Abstract
Trehalose (α-d-glucopyranosyl-1,1-α-d-glucopyranoside), a disaccharide widespread among microbes and lower invertebrates, is generally believed to be nonexistent in higher plants. However, the recent discovery of Arabidopsis genes whose products are involved in trehalose synthesis has renewed interest in the possibility of a function of trehalose in higher plants. We previously showed that trehalase, the enzyme that degrades trehalose, is present in nodules of soybean (Glycine max [L.] Merr.), and we characterized the enzyme as an apoplastic glycoprotein. Here we describe the purification of this trehalase to homogeneity and the cloning of a full-length cDNA encoding this enzyme, named GMTRE1 (G. max trehalase 1). The amino acid sequence derived from the open reading frame of GMTRE1 shows strong homology to known trehalases from bacteria, fungi, and animals. GMTRE1 is a single-copy gene and is expressed at a low but constant level in many tissues.
Trehalose (α-d-glucopyranosyl-1,1-α-d-glucopyranoside), a nonreducing disaccharide, is found in diverse organisms such as bacteria, fungi, and insects (Elbein, 1974). It is commonly considered a storage compound but more recently has been recognized to function mainly as a protectant for maintaining vital structures in the cytosol under stressful conditions such as extreme temperatures, drought, and desiccation (Wiemken, 1990; Ribeiro et al., 1997; Crowe et al., 1998). It might do so by stabilizing membranes and protecting enzymes (Crowe et al., 1984; Hottiger et al., 1994; Iwahasi et al., 1995). To date, trehalose has not been found in vascular plants, except for the two well-documented cases of the resurrection plant Selaginella lepidophylla and Myrothamnus flabellifolia. However, trehalase, the enzyme that specifically hydrolyzes trehalose, is widespread among higher plants and is found in multiple tissues, despite the apparent lack of its substrate (for review, see Müller et al., 1995a).
Higher plants, whether or not they produce trehalose, live together with a variety of microorganisms producing trehalose during most of their life span, entering mutual or antagonistic symbioses. Trehalose has been shown to be present in plants during interactions with various microorganisms, including antagonistic, endomycorrhizal, and ectomycorrhizal fungi and nitrogen-fixing bacteria (for review, see Müller et al., 1995a). Legume nodules have a much higher trehalase activity than normal roots (Müller et al., 1994). We previously partially purified and characterized trehalase from soybean (Glycine max L.) nodules as a 54-kD glycoprotein with both broad pH and high temperature optima (Müller et al., 1992). The same type of enzyme was also found in sterile soybean cell and tissue cultures, demonstrating that it is a plant enzyme.
Previously, we considered trehalase mainly in the context of plant symbioses (Müller et al., 1995a). However, recently, we and others (Blázquez et al., 1998; Vogel et al., 1998) showed that higher plants potentially have the capacity to synthesize trehalose. By functional complementation of yeast mutants that are devoid of one of the two trehalose-synthesizing enzymes, homologous activities for trehalose-6-P synthases and trehalose-6-P phosphatases from Arabidopsis were identified (Blázquez et al., 1998; Vogel et al., 1998). Therefore, higher plants may be capable of metabolizing trehalose, as highlighted recently (Goddijn and Smeekens, 1998). The apparent lack of accumulation of trehalose might be due to active trehalases that rapidly hydrolyze any synthesized trehalose.
Understanding the role trehalase plays in trehalose metabolism is imperative when trying to analyze the potential regulatory role of trehalose or its metabolites, both in symbiosis and in the plant's own development. We therefore decided to purify soybean trehalase and to clone its cDNA.
MATERIALS AND METHODS
Molecular Biology Techniques
If not otherwise mentioned, standard molecular biology techniques were performed according to the methods of Ausubel et al. (1992) and Sambrook et al. (1989).
Plant Material
Nodules were harvested from soybean (Glycine max [L.] Merr.) infected with Bradyrhizobium japonicum and grown in a field in Oberwil, Switzerland. For auxin-induction experiments, the soybean seeds were sterilized and axenically grown as described by Müller et al. (1995b). For nodulation studies, soybean plants were grown in Leonard jars and infected with B. japonicum 61-A-101 (Müller et al., 1992). Pseudonodules were obtained by cultivating soybean plants in the presence of nodulation (Nod) factors isolated from B. japonicum 61-A-101, as described previously (Staehelin et al., 1994).
Southern Analysis
DNA used for Southern analysis and total RNA for reverse transcription were prepared from soybean seedlings grown under sterile conditions. DNA was prepared using the Nucleon-Phytopure DNA isolation kit (Scotlab Limited, Lanarkshire, UK) with two additional CHCl3 extractions in the procedure. Total DNA (10 μg) was digested with appropriate restriction enzymes and separated in a 1% agarose gel. In the lane for undigested DNA only 5 μg of total DNA was loaded. The DNA was transferred to nylon membranes (Boehringer Mannheim) using the Southern technique. Hybridization and washings were done under moderate stringency conditions (40°C hybridization temperature; washings in 0.1× SSC and 0.2% SDS at 60°C). The probe was a fragment that had been amplified by PCR in the presence of digoxigenin-labeled deoxynucleotide triphosphates, and hybridization was done in DIG Easy Hyb buffer (Boehringer Mannheim). This 1.2-kb fragment was synthesized using the primers o221 (5′-TTCGAAATCGCTGTCAATTATG-3′) and o194 (5′GAACCTCCTCACATGTACTG3′), covering position 115 to position 1328 of the GMTRE1 cDNA. Detection of the signal was done using disodium 3-(4-methoxyspiro[1,2-dioxyetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan]-4-yl)phenyl phosphate substrate (Boehringer Mannheim). A chemiluminescense screen on a phosphor imager (Bio-Rad) was used for detection of the signal.
