Abstract
G-protein α subunits are involved in transmission of signals for development, pathogenicity, and secondary metabolism in plant pathogenic and saprophytic fungi. We cloned two G-protein α subunit genes, tgaA and tgaB, from the biocontrol fungus Trichoderma virens. tgaA belongs to the fungal Gαi class, while tgaB belongs to the class defined by gna-2 of Neurospora crassa. We compared loss-of-function mutants of tgaA and tgaB with the wild type for radial growth, conidiation, germination of conidia, the ability to overgrow colonies of Rhizoctonia solani and Sclerotium rolfsii in confrontation assays, and the ability to colonize the sclerotia of these pathogens in soil. Both mutants grew as well as the wild type, sporulated normally, did not sporulate in the dark, and responded to blue light by forming a conidial ring. The tgaA mutants germinated by straight unbranched germ tubes, while tgaB mutants, like the wild type, germinated by wavy and highly branched germ tubes. In confrontation assays, both tgaA and tgaB mutants and the wild type overgrew, coiled, and lysed the mycelia of R. solani, but tgaA mutants had reduced ability to colonize S. rolfsii colonies. In the soil plate assay, both mutants parasitized the sclerotia of R. solani, but tgaA mutants were unable to parasitize the sclerotia of S. rolfsii. Thus, tgaA is involved in antagonism against S. rolfsii, but neither G protein subunit is involved in antagonism against R. solani. T. virens, which has a wide host range, thus employs a G-protein pathway in a host-specific manner.
Trichoderma spp. are well established as plant disease biocontrol agents that can parasitize many plant-pathogenic fungi (7). Trichoderma virens, an important member of this group, produces several antifungal antibiotics and is a hyperparasite (or mycoparasite) of plant pathogens like Rhizoctonia solani, Sclerotium rolfsii, Sclerotinia sclerotiorum, and Pythium spp. (2, 9, 17). At least one commercial product (SOILGARD) has been formulated with T. virens (13). Mycoparasitism is, essentially, a host-parasite interaction. The interaction begins with recognition of the host or of molecules released by the host by enzymatic action of the mycoparasite. Such signals could be provided by fungal cell wall degradation products released upon contact with or approach to the host (4, 29).
Regardless of the chemical nature of the signals, they are likely to be perceived by conserved eukaryotic signal transduction pathways. Fungal G-protein subunits, mitogen-activated protein kinases (MAPKs), and the components of cyclic AMP (cAMP) signaling are required for virulence of plant pathogens (3, 14, 27). Signaling roles have been clearly defined for a class of fungal G-protein α subunit genes with homology to the mammalian Gi class that includes visual transducin. gna-1 of Neurospora crassa is involved in development and fertility (1). Activation of fadA of Aspergillus down-regulates mycotoxin production and conidiation but stimulates transcription of a gene required for penicillin production (26). The homolog in the rice blast fungus, Magnaporthe grisea, magB, is required for appressorium formation, mating, and virulence on rice (16). Additional genes, with no obvious homology to any particular mammalian Gα class, have been found for several species. In N. crassa, the function of gna-2 overlaps with that of gna-1, but loss of gna-2 alone has no discernible phenotype (1). Loss of the third Gα subunit gene of Neurospora, gna-3, leads to premature, dense conidiation and other developmental phenotypes, some of which are rescued by exogenous cAMP (12). Deletion of the M. grisea homolog of this gene, magA, prevents production of mature asci but has no apparent effect on virulence or development (16). A similar pattern is seen for Botrytis cinerea, for which the Gαi homolog bcg1, but not bcg2, the homolog of Neurospora gna-2 and Magnaporthe magC, is required for virulence (6). A recurring theme that has emerged from the study of fungal signal transduction is that a highly conserved signaling protein can perform a variety of tasks in different species (15).
Antagonistic fungal-fungal interactions, including those that could be applied for biocontrol, may provide novel modes of regulation by these conserved signaling elements. When accumulation of tga1, the Trichoderma atroviride Gαi homolog, was blocked by antisense expression, hyphal extension growth was inhibited and the mutant colonies underwent conidiation profusely (23). This phenotype is similar to that of Neurospora gna-3 (12). Signaling through fadA of Aspergillus also represses conidiation (28). Immunoblot analysis indicated that the amount of Gαi homolog tga1 was decreased in the T. atroviride antisense lines (23), while the level of a second Gα subunit was normal. Nevertheless, antisense technology has some limitations for obtaining loss-of-function mutants. More than one GTP-binding protein gene could be silenced completely or partially by the antisense RNA. We therefore obtained loss-of-function mutants by homologous integration.
We have recently shown (19) that a MAPK homolog of T. virens represses conidiation in the dark, is not involved in hyphal parasitism, and plays a role in the parasitism of sclerotia of R. solani and S. rolfsii. Thus, other members of the G-protein α subunit family might be required for mycoparasitism of different hosts or different stages in the life cycle. The objectives of this study were to isolate G-protein α subunit genes and to investigate their role in development and mycoparasitism. We have addressed, for the first time, the question of host-specific roles of a G-protein pathway in mycoparasitism.
