Human transthyretin, a hormone-binding protein of high abundance in blood and cerebrospinal fluid, is intrinsically amyloidogenic. Preliminary results from a neutron diffraction analysis of this protein that may shed light on the mechanism of tetramer dissociation and successive fatal aggregation are presented.
Keywords: transthyretin, TTR, amyloidosis, neutron crystallography, deuteration, perdeuteration
Abstract
Preliminary studies of perdeuterated crystals of human transthyretin (TTR) have been carried out using the LADI-III and D19 diffractometers at the Institut Laue–Langevin in Grenoble. The results demonstrate the feasibility of a full crystallographic analysis to a resolution of 2.0 Å using Laue diffraction and also illustrate the potential of using monochromatic instruments such as D19 for higher resolution studies where larger crystals having smaller unit cells are available. This study will yield important information on hydrogen bonding, amino-acid protonation states and hydration in the protein. Such information will be of general interest for an understanding of the factors that stabilize/destabilize TTR and for the design of ligands that may be used to counter TTR amyloid fibrillogenesis.
1. Introduction
Amyloidosis is a serious disease caused by extracellular deposition of insoluble abnormal fibrils (Pepys, 2006 ▶). Systemic amyloidosis, with deposits in the viscera, blood vessels and connective tissue, is usually fatal, causing about one death per thousand in developed countries. About 25 different unrelated human proteins form amyloid fibrils in vivo. Amyloid is deposited when there is (i) sustained exposure to either normal or increased concentrations of a normal, potentially amyloidogenic, protein; (ii) when an abnormal amyloidogenic protein is produced as a consequence of an acquired disease; or (iii) when a gene mutation encodes an amyloidogenic variant protein. Fibrillogenesis results from reduced stability of the native fold of the fibril precursor protein, which under physiological conditions populates partly unfolded intermediate states that subsequently aggregate as stable fibrils with the pathognomonic amyloid cross-β-sheet core structure (Sunde et al., 1997 ▶).
Wild-type transthyretin (TTR), the normal plasma protein which transports thyroid hormone and retinol-binding protein, is inherently amyloidogenic and forms microscopic amyloid deposits of uncertain clinical significance in all individuals aged over 80 years (Westermark et al., 1990 ▶). Massive deposits in the heart can also occur, causing fatal senile cardiac transthyretin amyloidosis. The inherent amyloidogenicity of wild-type transthyretin is markedly enhanced by most of the reported >100 different point mutations which encode single-residue substitutions in the transthyretin sequence (Connors et al., 2003 ▶). These mutations cause autosomal dominant adult-onset hereditary amyloidosis, a universally fatal condition affecting about 10 000 patients worldwide. The usual clinical presentation is familial amyloid polyneuropathy, with predominant peripheral and autonomic neuropathy, but there is commonly also serious involvement of the heart, kidneys and eyes. Transthyretin amyloidosis predominantly affecting the heart is particularly associated with the Val122Ile variant, which is very rare in Caucasians but is carried by 4% of African Americans: a total of 1.3 million people, including 13 000 individuals homozygous for the mutation (Jacobson et al., 1997 ▶).
There is no effective treatment for transthyretin amyloidosis other than orthotopic liver transplantation (Holmgren et al., 1991 ▶, 1993 ▶), which removes the hepatic source of circulating amyloidogenic variant transthyretin and replaces it with wild-type transthyretin. However, this is available only for a minority of patients and benefits only those with the common TTR Val30Met variant who are treated early in the disease. There is thus a very important unmet medical need for effective therapy.
A promising strategy is the use of small-molecule ligands which are specifically bound by TTR and stabilize its native fold, thereby preventing dissociation and fibrillogenic misfolding of the protomers. This approach, pioneered by Kelly’s group (Connelly et al., 2010 ▶), has produced the drug tafamidis, which is currently in clinical trials. Other compounds have been designed (Kolstoe et al., 2010 ▶) and are being developed in collaboration with GlaxoSmithKline.
Human TTR is a homotetramer of four noncovalently associated nonglycosylated 127-residue protomers, each containing an eight-stranded β-sheet and one short α-helix. The 1.8 Å resolution X-ray crystal structure of the protein was reported in 1978 (Blake et al., 1978 ▶) and there is a large amount of literature on the structure of TTR complexed with a wide range of different ligands. However, now that novel ligands are being designed for therapeutic use, more detailed structural information on the hydration and protonation states, which are pivotal for the stability of the native structure of TTR and its ligand binding, will be invaluable.
