The PYD domain of human NALP3 was crystallized. The crystals were found to belong to the primitive monoclinic space group P21, with unit-cell parameters a = 42.03, b = 60.14, c = 51.61 Å, β = 107.40°. The crystals were obtained at 293 K and diffracted to a resolution of 1.7 Å.
Keywords: inflammation, inflammasome, NALP3
Abstract
The NALP3 inflammasome is a macromolecular complex that is responsible for the innate immune response against infection by bacterial and viral pathogens. The NALP3 inflammasome is composed of three protein components: NALP3, ASC and caspase 1. Interaction between NALP3 and ASC via PYD domains is critical for the assembly of the NALP3 inflammasome. In this study, human NALP3 PYD, corresponding to amino acids 3–110, was overexpressed in Escherichia coli using engineered C-terminal His tags. NALP3 PYD was then purified to homogeneity and crystallized at 293 K. Finally, X-ray diffraction data were collected to a resolution of 1.7 Å from a crystal belonging to the primitive monoclinic space group P21, with unit-cell parameters a = 42.03, b = 60.14, c = 51.61 Å, β = 107.40°.
1. Introduction
NALPs are members of a family of proteins that are involved in innate immunity and are known as NOD-like receptors (NLRs; Martinon & Tschopp, 2005 ▶). Through a variety of danger signals, including infection and metabolic dysregulation, NALPs form intracellular multimolecular complexes with ASC and caspase 1 that are known as inflammasomes, creating a platform for the activation of caspase 1 and the promotion of IL-1b maturation (Schroder & Tschopp, 2010 ▶). IL-1b is a well known player in the process of inflammation and fever (Dinarello, 2004 ▶). The human genome contains 14 members of the NALP family, NALP1–NALP14, and it is likely that most of these can form inflammasomes. The inflammasomes function as a platform to recruit caspase 1, providing proximity for self-activation. Caspase 1 activated by the formation of inflammasome activates the inactive inflammatory cytokines pro-interleukin 1β and pro-interleukin 18, leading to NF-kB activation and elicitation of innate immunity.
Currently, four distinct inflammasomes have been identified: NALP1, NALP2, NALP3 and AIM2. Of the NALP inflammasomes, NALP3 has been the most thoroughly studied. This inflammasome is formed by several chemically and structurally diverse triggers that mimic danger signals, including ATP, multiple microbial toxins that disrupt the cell membrane and phagocytosed insoluble crystals (Martinon, 2008 ▶; Kanneganti et al., 2006 ▶; Martinon et al., 2006 ▶). The physiological role of the NALP3 inflammasome has been demonstrated by the observation of a tight association of mutations within the NALP3 gene with various autoinflammatory diseases such as Muckle–Wells syndrome (MWS), familial cold autoinflammatory syndrome (FCAS) and chronic infantile neurological cutaneous and articular syndrome (CINCA) (Church et al., 2006 ▶; Hoffman et al., 2001 ▶; Feldmann et al., 2002 ▶).
NALP3 (NACHT, LRR and PYD domains-containing protein 3), ASC (apoptosis-associated speck-like protein containing a caspase-recruitment domain) and caspase 1 are the three protein components that form the NALP3 inflammasome. The PYD domain-containing NALP3 plays an important role in signal sensing and oligomerization. ASC is a bipartite adaptor protein that contains an N-terminal PYD domain and a C-terminal CARD domain and plays a role in connecting NALP3 and caspase 1. The CARD domain-containing caspase 1 is a prototypical inflammatory caspase that is responsible for maturation of the inflammatory cytokines interleukin 1β and interleukin 18 (Thornberry et al., 1992 ▶; Ghayur et al., 1997 ▶). The assembly of the NALP3 inflammasome depends on the protein-interacting domain known as the death domain superfamily, which is composed of four subfamilies, death domains (DDs), death effector domains (DEDs), caspase-recruitment domains (CARDs) and pyrin domains (PYDs) (Park, 2011 ▶; Jang et al., 2010 ▶). The NALP3 inflammasome is assembled via a PYD–PYD interaction between ASC and NALP3 and a CARD–CARD interaction between ASC and caspase 1 (Park et al., 2007 ▶). Although several PYD structures (ASC2 PYD, PDB entry 2hm2; NALP1 PYD, 1pn5; ASC PYD, 1ucp; NALP7 PYD, 2km6) have been elucidated, limited structural information is available on the NALP3 inflammasome (Hiller et al., 2003 ▶; Pinheiro et al., 2010 ▶; Liepinsh et al., 2003 ▶; Natarajan et al., 2006 ▶; Jang & Park, 2011 ▶).