MATERIALS AND METHODS
Fungal strains and culture conditions.
T. virens IMI 304061 was isolated from soil of Pantnagar, India, with S. rolfsii as bait (18), and is a mycoparasite on the sclerotia of S. rolfsii and R. solani and the hyphae of R. solani (17). S. rolfsii was isolated from a ginger rhizome and has been deposited at the Microbial Type Culture Collection of India (20), and R. solani (ITCC 4110) was obtained from the Indian Type Culture Collection, New Delhi, India. The fungi were routinely maintained on potato dextrose agar (PDA; Difco) at room temperature and stored as glycerol (20%) stocks at −80°C for long-term storage.
Genomic and cDNA clones, nucleic acid manipulations, constructs, and transformation.
Degenerate primers were designed based on conserved motifs (oMP19 and oMP20) in the GTP-binding site (25) and were used to amplify a DNA fragment encoding part of the G-protein α subunit to be used as a probe. The template was genomic DNA of the wild-type strain. These partial sequences were used to screen a cDNA library (19). Using the cDNA clones as probes, we screened a cosmid library (19), and cosmid clones with inserts of >45 kb were obtained. tgaA was subcloned in two pieces as an approximately 3.5-kb EcoRV fragment and a 5-kb SacI fragment. tgaB was subcloned as an approximately 5-kb SacI fragment (the 5′ SacI site came from the cosmid vector). Southern and Northern blot analyses were performed according to standard methods (24); hybridization was done in 7% sodium dodecyl sulfate-0.25 M phosphate buffer (pH 7) on a Hybond N+ membrane (Amersham), according to the manufacturer's instructions. PCR products were cloned in pCR-ScriptAmp SK(+) (Stratagene). Genomic DNA and total RNA were isolated from T. virens as described previously (19).
The deletion construct for tgaA was made by replacing the XbaI fragment from the coding region with the selection marker gene for hygromycin resistance (hph under the control of the Aspergillus nidulans trpC promoter and trpC transcription termination signals). The disruption construct for tgaB was made by insertion of the hph gene with the trpC promoter (this cassette lacked the trpC termination signals) at the BglII site inside the coding region. Protoplast transformation was performed as previously described (19); DNA for the transformation was prepared by PCR amplification from the constructs with standard primers T3 and T7.
Growth rate, confrontation assay, coiling, and sclerotial parasitism.
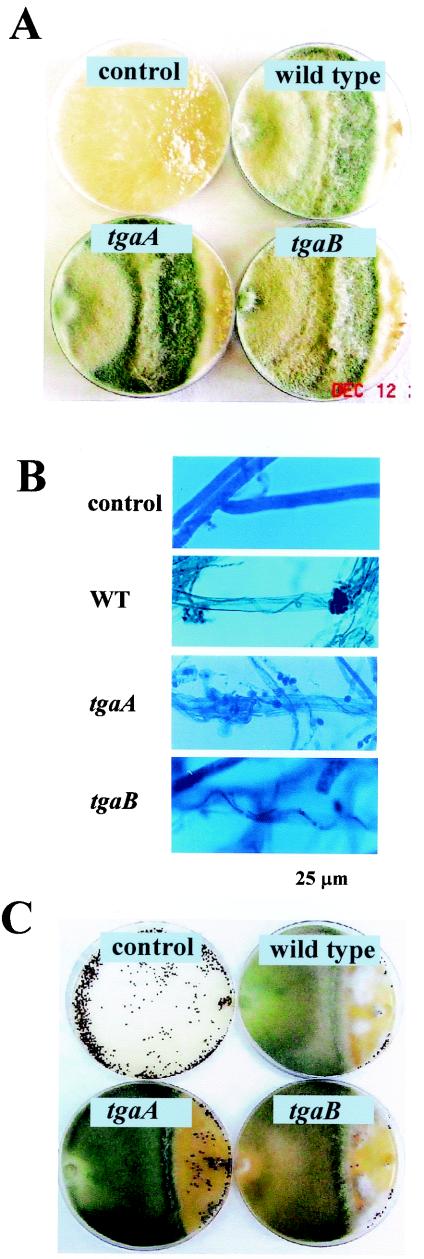
The growth rate of the mutants relative to the wild type was determined by placing a 5-mm-diameter mycelial disk of the fungus in the center of a PDA plate and measuring the colony diameter every 24 h. Confrontation assays to assess the ability of T. virens to overgrow S. rolfsii and R. solani were done as described previously (17). The ability of T. virens strains to parasitize R. solani hyphae was studied after staining of the interacting fungi with cotton blue in lactophenol. The ability of T. virens to parasitize the sclerotia of R. solani and S. rolfsii in soil was assessed by using the soil plate assay, as described previously (17), except that autoclaved soil and 109 Trichoderma conidia were used. Sclerotia of the pathogens were put in the middle of the soil plate after mixing of the conidia with the soil. For monitoring of germination and early development, conidia were suspended in sterile distilled water and incubated on a glass slide in a moist chamber at room temperature. The germinated conidia were stained with cotton blue in lactophenol and photographed after 18 h of incubation.