Enzyme Assays and Purification
Trehalase was assayed at pH 6.3 and soluble proteins were determined as previously described (Müller et al., 1995b). Nodule trehalase was extracted as described by Müller et al. (1992). For ion-exchange chromatography on DEAE-Trisacryl (IBF Biotechnics, Villeneuve-la-Garenne, France), the column (100 mL, 5 cm in diameter) was equilibrated with Bis-Tris (Cl−) buffer (50 mm, pH 6.5). Trehalase was eluted with 100 mm NaCl in the same buffer. Affinity chromatography on concanavalin A-Sepharose and gel filtration on Superose 12 were performed as previously described (Müller et al., 1992). For affinity chromatography on hydroxyapatite, purified hydroxyapatite (Bio-Rad) was equilibrated with Bis-Tris (Cl−) buffer (20 mm, pH 6.5). Trehalase was eluted using the same buffer containing 80 mm potassium phosphate. The final ion-exchange chromatography was performed on Hi-Trap Q (Pharmacia) equilibrated as described for the DEAE-Trisacryl column. The protein used for the subsequent steps was eluted with 120 mm NaCl.
Protein and DNA Sequencing
The purified protein (usually 10–18 μg) was separated on a PAGE gel (Laemmli et al., 1970), electroblotted to a PVDF membrane (Immobilon, Millipore), and stained with Coomassie blue R. The trehalase band was cut out and cleaved with trypsin. Tryptic peptides were separated by reverse-phased HPLC and sequenced by automated Edman degradation (Sprenger et al., 1995). Degenerate oligonucleotides of about 20 bases were synthesized at a degeneracy of no more than 256-fold. Total RNA was extracted from soybean nodules and roots treated with auxin (1 μm IAA for 4 d) using the RNeasy kit (Qiagen, Basel, Switzerland). The RNA in this preparation was reverse transcribed using a kit (Boehringer Mannheim) and an oligo(dT) primer. PCR amplification was done using pairs of one forward and one reverse degenerate primer on this first-strand cDNA.
A specific fragment of about 500 bp was obtained using the primers o128 5′-GARRAYGARTTYTGGAAYTC-3′and o130 5′-GCRAANACRTTYTGRTTYTG-3′, derived originally from the peptides EYEFWNSDIHK and XEXQNQNVFAXNF. In the succesful amplifications we used an excess of primers (100 μm) and 40 cycles. This specific 500-bp fragment was identified in nodules as well as in auxin-treated root cDNA. Both fragments were sequenced and found to be identical. DNA sequencing was done on an automatic sequencer (Perkin-Elmer). Specific primers were synthesized and used to amplify both the 5′ and 3′ ends of the cDNA by RACE experiments (Frohman et al., 1988). We used oligo(dT) primers to synthesize the cDNA for the RACE reactions and tailed the 5′ end with terminal transferase using deoxyguanidine triphosphates, as described previously (Aeschbacher et al., 1995). PCR reactions were then done on a fraction of the cDNA using internal primers and a synthetic oligonucleotide that hybridizes to deoxyguanosine stretches. The 5′ end of the cDNA was reached after three successive rounds of 5′ RACE. Finally, primers were used that map to the very 5′ end of the cDNA and to a position immediately 5′ to the poly(A+) tail. Parallel PCR amplifications were performed with these primers using a mixture of Pfu DNA polymerase (Stratagene) and Taq polymerase I (Pharmacia; ratio 5:1). One cDNA of 2.3 kb and, using a more internal primer, a cDNA of 2.2 kb were amplified that were homologous in the 2.2-kb overlapping part. For positions 1 to 46 of the GMTRE1 cDNA only sequences obtained through RACE-PCR are available.
Sequence Analysis and Comparison
Sequence analysis was done using Genetics Computer Group (Madison, WI) software. The homology comparisons were done with version 9.0 of the program “PileUp” using the default conditions (gap creation penalty, 12; gap extension penalty, 4).
