to anisotropic behaviour, data were only processed to 3.9 Å resolution (Fig. 2 ▶). The crystals belonged to space group C2, with unit-cell parameters a = 745.4, b = 149.6, c = 80.5 Å, β = 94.7° (Table 3 ▶). Alternately, Hammel et al. (2011 ▶) produced crystals that belonged to space group P6522, which may be the result of using a shorter XRCC4 truncation (1–140). However, these crystals also exhibited a long unit-cell axis of 764 Å. The extremely long unit-cell axis accounts for the anisotropy and is the result of limited lateral crystal contacts in the extended repeating unit (the structure will be discussed elsewhere; PDB entry 3rwr; Sheriff & Hendrickson, 1987 ▶).
Figure 2.
Diffraction image of XRCC4Δ157–XLFΔ224 crystals, illustrating the anisotropy.
Table 3. Crystallographic data statistics.
Values in parentheses are for the highest resolution data shell.
| Space group | C2 |
| Wavelength (Å) | 1.25 |
| Unit-cell parameters (Å, °) | a = 745.4, b = 149.6, c = 80.5, α = γ = 90, β = 94.7 |
| Molecules in asymmetric unit | 24 |
| Solvent content (%) | 71 |
| Resolution range (Å) | 50.0–3.9 (4.2–3.9) |
| Unique reflections | 78630 |
| Data multiplicity | 6.7 (6.6) |
| Completeness (%) | 97.6 (98.4) |
| 〈I/σ(I)〉 | 11.7 (2.1) |
| Rmerge (%) | 15.2 (96.3) |
| Mosaicity (°) | 0.35 |
| Wilson scaling B factor (Å2) | 168.55 |
Even though individual structures of XRCC4 (Junop et al., 2000 ▶; PDB entry 1fu1) and XLF (Andres et al., 2007 ▶, PDB entry 2r9a; Li et al., 2008 ▶, PDB entry 2qm4) have been solved, molecular replacement alone was unable to provide sufficient phasing. This may reflect the very large asymmetric unit and the high degree of structural similarity between XRCC4 and XLF. Tantalum bromide has been well documented as a heavy-metal cluster that is suitable for determining the phase of low-resolution structures (Knäblein et al., 1997 ▶; Ban et al., 1999 ▶; Banumathi et al., 2002 ▶; Pomeranz Krummel et al., 2009 ▶). Therefore, we attempted to obtain tantalum bromide derivatives by soaking crystals at varying concentrations and for different lengths of time with and without back-soaking. Soaking for less than 30 min did not produce a useful derivative, while soaking for >12 h and/or back-soaking significantly decreased the diffraction. Only direct soaking of crystals for ∼3 h generated sufficient phasing without a significant loss in resolution.
The combination of low-resolution data, high solvent content and large unit cell required phasing using both molecular replacement and the anomalous signal from tantalum bromide. An initial search model of XRCC4–XLF was generated based upon mutational analysis and docking (Malivert et al., 2010 ▶). Phases from molecular replacement were greatly improved by the additional phasing information from the tantalum bromide clusters (eight sites per asymmetric unit). Phasing from molecular replacement with single-wavelength anomalous diffraction in PHENIX produced an FOM of 0.452 and an LLG of −334 048 (Adams et al., 2010 ▶) and a structural solution was obtained (PDB entry 3rwr).
Acknowledgments
We thank the helpful staff at beamline X25, NSLS, Brookhaven National Laboratory. This work was supported by a Canadian Institute for Health Research grant to MSJ (MOP-89903) and an NSERC–CGS fellowship to SNA.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Ahnesorg, P., Smith, P. & Jackson, S. P. (2006). Cell, 124, 301–313. [DOI] [PubMed]
- Andres, S. N., Modesti, M., Tsai, C. J., Chu, G. & Junop, M. S. (2007). Mol. Cell, 28, 1093–1101. [DOI] [PubMed]
- Ban, N., Nissen, P., Hansen, J., Capel, M., Moore, P. B. & Steitz, T. A. (1999). Nature (London), 400, 841–847. [DOI] [PubMed]
- Banumathi, S., Dauter, M. & Dauter, Z. (2002). Acta Cryst. A58, C81.
- Giaccia, A. J., Denko, N., MacLaren, R., Mirman, D., Waldren, C., Hart, I. & Stamato, T. D. (1990). Am. J. Hum. Genet. 47, 459–469. [PMC free article] [PubMed]
- Hammel, M., Rey, M., Yu, Y., Mani, R. S., Classen, S., Liu, M., Pique, M. E., Fang, S., Mahaney, B., Weinfeld, M., Schreimer, D. C., Lees-Miller, S. P. & Tainer, J. A. (2011). J. Biol. Chem., doi:10.1074/jbc.M111.272641. [DOI] [PMC free article] [PubMed]
- Hammel, M., Yu, Y., Fang, S., Lees-Miller, S. P. & Tainer, J. A. (2010). Structure, 18, 1431–1442. [DOI] [PMC free article] [PubMed]
- Heras, B., Edeling, M. A., Byriel, K. A., Jones, A., Raina, S. & Martin, J. L. (2003). Structure, 11, 139–145. [DOI] [PubMed]
- Junop, M. S., Modesti, M., Guarné, A., Ghirlando, R., Gellert, M. & Yang, W. (2000). EMBO J. 19, 5962–5970. [DOI] [PMC free article] [PubMed]
- Knäblein, J., Neuefeind, T., Schneider, F., Bergner, A., Messerschmidt, A., Löwe, J., Steipe, B. & Huber, R. (1997). J. Mol. Biol. 270, 1–7. [DOI] [PubMed]
- Li, Y., Chirgadze, D. Y., Bolanos-Garcia, V. M., Sibanda, B. L., Davies, O. R., Ahnesorg, P., Jackson, S. P. & Blundell, T. L. (2008). EMBO J. 27, 290–300. [DOI] [PMC free article] [PubMed]
- Lieber, M. R., Gu, J., Lu, H., Shimazaki, N. & Tsai, A. G. (2010). Subcell. Biochem. 50, 279–296. [DOI] [PMC free article] [PubMed]
- Malivert, L., Ropars, V., Nunez, M., Drevet, P., Miron, S., Faure, G., Guerois, R., Mornon, J. P., Revy, P., Charbonnier, J. B., Callebaut, I. & de Villartay, J. P. (2010). J. Biol. Chem. 285, 26475–26483. [DOI] [PMC free article] [PubMed]
- Modesti, M., Hesse, J. E. & Gellert, M. (1999). EMBO J. 18, 2008–2018. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pomeranz Krummel, D. A., Oubridge, C., Leung, A. K. W., Li, J. & Nagai, K. (2009). Nature (London), 458, 475–480. [DOI] [PMC free article] [PubMed]
- Sheriff, S. & Hendrickson, W. A. (1987). Acta Cryst. A43, 118–121.
- Tsai, C. J., Kim, S. A. & Chu, G. (2007). Proc. Natl Acad. Sci. USA, 104, 7851–7856. [DOI] [PMC free article] [PubMed]
- Wu, P.-Y., Frit, P., Meesala, S., Dauvillier, S., Modesti, M., Andres, S. N., Huang, Y., Sekiguchi, J., Calsou, P., Salles, B. & Junop, M. S. (2009). Mol. Cell. Biol. 29, 3163–3172. [DOI] [PMC free article] [PubMed]
- Zha, S., Alt, F. W., Cheng, H.-L., Brush, J. W. & Li, G. (2007). Proc. Natl Acad. Sci. USA, 104, 4518–4523. [DOI] [PMC free article] [PubMed]