Photoinduction.
Mycelial colonies were grown and then photoinduced (23). Cultures were inoculated in 9-cm-diameter petri dishes containing 3 ml of PD broth (Difco), at the center of an 8-cm-diameter disk of Whatman no. 3 filter paper. After 36 h of growth, the cultures were exposed to blue light at 480 μmol m−2 and fixed in ethanol 24 h later.
Phylogenetic analysis.
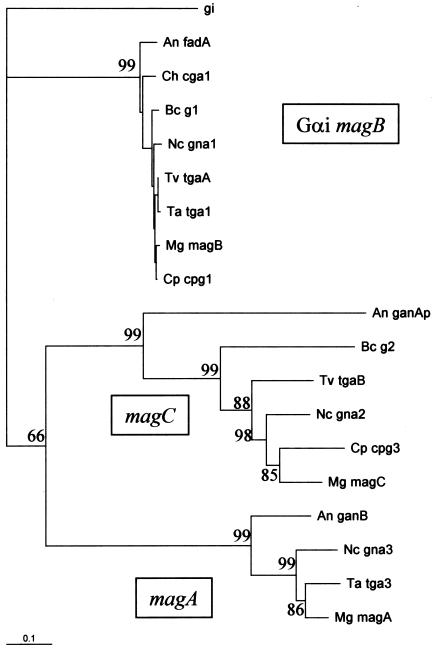
Protein sequences were aligned by use of CLUSTALW 1.7 (http://npsa-pbil.ibcp.fr). A distance matrix was computed from this alignment by PROTDIST from the PHYLIP package (5) (http://bioweb.pasteur.fr). The model used by PROTDIST is empirical and is based on the probabilities of change from one amino acid to another (11). Bootstrapping was used to create 100 data sets. The resulting distance was scaled in units of the expected fraction of amino acids changed. The FITCH program (PHYLIP; Fitch-Margoliash method, with bootstrapping; 100 data sets) was used to generate a phylogenetic tree; a human Gαi was chosen as the outgroup species. One of the possible trees is plotted in Fig. 1, and the percentages indicated on the major branches are derived from a consensus tree generated by the program CONSENSE (PHYLIP package). The tree was plotted with TREEVIEW (22).
FIG. 1.
Phylogenetic relationships of tgaA and tgaB to other G-protein α subunits based on protein sequences. Binomial names are indicated by a two-letter code, followed by the gene name, as follows (the corresponding protein database accession numbers are also indicated): Mg, M. grisea (magB, O13315; magA, AAB65425; magC, AAB65427); Cp, Cryphonectria parasitica (cpg1, Q00580; cpg3, AAM14395); Nc, N. crassa (gna1, QO5425; gna2, Q05424; gna3, Q9HFW7); Ta, T. atroviride (tga1, AAK74191; tga3, AAM69919); Tv, T. virens (tgaA, AAO18659; tgaB, AAN65182); Bc, B. cinerea (G1, CAC19871; G2, CAC19872); Ch, C. heterostrophus (cga1, O74227); An, A. nidulans (fadA, Q00743; ganA, AAD34893; ganB, AAF12813). Gi, a human gene encoding Gαi subunit 2 (accession no. NP_006487). The scale bar indicates 0.1 nucleotide substitutions per site, and the numbers at the forks indicate the number of times the group consisting of the species which are to the right of that fork occurred among the trees, of 100 trees generated from the distance matrix. The groups have not yet been assigned standard names, so they are labeled here, arbitrarily, according to the nomenclature of M. grisea, a plant pathogen for which all three genes have been characterized.
Nucleotide sequence accession numbers.
The sequences for tgaA and tgaB have been deposited under GenBank accession numbers AY186729 (tgaA) and AY168002 (tgaB).
RESULTS
Isolation of tgaA and tgaB.
Using degenerate primers, we obtained two products, of 189 and 235 bp, which showed high levels of sequence similarity to N. crassa gna1 and gna2, respectively. Full-length cDNA clones named pG1204 (1,836 bp, for tgaA) and pG1602 (2,246 bp, for tgaB) were identified after screening of a T. virens cDNA library (19) with the probes. tgaA cDNA had a 273-bp 5′ untranslated region (UTR) and a 501-bp 3′ UTR, while the tgaB cDNA had a 449-bp 5′ UTR and a 732-bp 3′ UTR. tgaA and tgaB code for 353 and 354 amino acids, respectively. tgaA is almost identical to tga1 from T. atroviride (differs by two amino acids), while it differs by nine amino acids from its N. crassa homolog. tgaB, on the other hand, is quite different from its N. crassa homolog (77% amino acid identity). tgaA and tgaB of T. virens have about 52% similarity to each other (183 of 354 amino acids are identical). tgaA belongs to a very highly conserved group of fungal Gα subunits (Gαi group) (Fig. 1). A mammalian Gαi (human Gi2) included in the calculation as an outgroup falls closer to the ascomycete Gi class than to the other ascomycete sequences. tgaB belongs to the second group of fungal Gα subunits defined in Fig. 1, which has been tentatively related to the Gs class based on sequence similarity to Drosophila Gs (8). Comparison of cDNA and genomic sequences showed that tgaA has a long (292 bp) intron in the 5′ UTR and that the coding region is interrupted by three introns, of 100, 78, and 63 bp. tgaB, however, does not have an intron in the 5′ UTR, and the coding region is interrupted by three introns, of 207, 56, and 63 bp. The intron positions are conserved, in part, between each T. virens gene and its close fungal homologs. The locations with respect to the amino acid sequence of all three introns of tgaA are identical to those of N. crassa gna-1, and their lengths are quite similar. All three tgaA introns are conserved in Cochliobolus heterostrophus cgal, but the latter has an additional intron preceding the last, conserved one. Likewise, all three introns of tgaB are found at the same positions in magC of M. grisea, but M. grisea has an additional intron near the 5′ end.