2. Materials and methods
2.1. Production of perdeuterated transthyretin and crystallization
Mature human transthyretin contains 127 amino acids [UniProtKB entry P02766(21–147)]. The N-terminal (20-amino-acid) signalling peptide is removed upon secretion (Kanda et al., 1974 ▶). The corresponding cDNA for the gene coding for human transthyretin was cloned into a pET-M11 bacterial expression vector with an N-terminal His6 tag and a TEV cleavage site (EMBL Protein and Purification Facility, Germany) via NcoI and Acc65I restriction sites. For expression, the plasmid was transformed into Escherichia coli BL21 (DE3) cells (Invitrogen). The cells were adapted to perdeuterated Enfors minimal medium and grown in a fed-batch fermentation process at 303 K in 4 l Infors fermenters. 50 g d8-glycerol (99% deuterium; Euriso-top) was added as the only carbon source and yielded 58.54 g wet cell paste.
Wet cell paste was dissolved in 130 ml lysis buffer with the addition of four tablets of EDTA-free complete protease inhibitor cocktail (Roche), 14 µl β-mercaptoethanol and DNaseI. Readily suspended cells were broken by sonication and cleared cell lysate was loaded onto an Ni2+-saturated IMAC HP HiTrap column (GE Healthcare). TEV cleavage was carried out overnight at room temperature. The flowthrough from the Ni2+ IMAC HP HiTrap column was concentrated and loaded onto a Superdex 200 (10/300 GL) gel-filtration column (Amersham). Peak fractions were pooled and concentrated using Amicon Ultra centrifugal filter units (Millipore). The concentrated sample was diluted ten times with perdeuterated buffer and concentrated again to replace hydrogen that had been introduced during the purification process by deuterium. This step was repeated three times. The total yield was 38 mg pure perdeuterated protein. Analysis of the protein by MALDI–TOF mass spectrometry showed deuteration of 100% of all non-exchangeable H atoms.
Large single crystals of ≥3 mm3 (linear dimensions of approximately 2.2 × 1.4 × 1.1 mm) were grown by the vapour-diffusion method in sitting drops on bridges (Hampton Research) in 2.15 M malonate buffer pD 6.0 using a protein concentration of 23 mg ml−1 (Fig. 1 ▶). For buffer preparation, malonic acid [CH2(COOH)2] was dissolved in D2O followed by subsequent removal of the solvent with a rotary evaporator. This step was repeated three times. The malonic acid was adjusted to 2.4 M and sterile filtered. Sodium malonate [CH2(COONa)2] was directly dissolved in D2O and sterile filtered. A layer of an 80:20 mixture of paraffin and silicon oil in the reservoir was used to enhance the quality of the crystals.
Figure 1.
The large crystal of human transthyretin used in this neutron crystallographic study.
2.2. Neutron diffraction
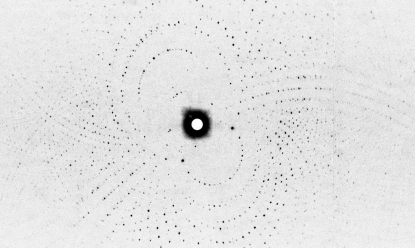
Neutron Laue diffraction data were collected at room temperature using the LADI-III instrument installed on cold neutron guide H142 at the ILL. The LADI-III instrument (Blakeley et al., 2010 ▶) uses a large neutron image-plate detector and exploits the large radius of the drum to improve the signal-to-noise ratio of the recorded reflections and to decrease spatial overlap. An Ni/Ti multi-layer bandpass filter was used to select a restricted neutron wavelength range (Δλ/λ ≃ 25%) centred at 3.75 Å and extending from 3.2 to 4.2 Å. Data were recorded in two passes, each with a ϕ step between successive images (Δϕ) of 7° around the vertical rotation axis of the detector. A series of 12 contiguous Laue images were first collected using an exposure time of 8 h per image in order to collect the high-resolution data and a series of 11 contiguous Laue images were then collected using an exposure time of 2 h per image in order to measure the low-resolution data. Data were processed to 2.0 Å resolution (Fig. 2 ▶). Neutron Laue data were indexed and integrated using the Daresbury Laboratory LAUEGEN software suite (Helliwell et al., 1989 ▶; Campbell, 1995 ▶) modified for the cylindrical geometry of the LADI-III detector (Campbell et al., 1998 ▶). The LSCALE program (Arzt et al., 1999 ▶) was used to derive the wavelength-normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths. No provision was needed for damage and no explicit absorption corrections were applied. SCALA (Winn et al., 2011 ▶) was used to combine and merge the 69 034 observed reflections. Relevant statistics of diffraction data are shown in Table 1 ▶. The multiplicity and completeness figures relevant to the neutron and X-ray data (see §2.3) reflect the different instrument geometries and data-collection regimes used; these issues have been discussed elsewhere (Blakeley, 2009 ▶; Blakeley et al., 2010 ▶).