As a first step towards elucidating the molecular structure of the NALP3 inflammasome and further understanding the homotypic interaction of PYD in the inflammation signalling pathway, we overexpressed, purified and crystallized human NALP3 PYD. Overexpressed NALP3 PYD was solubilized at pH 5.0 and then purified by affinity chromatography followed by gel-filtration chromatography. Initial NALP3 PYD crystals were obtained in a solution consisting of 32% PEG 2000 MME, 0.2 M ammonium sulfate and 0.1 M sodium acetate pH 4.4. The crystals belonged to space group P21, with unit-cell parameters a = 42.03, b = 60.14, c = 51.61 Å, β = 107.40°. The crystals were obtained at 293 K and diffracted to a resolution of 1.7 Å. Details regarding the structure of NALP3 PYD should enable understanding of the mechanism of assembly of the NALP3 inflammasome via the PYD–PYD interaction.
2. Materials and methods
2.1. Expression and purification
To express C-terminally His-tagged protein, human NALP3 PYD corresponding to amino acids 3–110 was amplified by PCR using forward (5′-GGGCATATGATGGCAAGCACCCGCTG-3′) and reverse (5′-GGGCTCGAGGCTGTCTTCCTGGCATATCACAG-3′) primers. The PCR product was then digested with the restriction enzymes NdeI and XhoI (New England Biolabs, USA), after which it was inserted into pOKD vector which had been cut with the same restriction enzymes. The pOKD vector is a home-made vector that was modified from commercially available pET26b (Novagen, USA; Dzivenu et al., 2004 ▶).
The plasmid was transformed into Escherichia coli BL21 (DE3) competent cells and expression was induced by treating the bacteria with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K. Cells expressing NALP3 PYD were pelleted by centrifugation, resuspended in 50 ml lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl and 5 mM imidazole) and lysed by sonication. The lysate was then centrifuged at 16 000 rev min−1 for 1 h at 277 K, after which the supernatant fractions were applied onto a gravity-flow column (Bio-Rad) packed with Ni–NTA affinity resin (Qiagen). The unbound bacterial proteins were subsequently removed from the column using wash buffer (20 mM Tris pH 8.0, 500 mM NaCl, 60 mM imidazole). The C-terminally His-tagged NALP3 PYD was eluted from the column using elution buffer (20 mM Tris buffer pH 8.0, 500 mM NaCl, 250 mM imidazole).
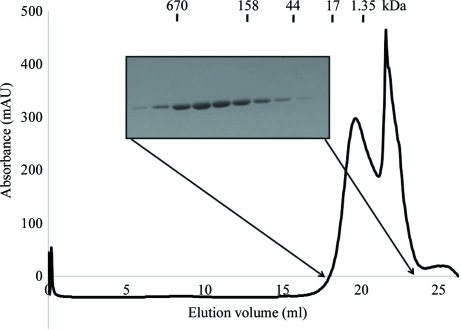
The elution fractions were collected on a 0.5 ml scale to 3 ml. Fractions containing more than 95% homogeneous NALP3 PYD, as indicated by SDS–PAGE, were selected, combined and concentrated to 20–25 mg ml−1 using a concentration kit (Millipore). The concentrated protein was then applied onto a Superose 6 gel-filtration column (GE Healthcare) pre-equilibrated with a solution consisting of 20 mM sodium citrate pH 5.0 and 150 mM NaCl.
NALP3 PYD (molecular weight 12 685 Da), which eluted at around 19 ml, was collected and concentrated to 6–8 mg ml−1. The peak was confirmed to contain NALP3 PYD by SDS–PAGE (Fig. 1 ▶). Purified NALP3 PYD contained the C-terminal extra residues LEHHHHHH, which were not removed. Selenomethionine-substituted NALP3 PYD was expressed in the methionine-auxotrophic strain B834 (Novagen) grown in minimal medium supplemented with seleno-l-methionine (Sigma) and other nutrients. It was purified and crystallized using the same procedure as used for the native protein.
Figure 1.
Purification of human NALP3 PYD: gel-filtration chromatography and SDS–PAGE of human NALP3 PYD.