Isolation of TgaA and TgaB loss-of-function mutants.
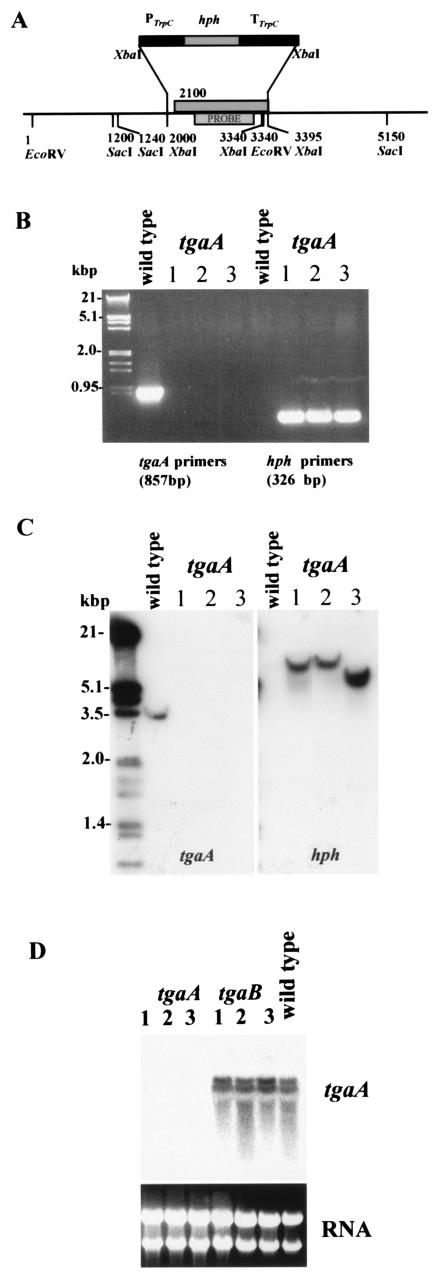
Loss-of-function mutants were obtained by homologous integration through double-crossover recombination (Fig. 2 and 3). After the transformation of protoplasts, we obtained three stable mutants each for tgaA and tgaB. The homologous recombination events were confirmed by both PCR and Southern hybridization (Fig. 2). Primer pairs designed to amplify a fragment internal to the tgaA coding region gave no products when DNA from the homologous recombinants was used as template, while a fragment of the hph gene could be amplified from the same samples (Fig. 2B). A probe corresponding to part of the tgaA coding region gave no signal on a Southern blot of the tgaA deletion mutants (Fig. 2C), while the hygromycin resistance cassette was detected for the mutants but not the wild type (Fig. 2C). As shown in Fig. 2C, two differently sized bands were obtained for the transformants. The reason for this is not clear and would require additional mapping of several more kilobases of sequence downstream from the gene, but it may be related to a rearrangement following integration of the construct. Northern blot analysis detected two transcripts, perhaps differing in size as a result of alternative splicing of the large intron in the 5′ UTR. Both signals were completely absent from all three tgaA mutants (Fig. 2D).
FIG. 2.
Deletion of tgaA by gene replacement. (A) Strategy for double-crossover integration. (B) PCR amplification showing absence of the wild-type band for the tgaA mutant strains GAT6, GAT38, and GAT39 (lanes 1 to 3). The amplification was performed with theprimer pair g12for (GGA AAG TCA ACC ATT CTC AAG) and g12intas (TCAGGATGTAGTCACACGCG) or hphfor (GAGGGCGAAGAATCTCGTGC) and hphrev (CACTGACGGTGTCGTCCATC). (C) Southern blot analysis of transformants using the tgaA probe indicated in panel A or hph as probe. Map positions are labeled, starting with the first EcoRV site. Genomic DNA was digested with EcoRV, and the blot was hybridized with the labeled fragments amplified from the genomic clone (for tgaA) or the transformation vector (for hph), using the primer pairs indicated above. Size markers are from a λ HindIII/EcoRI digest. (D) Northern blot of total RNA, hybridized to a probe generated by PCR with primers g12for and g12intas and the tgaA cDNA clone as template. Lanes 1 to 3, tgaA transformant strains GAT6, GAT38, and GAT39, respectively.