Figure 2.
Neutron Laue diffraction pattern recorded from the transthyretin crystal using the LADI-III diffractometer at the ILL. The exposure time was 8 h.
Table 1. Data-collection and structure-solution statistics.
Values in parentheses are for the outer shell.
| Data set | Neutron | X-ray |
|---|---|---|
| Beamline | LADI-III, ILL | ID23-1, ESRF |
| Wavelength (Å) | 3.24–4.18 | 0.9791 |
| No. of images | 23 | 180 |
| Oscillation angle (°) | 7 [step angle] | 0.5 |
| Exposure time per image (s) | 18470 [5.13 h per image] | 0.1 [beam attenuated to 0.74%] |
| Space group | P21212 | P21212 |
| Unit-cell parameters (Å) | a = 43.68, b = 86.26, c = 65.72 | a = 43.68, b = 86.26, c = 65.72 |
| Resolution range (Å) | 43.68–2.00 (2.11–2.00) | 52.28–1.85 (1.95–1.85) |
| No. of unique reflections | 13480 (1524) | 21718 (3060) |
| Completeness (%) | 78.3 (62.1) | 99.4 (97.9) |
| Rmerge | 0.182 (0.337) | 0.093 (0.420) |
| Multiplicity | 5.1 (4.0) | 3.53 (3.49) |
| Mean I/σ(I) | 7.3 (3.4) | 7.49 (2.67) |
| Wilson B factor (Å2) | 19.8 |
Monochromatic data were also recorded during a test on the D19 instrument. The wavelength used was 2.42 Å, obtained through the use of a pyrolytic graphite monochromator. A narrow rotation range of 2.9° was recorded over an exposure time of 14 min. No data statistics are shown in Table 1 ▶ for this feasibility test. A typical data slice is shown in Fig. 3 ▶ and extends to essentially the same resolution as that observed during the LADI-III study.
Figure 3.
Monochromatic neutron diffraction pattern recorded from the transthyretin crystal using the D19 diffractometer. The data were recorded over a period of 14 min and cover a rotation range of 2.9°. They extend to a resolution approximately equivalent to that recorded using the LADI-III diffractometer.
2.3. X-ray diffraction
X-ray diffraction data were recorded on beamline ID23-1 (Flot et al., 2010 ▶) at the European Synchrotron Radiation Facility (ESRF) using a heavily attenuated X-ray beam of wavelength 0.9791 Å. Data were recorded using a capillary-mounted sample at room temperature and a MAR 225 CCD-based detector. Integrated and scaled reflection intensities were extracted using MOSFLM (Leslie, 2006 ▶) and SCALA (Winn et al., 2011 ▶) and were converted to structure factors using TRUNCATE (Winn et al., 2011 ▶). Radiation-damage effects were monitored using the inter-frame merging R factors and B factors.