2.2. Crystallization
The crystallization conditions were initially screened at 293 K by the hanging-drop vapour-diffusion method using screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, Natrix, MembFac, Index, Crystal Screen Cryo and Crystal Screen Lite) and from the deCODE Biostructures Group (Wizard I, II, III and IV). Initial crystals were grown on plates by equilibrating a mixture consisting of 1 µl protein solution (6–8 mg ml−1 protein in 20 mM sodium citrate pH 5.0, 150 mM NaCl) and 1 µl reservoir solution (condition No. 13 of Crystal Screen 2; 0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate pH 4.6, 30% polyethylene glycol monomethyl ether 2000) against 0.4 ml reservoir solution. Crystallization was further optimized by searching over a range of concentrations of protein, PEG MME 2000 and ammonium sulfate. Crystals appeared within 2 d and grew to maximum dimensions of 0.1 × 0.1 × 0.4 mm (Fig. 2 ▶) in the presence of 32% PEG 2000 MME, 0.2 M ammonium sulfate and 0.1 M sodium acetate pH 4.4. The crystals diffracted to a resolution of 1.7 Å.
Figure 2.
Crystals of human NALP3 PYD. The crystals were grown in 2 d in the presence of 32% PEG 2000 MME, 0.2 M ammonium sulfate and 0.1 M sodium acetate pH 4.4. The approximate dimensions of the crystals were 0.1 × 0.1 × 0.4 mm.
2.3. Crystallographic data collection
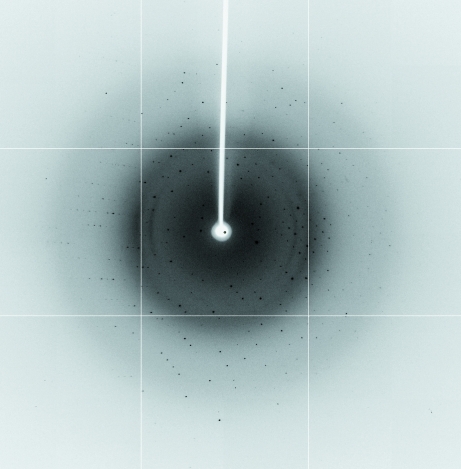
For data collection, the crystals were briefly soaked in a solution corresponding to the reservoir solution supplemented with 35%(v/v) glycerol. The soaked crystals were then frozen in liquid nitrogen. A native diffraction data set and a single-wavelength anomalous diffraction (SAD) data set were collected to 1.7 Å resolution on beamline BL-4A at the Pohang Accelerator Laboratory (PAL), Republic of Korea (Fig. 3 ▶). The SAD data set was collected at the selenium peak wavelength (E = 12 669 eV, λ = 0.979 Å). The data sets were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Selenomethionines were found and the structure was phased using the SOLVE/RESOLVE (Terwilliger, 2004 ▶) or SHELX (Sheldrick, 2008 ▶) programs. The data-collection statistics are summarized in Table 1 ▶.
Figure 3.
Diffraction image (1° oscillation) of a NALP3 PYD crystal with a 1.7 Å resolution limit.
Table 1. Diffraction data statistics of NALP3 PYD crystals.
Values in parentheses are for the highest resolution shell.
| Native | SeMet | |
|---|---|---|
| X-ray source | BL-4A, PAL | BL-4A, PAL |
| Wavelength (Å) | 1.0000 | 0.979 |
| Space group | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 42.03, b = 60.14, c = 51.61, β = 107.40 | a = 42.04, b = 60.21, c = 51.40, β = 107.20 |
| Resolution limits (Å) | 50–1.7 | 30–1.7 |
| No. of observations | 46584 | 100286 |
| No. of unique reflections | 23814 | 26472 |
| Mean I/σ(I) | 46.21 (27.47) | 34.89 (6.72) |
| Completeness (%) | 97.5 (96.0) | 98.3 (97.1) |
| Rmerge | 3.5 (6.6) | 9.2 (30.2) |
3. Results and discussion
NALP3 PYD was purified using two chromatographic steps, His-tag affinity chromatography and gel-filtration chromatography, which produced 98% pure target protein. The generated protein was subsequently analyzed by SDS–PAGE (Fig. 1 ▶). No contaminating bands were visible. The calculated monomeric molecular weight of NALP3 PYD including the C-terminal His tag was 12 685 Da and it eluted at approximately 15 kDa, suggesting that it may exist as a monomer in solution.
The success in crystallizing NALP3 PYD was the result of several factors. Firstly, many different constructs were used, most of which either led to no crystals or poorly shaped crystals that could not be optimized. One of the critical features of NALP3 PYD was that it crystallized and diffracted only when purified under acidic conditions such as pH 5.0.