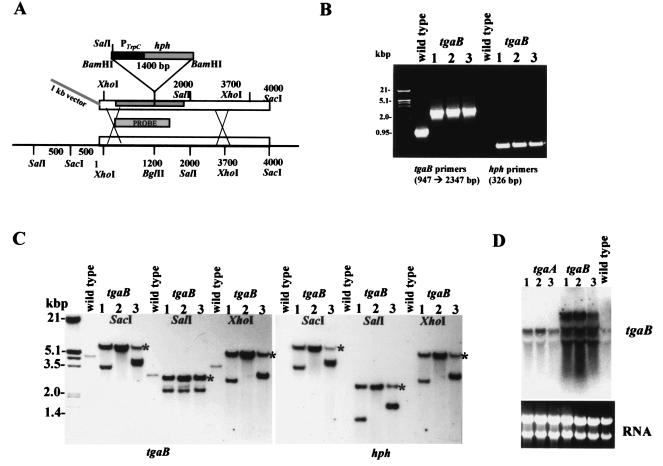
FIG. 3.
Disruption of tgaB by homologous integration of an insertion construct. (A) Strategy for double-crossover integration. (B) PCR amplification showing absence of a wild-type band for the tgaB mutant strains GBT26, GBT59, and GBT60 (lanes 1 to 3). The amplification was performed with the primer pair ga2ndeI (AACGCCCATATGTGCTTCGGCGC) and ga2as (CAACGAGGAAGAGCAGGC) or hphfor and hphrev. (C) Southern blot analysis of transformants, with tgaB and hph as probes. Map positions are labeled, starting with the first SalI site. Genomic DNA was digested with the indicated enzymes, and the blot was hybridized with the labeled fragments amplified from the genomic clone (for tgaB) or the transformation vector, using the primer pairs indicated above. Predicted mutant band sizes (see text) are indicated (*). (D) Northern blot analysis of total RNA with a probe generated by the primer pair ga2ndeI and ga2as, with the tgaB cDNA as template. Lanes 1 to 3, tgaB transformant strains GBT26, GBT59, and GBT60, respectively.
tgaB mutants resulting from double-crossover replacement events have the hygromycin resistance cassette inserted into the BglII site (Fig. 3A). The 947-bp wild-type product was absent from all three mutants and was replaced by a 2.35-kb band, the size expected when the 1.4-kb hygromycin resistance cassette is inserted. The three transformants gave the expected PCR product with primers for the hygromycin resistance cassette (Fig. 3B). These transformants were further analyzed by Southern hybridization (Fig. 3C). The wild-type bands are 3.7, 3.0, and 4.5 kb for XhoI, SalI, and SacI, respectively. For all three transformants, these single bands corresponding to the wild-type tgaB allele in the SacI, SalI, and XhoI digests were missing. Single-crossover integration would preserve one intact wild-type copy, which also would have been detected by PCR. In the XhoI and SacI digests, a new band consistent with the expected increase of 1.4 kb appeared. With SalI, introduction of a new site should produce two new bands, both of 2.2 kb. A signal is indeed observed which is consistent with 2.2 kb. Additional, non-wild-type bands appeared in all three digests, suggesting multiple integration (Fig. 3C). This result is consistent with the high intensity of the signals, since approximately equal amounts of DNA were analyzed. All three transformants have the same phenotype, so the additional copies, if expressed, have no effect on growth, morphology, conidiation, or biocontrol capability. As expected from the properties of the construct (in which the hygromycin resistance cassette has no termination signal) and the results of Southern blot analysis, Northern blot analysis detected four transcripts of different sizes hybridizing to the tgaB probe; none appeared to be identical to the wild-type transcript size. The largest of these was of several kilobases and could span the entire region shown in Fig. 3A, including the tgaB gene and the inserted selection marker. tgaB transcripts were not altered in tgaA mutants (Fig. 3D). The insertion in the tgaB mutants is located in the coding sequence, in a well-conserved domain of the Gα subunit involved in nucleotide binding. Should any stable protein be produced from the transcripts in the tgaB mutants, it is very unlikely that it could function in the finely tuned G-protein heterotrimer. We note, furthermore, that insertions and deletions at cga1 of C. heterostrophus (the homolog of tgaA) resulted in identical phenotypes (8; B.A.H., unpublished data).
Growth, colony morphology, conidiation, and conidial germination.