3. Results and discussion
These preliminary crystallographic results on TTR demonstrate the feasibility of a full structural analysis driven by a joint neutron/X-ray target function. The analysis will be carried out with the aid of the PHENIX software package (Adams et al., 2009 ▶, 2010 ▶) optimized for joint refinements (Afonine et al., 2010 ▶) and can be expected to yield detailed information on the molecular structure of TTR, including information relating to amino-acid protonation states and hydration. There is now ample evidence demonstrating that information of this type almost invariably provides new insights into functional aspects of protein structure (Blakeley et al., 2008 ▶; Gardberg et al., 2009 ▶, 2010 ▶; Howard et al., 2011 ▶; Kovalevsky et al., 2010 ▶, 2011 ▶; Leal et al., 2009 ▶, 2010 ▶; Teixeira et al., 2008 ▶, 2010 ▶; Tomanicek et al., 2010 ▶, 2011 ▶). This analysis will be of importance for an understanding of monomer and dimer interactions and the role that structured water plays in these interactions. This information may be valuable in studying the factors underlying the stability of TTR amyloid fibrils as well as the aggregation processes leading to their formation. Neutron diffraction studies of TTR fibrils themselves may also prove to be useful: the first high-angle neutron fibre diffraction studies of an amyloid system have recently been described by Tiggelaar et al. (2011 ▶) and demonstrate the feasibility of using partially ordered samples to probe issues such as hydration and water-driven conformational change. Additionally, this study will provide structural data that may be of value in optimizing small-molecule compounds designed to stabilize the native fold of TTR. This study further demonstrates the feasibility of using the D19 diffractometer at the ILL for crystallographic studies of smaller proteins where large crystals are available and re-emphasises the value of using perdeuterated protein for both Laue and monochromatic macromolecular crystallography.
Acknowledgments
VTF, JBC and SAM acknowledge funding from the EPSRC under grants GR/R47950/01, GR/R99393/01 and EP/C015452/1. VTF also acknowledges support from the EU under contracts RII3-CT-2003-505925 and NMP4-CT-2006-033256. We acknowledge the ILL for the provision of beam time on LADI-III and D19 and the ESRF for beam time on ID23-1. We thank Stuart Fisher for help and useful discussions. We thank Luca Signor from the IBS platform of the Partnership for Structural Biology and the Institut de Biologie Structurale in Grenoble (PSB/IBS) for assistance and for access to the mass-spectrometry facility. We thank Professor M. B. Pepys of University College London for commenting on the manuscript.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Adams, P. D., Mustyakimov, M., Afonine, P. V. & Langan, P. (2009). Acta Cryst. D65, 567–573. [DOI] [PMC free article] [PubMed]
- Afonine, P. V., Mustyakimov, M., Grosse-Kunstleve, R. W., Moriarty, N. W., Langan, P. & Adams, P. D. (2010). Acta Cryst. D66, 1153–1163. [DOI] [PMC free article] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst. 32, 554–562.
- Blake, C. C., Geisow, M. J., Oatley, S. J., Rérat, B. & Rérat, C. (1978). J. Mol. Biol. 121, 339–356. [DOI] [PubMed]
- Blakeley, M. P. (2009). Crystallogr. Rev. 15, 157–218.
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Teixeira, S. C. M., Petit-Haertlein, I., Hazemann, I., Mitschler, A., Haertlein, M., Howard, E. & Podjarny, A. D. (2010). Acta Cryst. D66, 1198–1205. [DOI] [PubMed]
- Campbell, J. W. (1995). J. Appl. Cryst. 28, 228–236.
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst. 31, 496–502.
- Connelly, S., Choi, S., Johnson, S. M., Kelly, J. W. & Wilson, I. A. (2010). Curr. Opin. Struct. Biol. 20, 54–62. [DOI] [PMC free article] [PubMed]
- Connors, L. H., Lim, A., Prokaeva, T., Roskens, V. A. & Costello, C. E. (2003). Amyloid, 10, 160–184. [DOI] [PubMed]
- Flot, D., Mairs, T., Giraud, T., Guijarro, M., Lesourd, M., Rey, V., van Brussel, D., Morawe, C., Borel, C., Hignette, O., Chavanne, J., Nurizzo, D., McSweeney, S. & Mitchell, E. (2010). J. Synchrotron Rad. 17, 107–118. [DOI] [PMC free article] [PubMed]
- Gardberg, A. S., Blakeley, M. P. & Myles, D. A. A. (2009). Acta Cryst. F65, 184–187. [DOI] [PMC free article] [PubMed]
- Gardberg, A. S., Del Castillo, A. R., Weiss, K. L., Meilleur, F., Blakeley, M. P. & Myles, D. A. A. (2010). Acta Cryst. D66, 558–567. [DOI] [PubMed]
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst. 22, 483–497.