The crystals belonged to space group P21, with unit-cell parameters a = 42.03, b = 60.14, c = 51.61 Å, β = 107.40°. Assuming the presence of one dimer or two monomers in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.53 Å3 Da−1, corresponding to a solvent content of 51.39% (Matthews, 1968 ▶). Diffraction data statistics are given in Table 1 ▶. The data set was indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The molecular-replacement phasing method was conducted with the CNS program (Brünger et al., 1998 ▶) using the ASC PYD structure (PDB entry 1ucp; Liepinsh et al., 2003 ▶), which has 22% amino-acid sequence identity to NALP3 PYD, as a search model. Since solving the structure of a member of the death domain superfamily using the molecular-replacement method is difficult in our experience, we are also conducting SAD phasing using SOLVE/RESOLVE (Terwilliger, 2004 ▶). Three methionines, not including the N-terminal methinione, will be expected to produce sufficient phasing power to solve the structure.
Acknowledgments
We are grateful to Dr Yeon Gil Kim of BL-4A at the Pohang Accelerator Laboratory. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0003406 and 2011-0025697).
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Church, L. D., Churchman, S. M., Hawkins, P. N. & McDermott, M. F. (2006). Springer Semin. Immunopathol. 27, 494–508. [DOI] [PubMed]
- Dinarello, C. A. (2004). J. Endotoxin Res. 10, 201–222. [DOI] [PubMed]
- Dzivenu, O. K., Park, H. H. & Wu, H. (2004). Protein Expr. Purif. 38, 1–8. [DOI] [PubMed]
- Feldmann, J., Prieur, A. M., Quartier, P., Berquin, P., Certain, S., Cortis, E., Teillac-Hamel, D., Fischer, A. & de Saint Basile, G. (2002). Am. J. Hum. Genet. 71, 198–203. [DOI] [PMC free article] [PubMed]
- Ghayur, T., Banerjee, S., Hugunin, M., Butler, D., Herzog, L., Carter, A., Quintal, L., Sekut, L., Talanian, R., Paskind, M., Wong, W., Kamen, R., Tracey, D. & Allen, H. (1997). Nature (London), 386, 619–623. [DOI] [PubMed]
- Hiller, S., Kohl, A., Fiorito, F., Herrmann, T., Wider, G., Tschopp, J., Grütter, M. G. & Wüthrich, K. (2003). Structure, 11, 1199–1205. [DOI] [PubMed]
- Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A. & Kolodner, R. D. (2001). Nature Genet. 29, 301–305. [DOI] [PMC free article] [PubMed]
- Jang, T.-H. & Park, H. H. (2011). J. Biotechnol. 151, 335–342. [DOI] [PubMed]
- Jang, T.-H., Zheng, C., Wu, H., Jeon, J.-H. & Park, H. H. (2010). Apoptosis, 15, 1444–1452. [DOI] [PMC free article] [PubMed]
- Kanneganti, T.-D., Özören, N., Body-Malapel, M., Am, A., Park, J.-H., Franchi, L., Whitfield, J., Barchet, W., Colonna, M., Vandenabeele, P., Bertin, J., Coyle, A., Grant, E. P., Akira, S. & Núñez, G. (2006). Nature (London), 440, 233–236. [DOI] [PubMed]
- Liepinsh, E., Barbals, R., Dahl, E., Sharipo, A., Staub, E. & Otting, G. (2003). J. Mol. Biol. 332, 1155–1163. [DOI] [PubMed]
- Martinon, F. (2008). J. Leukoc. Biol. 83, 507–511. [DOI] [PubMed]
- Martinon, F., Pétrilli, V., Mayor, A., Tardivel, A. & Tschopp, J. (2006). Nature (London), 440, 237–241. [DOI] [PubMed]
- Martinon, F. & Tschopp, J. (2005). Trends Immunol. 26, 447–454. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Natarajan, A., Ghose, R. & Hill, J. M. (2006). J. Biol. Chem. 281, 31863–31875. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Park, H. H. (2011). Apoptosis, 16, 209–220. [DOI] [PubMed]
- Park, H. H., Lo, Y.-C., Lin, S.-C., Wang, L., Yang, J. K. & Wu, H. (2007). Annu. Rev. Immunol. 25, 561–586. [DOI] [PMC free article] [PubMed]
- Pinheiro, A. S., Proell, M., Eibl, C., Page, R., Schwarzenbacher, R. & Peti, W. (2010). J. Biol. Chem. 285, 27402–27410. [DOI] [PMC free article] [PubMed]
- Schroder, K. & Tschopp, J. (2010). Cell, 140, 821–832. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Terwilliger, T. (2004). J. Synchrotron Rad. 11, 49–52. [DOI] [PubMed]
- Thornberry, N. A. et al. (1992). Nature (London), 356, 768–774. [DOI] [PubMed]