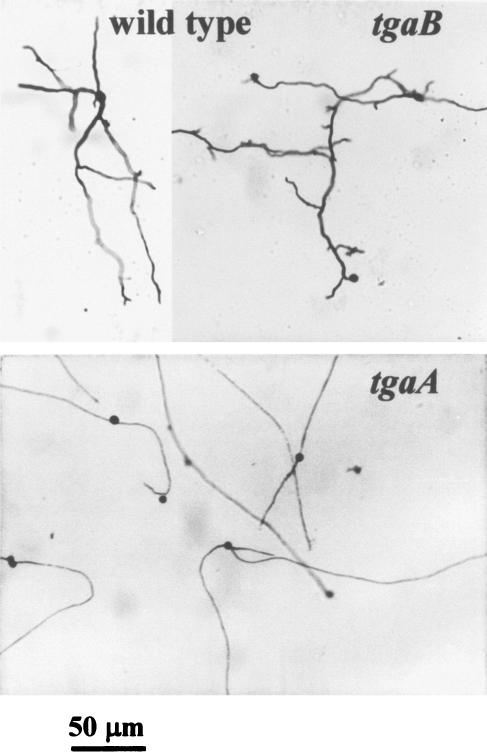
The wild type and the tgaA and tgaB mutants were similar in growth rate and appearance, except that the colonies of tgaA mutants appeared greener than those of the wild type or the tgaB mutants. Neither tgaA nor tgaB mutants underwent conidiation in the dark, and both responded to blue light by forming a typical conidial ring, like the wild type (data not shown). The conidia of the wild type and the tgaB mutants germinated in distilled water by producing typically wavy, highly branched germ tubes, but conidia from tgaA mutants germinated by producing straight unbranched germ tubes (Fig. 4).
FIG. 4.
Germination of conidia of the wild type and the tgaA or tgaB mutant. Note the long, straight, unbranched germ tubes of the tgaA mutant compared to the wavy, highly branched germ tubes of the wild type and the tgaB mutant. Spores were incubated in sterile distilled water and photographed after 18 h.
Antagonism in confrontation assays.
When confronted with R. solani or S. rolfsii, both the tgaA and tgaB loss-of-function mutants could overgrow the R. solani colonies and reached the R. solani inoculum plug in 5 days. In the confrontation assays, the rates of growth of the wild type and all of the mutants were very similar (Fig. 5A). Typical mycoparasitic behavior of T. virens, i.e., attachment, coiling, and lysis of Rhizoctonia hyphae, occurred in the zone of interaction between all of the Trichoderma strains and the test pathogen (Fig. 5B). Lysis of the host is indicated by loss of cotton blue staining (Fig. 5B). In coculture with S. rolfsii, none of the T. virens isolates could grow over S. rolfsii colonies until 9 days, after which profuse growth of the T. virens wild type and the tgaB mutant on the S. rolfsii colony could be seen. The growth of the tgaA mutant was less prolific on the S. rolfsii colony than either the wild type or the tgaB mutant (Fig. 5C).
FIG. 5.
Confrontation assays with T. virens and host plant pathogenic fungi. The host was inoculated at the right-hand side of the plate.(A) Colony interaction between R. solani and T. virens 5 days after inoculation. Control, host only. (B) Wild type and mutants on R. solani hyphae (after 5 days of inoculation). Note the lysis of host hyphae from confrontation plates, indicated by the absence of stain. Control, R. solani hyphae only. (C) Growth of T. virens in confrontation plates with S. rolfsii.
Parasitism of sclerotia.
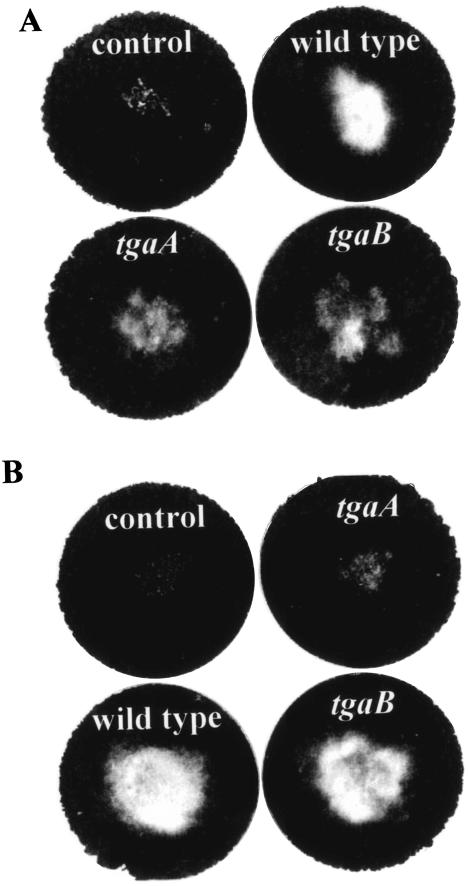
When the sclerotia of R. solani were placed on soil seeded with conidia of the T. virens wild type or the tgaA or tgaB mutants, all the isolates began colonizing the sclerotia within 2 days, and profuse growth of T. virens on the sclerotia could be seen after 6 days (Fig. 6). Colonization of the sclerotia by the tgaA mutant appeared to be slightly delayed, but this delay is difficult to quantitate in our assay system. In contrast, S. rolfsii sclerotia, although readily colonized by the wild type and the tgaB mutants in 5 days, were resistant to colonization by the tgaA mutants. The tgaA mutants failed to parasitize the sclerotia of S. rolfsii, as revealed by the integrity of the sclerotia and the ability of the colonized sclerotia to germinate on PDA amended with 10 mg of benomyl per liter (to selectively inhibit Trichoderma, while allowing the S. rolfsii sclerotia to germinate).
FIG. 6.