- Holmgren, G., Steen, L., Ekstedt, J., Groth, C.-G., Ericzon, B.-G., Eriksson, S., Andersen, O., Karlberg, I., Nordén, G., Nakazato, M., Hawkins, P., Richardson, S. & Pepys, M. (1991). Clin. Genet. 40, 242–246. [DOI] [PubMed]
- Holmgren, G., Steen, L., Suhr, O., Ericzon, B.-G., Groth, C.-G., Andersen, O., Wallin, B. G., Seymour, A., Richardson, S., Hawkins, P. N. & Pepys, M. B. (1993). Lancet, 341, 1113–1116. [DOI] [PubMed]
- Howard, E. I., Blakeley, M. P., Haertlein, M., Petit-Haertlein, I., Mitschler, A., Fisher, S. J., Cousido-Siah, A., Salvay, A. G., Popov, A., Muller-Dieckmann, C., Petrova, T. & Podjarny, A. (2011). J. Mol. Recognit. 24, 724–732. [DOI] [PubMed]
- Jacobson, D. R., Pastore, R. D., Yaghoubian, R., Kane, I., Gallo, G., Buck, F. S. & Buxbaum, J. N. (1997). N. Engl. J. Med. 336, 466–473. [DOI] [PubMed]
- Kanda, Y., Goodman, D. S., Canfield, R. E. & Morgan, F. J. (1974). J. Biol. Chem. 249, 6796–6805. [PubMed]
- Kolstoe, S. E. et al. (2010). Proc. Natl Acad. Sci. USA, 107, 20483–20488.
- Kovalevsky, A. Y., Hanson, L., Fisher, S. Z., Mustyakimov, M., Mason, S. A., Forsyth, V. T., Blakeley, M. P., Keen, D. A., Wagner, T., Carrell, H. L., Katz, A. K., Glusker, J. P. & Langan, P. (2010). Structure, 18, 688–699. [DOI] [PMC free article] [PubMed]
- Kovalevsky, A. Y., Hanson, B. L., Mason, S. A., Yoshida, T., Fisher, S. Z., Mustyakimov, M., Forsyth, V. T., Blakeley, M. P., Keen, D. A. & Langan, P. (2011). Angew. Chem. Int. Ed. Engl. 50, 7520–7523. [DOI] [PubMed]
- Leal, R. M. F., Callow, S., Callow, P., Blakeley, M. P., Cardin, C. J., Denny, W. A., Teixeira, S. C. M., Mitchell, E. P. & Forsyth, V. T. (2010). Acta Cryst. D66, 1244–1248. [DOI] [PubMed]
- Leal, R. M. F., Teixeira, S. C. M., Blakeley, M. P., Mitchell, E. P. & Forsyth, V. T. (2009). Acta Cryst. F65, 232–235. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Pepys, M. B. (2006). Annu. Rev. Med. 57, 223–241. [DOI] [PubMed]
- Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997). J. Mol. Biol. 273, 729–739. [DOI] [PubMed]
- Teixeira, S. C. M., Blakeley, M. P., Leal, R. M. F., Gillespie, S. M., Mitchell, E. P. & Forsyth, V. T. (2010). Acta Cryst. D66, 1139–1143. [DOI] [PubMed]
- Teixeira, S. C. M., Blakeley, M. P., Leal, R. M. F., Mitchell, E. P. & Forsyth, V. T. (2008). Acta Cryst. F64, 378–381. [DOI] [PMC free article] [PubMed]
- Tiggelaar, S. M., Mossou, E., Callow, P., Callow, S., Teixeira, S. C. M., Mitchell, E. P., Mitraki, A. & Forsyth, V. T. (2011). Acta Cryst. F67, 332–335. [DOI] [PMC free article] [PubMed]
- Tomanicek, S. J., Blakeley, M. P., Cooper, J., Chen, Y., Afonine, P. V. & Coates, L. (2010). J. Mol. Biol. 396, 1070–1080. [DOI] [PubMed]
- Tomanicek, S. J., Wang, K. K., Weiss, K. L., Blakeley, M. P., Cooper, J., Chen, Y. & Coates, L. (2011). FEBS Lett. 585, 364–368. [DOI] [PubMed]
- Westermark, P., Sletten, K., Johansson, B. & Cornwell, G. G. III (1990). Proc. Natl Acad. Sci. USA, 87, 2843–2845. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.