Parasitism of sclerotia by T. virens. The sclerotia were put in the middle of a soil plate after mixing of conidia into the soil. Control plates, sclerotia only, with no Trichoderma conidia. Plates were photographed after 7 days of incubation. Mycelial growth is visible as a light-colored colony contrasting with the dark background of the soil. (A) Growth on R. solani sclerotia. (B) Growth on S. rolfsii sclerotia.
DISCUSSION
Trichoderma spp. respond to external stimuli during growth, sporulation, and mycoparasitism. Growth, conidiation, and mycoparasitism are all important attributes contributing to the process of development of these organisms as biofungicides. The role of signal transduction proteins (especially MAPK and G proteins) in transmission of external stimuli has been studied in detail in genetic model fungi, e.g., Neurospora and Aspergillus, and in a variety of plant and human pathogens. Earlier biochemical and genetic studies with T. atroviride provided evidence for host-related signaling and for the importance of a G-protein α subunit, tga1, in the parasitism of R. solani hyphae (10, 21, 23). Loss of many (3, 14, 27), but not all (8), signaling functions drastically impairs virulence in plant-pathogen interactions. The gray mold Botrytis has a wide host range. Different signaling pathways might be required to detect and attack different hosts. The disruption of genes encoding a MAPK (bmp1) and a Gαi protein (bcg1), however, affected virulence against host plants belonging to different dicot families (6, 30), so there is no evidence for host-specific use of signaling pathways in this pathogen. Mycoparasitism, being a fungal-fungal interaction, provides a different type of host-pathogen interaction. Trichoderma species are effective against a wide range of soilborne plant pathogens, from oomycetes to ascomycetes (7). These hosts probably differ in their surface properties and may provide chemically distinct signals to the advancing mycoparasite. A MAPK of T. virens, tmkA, was not involved in hyphal parasitism of R. solani, but was essential for the parasitism of sclerotia of this fungus and of S. rolfsii (19).
We cloned two G-protein α subunits from T. virens. tgaA or tgaB loss-of-function mutants had normal growth and conidiation, did not undergo conidiation in the dark, and were mycoparasitic on R. solani hyphae. In contrast, T. atroviride mutants in which the level of tga1 (the homolog of tgaA) (Fig. 1) was greatly reduced through antisense technology grew very slowly, sporulated in the dark, and were deficient in hyphal parasitism (23). Loss of the homologs of tgaA from two plant pathogens, C. heterostrophus and M. grisea, resulted in abnormally straight growth of germ tubes that appear to branch less (8, 16), which is similar to the ΔtgaA phenotype (Fig. 4). Cochliobolus and Magnaporthe have large multicellular conidia that attach to the host and other surfaces. The observation of a similar phenotype for Trichoderma, which has small unicellular conidia that do not attach tightly to the substrate, suggests that the Gαi class has a general role in controlling hyphal growth pattern. No phenotypes related to tgaB were detected in this study. Disruption or deletion of the homologs of tgaB in most other fungi, likewise, conferred no major phenotypic defects (for examples, see references 6 and 16).
The MAPK (tmkA) loss-of-function mutants of T. virens, in contrast to mutants of tgaA, had near normal growth but underwent conidiation in the dark. They also had normal mycoparasitic behavior on R. solani hyphae. The MAPK loss-of-function mutants, however, were less effective in parasitization of the sclerotia of R. solani and could not parasitize the sclerotia of S. rolfsii (19). The role of tga1 of T. atroviride in the parasitism of sclerotia is not yet known. The present study clearly establishes the involvement of tgaA-mediated pathways in the parasitism of sclerotia of S. rolfsii, but not of R. solani. tgaA and its homolog in T. atroviride, which belongs to the same genus, apparently have different functions.
T. virens thus employs both a MAPK pathway (19) and a G-protein pathway (this study) in a host-specific manner. The basic structural differences between the sclerotia of S. rolfsii and R. solani might explain the differential mycoparasitic behavior against S. rolfsii and R. solani sclerotia. The sclerotia of S. rolfsii are highly compact and well-differentiated structures (comprised of cortex, medulla, and rind), while those of R. solani are aggregates of a rather loose mass of mycelia (17). Since R. solani hyphae are readily parasitized by tmkA, tgaA, and tgaB mutants, these signal transduction mutants are also capable of colonizing (though at a reduced level in the case of tmkA mutants) the sclerotia. On the other hand, since the sclerotia of S. rolfsii are highly melanized specialized structures that are difficult to penetrate, some specific enzyme(s) is probably required for the penetration and degradation of these structures. A gene(s) for this enzyme could be the downstream target of the tmkA and tgaA pathways. The sclerotia are important survival structures of these pathogens, and their destruction in soil is of utmost importance for obtaining effective biocontrol of these highly damaging plant pathogens with a very broad host range. Therefore, in addition to tmkA, tgaA could also be a target for gene manipulation (overexpression and/or expression of the constitutively active allele) of T. virens for improved biocontrol potential. Furthermore, identification of the genes that are regulated downstream of the signaling pathways should identify the enzymes that degrade the host fungi and the transcriptional regulators that act between the signal transducers and their ultimate downstream targets.
Acknowledgments
P.K.M. thanks the Department of Science and Technology, Government of India, for a BOYSCAST Visiting Scientist Fellowship (1999-2000), during which this work was initiated. This study was supported in part by grant 233/00-2 from the ISF (Israel Academy of Sciences) and grants from the Department of Biotechnology, Government of India, and the Israel Ministry of Sciences, in the form of a joint Indo-Israel research project.
We thank S. F. D’Souza, Head, Nuclear Agriculture and Biotechnology Division, Bhabha Atomic Research Centre, Mumbai, India, for encouragement and support.
REFERENCES
- 1.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 147:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek, J. M., C. R. Howell, and C. M. Kenerley. 1999. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr. Genet. 35:41-50. [DOI] [PubMed] [Google Scholar]
- 3.Bölker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fung. Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 4.Cortés, C., A. Gutierrez, V. Olmedo, J. Inbar, I. Chet, and A. Herrera-Estrella. 1998. The expression of genes involved in parasitism by Trichoderma is triggered by a diffusible factor. Mol. Gen. Genet. 260:218-225. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1993. PHYLIP (phylogeny inference package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- 6.Gronover, C. S., D. Kasulke, P. Tudzynski, and B. Tudzynski. 2001. The role of G protein α subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 14:1293-1302. [DOI] [PubMed] [Google Scholar]
- 7.Herrera-Estrella, A., and I. Chet. 1998. Biocontrol of bacteria and phytopathogenic fungi, p. 263-282. In A. Altman (ed.), Agricultural bio/technology. Marcel Dekker, Inc., New York, N.Y.
- 8.Horwitz, B. A., A. Sharon., S.-W. Lu, V. Ritter, T. Sandrock, B. G. Turgeon, and O. C. Yoder. 1999. A G protein α subunit gene of Cochliobolus heterostrophus involved in mating and appressorium formation. Fung. Genet. Biol. 26:19-32. [DOI] [PubMed] [Google Scholar]
- 9.Howell, C., R. Stipanovic, and R. Lumsden. 1993. Antibiotic production by strains of Gliocladium virens and its relation to biocontrol of cotton seedling diseases. Biocontrol Sci. Technol. 3:435-441. [Google Scholar]
- 10.Inbar, J., and I. Chet. 1992. Biomimics of fungal cell-cell recognition by use of lectin-coated nylon fibers. J. Bacteriol. 174:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 12.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch, E. 1999. Evaluation of commercial products for microbial control of soil-borne plant pathogens. Crop Prot. 18:119-125. [Google Scholar]
- 14.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96:13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, S., and R. Dean. 1997. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee, P. K., A. N. Mukhopadhyay, D. K. Sarmah, and S. M. Shreshtha. 1995. Comparative antagonistic properties of Gliocladium virens and Trichoderma harzianum on Sclerotium rolfsii and Rhizoctonia solani—its relevance to understanding the mechanisms of biocontrol. J. Phytopathol. 143:275-279. [Google Scholar]
- 18.Mukherjee, P. K., S. M. Shreshtha, and A. N. Mukhopadhyay. 1993. Baiting with Sclerotium rolfsii for selective isolation of Gliocladium virens from natural soil. Biocontrol Sci. Technol. 3:101-104. [Google Scholar]
- 19.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. tmkA, a MAP kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee, P. K., P. Thomas, and K. Raghu. 1995. Shelf-life enhancement of fresh ginger rhizomes at ambient temperatures by combination of gamma-irradiation, biocontrol and closed polyethylene bag storage. Ann. Appl. Biol. 127:375-384. [Google Scholar]
- 21.Omero, C., J. Inbar, V. Rocha-Ramírez, A. Herrera-Estrella, I. Chet, and B. A. Horwitz. 1999. G protein activators and cAMP promote mycoparasitic behaviour in Trichoderma harzianum. Mycol. Res. 103:1637-1642. [Google Scholar]
- 22.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 23.Rocha-Ramírez, V., C. Omero, I. Chet, B. A. Horwitz, and A. Herrera-Estrella. 2002. Trichoderma atroviride G-protein α-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 1:594-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Strathmann, M., and M. I. Simon. 1990. G protein diversity: a distinct class of α subunits is present in vertebrates and invertebrates. Proc. Natl. Acad. Sci. USA 87:9113-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tag, A., J. Hicks, G. Garifullina, C. Ake, Jr., T. D. Phillips, M. Beremand, and N. Keller. 2000. G-protein signaling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38:658-665. [DOI] [PubMed] [Google Scholar]
- 27.Xu, J.-R. 2000. MAP kinases in fungal pathogens. Fung. Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 28.Yu, J. H., S. Rosen, and T. H. Adams. 1999. Extragenic suppressors of loss-of-function mutations in the Aspergillus flbA regulator of G-protein signaling domain protein. Genetics 151:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeilinger, S., C. Galhaup, K. Payer, S. L. Woo, R. L. Mach, C. Fekete, M. Lorito, and C. P. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fung. Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, L., M. Campbell, J. Murphy, S. Lam, and J.-R. Xu. 2000. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13:724-732. [DOI] [PubMed] [Google Scholar]