RT-PCR
Tissue material and total RNA were prepared and cDNA was synthesized as described above. cDNA preparations (1 μL) were used per PCR reaction in a total volume of 30 μL, and 36 cycles were performed. For amplification of actin, 32 cycles were performed. Actin showed an identical expression pattern when cycled for 27 cycles only, although the amounts obtained were lower. Primers used for the amplification were designed to have similar annealing temperatures and to span at least one intron to be able to distinguish the amplified cDNA from any potential genomic DNA contaminants. Primers and cDNA fragment sizes and accession numbers are: GMTRE1, primer o221 5′-TTCGAAATCGCTGTCAATTATG-3′ and o205 5′-GGTGGTTCACCTTGGGCAAGAA-3′(344 bp; this publication); Nod26, o11 5′-CAATCCTGCTGTCACCATTG-3′ and primer o12 5′-CACTCTTGGTAGTCTCACTC -3′ (494 bp; accession no. X04782); and Actin, o222 5′-GTTCTCTCCTTGTATGCAAGTG-3′ and o223 5′-CCAGACTCATCATATTCACCTTTAG-3′ (683 bp; accession no. V00450).
RESULTS
Trehalase Activity in Nodules and Pseudonodules
We measured trehalase activity in soybean nodules and pseudonodules. As shown previously (Müller et al., 1992), a strong trehalase activity was found in nodules colonized by B. japonicum 61-A-101 (442 ± 51 μkat g−1 protein). A similar high activity (353 ± 85 μkat g−1 protein) was found in ineffective pseudonodules (fix−) obtained by cultivating soybean plants in the presence of Nod factors isolated from B. japonicum 61-A-101. The activity in nodules was 1.3-fold higher than in pseudonodules.
Cloning of the Soybean Trehalase cDNA
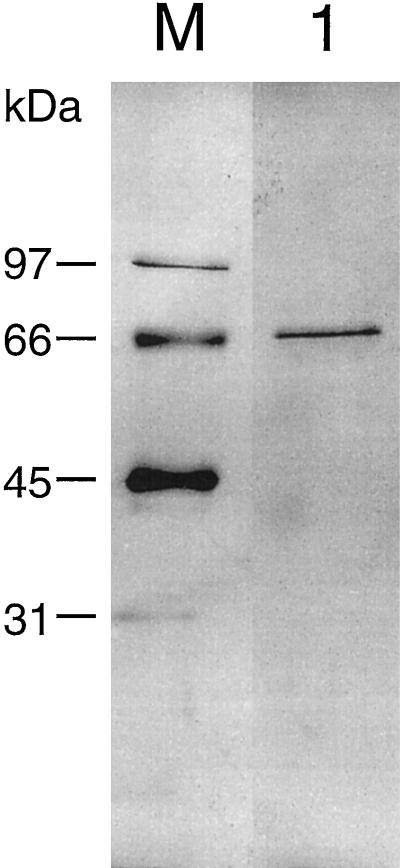
Since a strong trehalase activity was found in soybean nodules, we decided to purify this activity and to clone the trehalase cDNA on the basis of partial amino acid sequences by RT-PCR. We purified the nodules' trehalase activity using an optimized purification scheme that is based on a partial purification of trehalase reported earlier (Müller et al., 1992). By including a hydroxylapatite-affinity chromatography step and a Hi-Trap Q ion-exchange chromatography step, we were able to purify the trehalase 2600-fold (Table I). With this level of purity, a single band was visible on silver-stained SDS gels at about 66 kD (Fig. 1). The band was excised from the gel and further processed by tryptic digestion. The obtained peptides were sequenced and several internal peptide sequences were obtained. Degenerate primers corresponding to these peptides were used for PCR amplifications of reverse-transcribed RNA from nodules. A cDNA fragment of 500 bp was identified that contained a single open reading frame and encoded additional peptides of the purified trehalase protein. The complete cDNA sequence was obtained by RACE-PCR (Frohman et al., 1988). To reduce the probability of errors in the cDNA sequence, we reamplified the complete coding region in two independent PCR reactions using primers that lie outside of the open reading frame. The sequences of both cDNAs were identical and we concluded that the identified cDNA sequence is authentic.
Table I.
Purification procedure of nodule trehalase from soybean
| Step | Total Protein | Trehalase Activity | Specific Trehalase Activity | Purification Factor |
|---|---|---|---|---|
| mg | nkat | nkat mg−1 protein | ||
| 1. Crude nodule extract (pH 3.7) | 5050 | 9682 | 1.917 | 1 |
| 2. Ion-exchange chromatography DEAE- Trisacryl (pH 6.5, 100 mm NaCl) | 575 | 7786 | 13.54 | 7 |
| 3. Concanavalin A-affinity chromatography | 47 | 4776 | 101.6 | 53 |
| 4. Gel filtration (Superose 12) | 3 | 1647 | 549.0 | 286 |
| 5. Hydroxylapatite-affinity chromatography | 0.55 | 661 | 1201 | 626 |
| 6. Ion-exchange chromatography (Hi-Trap Q, pH 6.5, 120 mm NaCl) | 0.018 | 90 | 5000 | 2608 |
The flow chart shows the optimized procedure for trehalase purification. As starting material, 888 g of soybean nodules was extracted as described by Müller et al. (1992). The individual steps of the purification procedure are given with the amount of total protein, total trehalase activity, specific trehalase activity, and the purification factor obtained at that step.
Figure 1.
Purified trehalase from soybean nodules. One hundred nanograms of the the final elution (120 mm NaCl) from the Hi-Trap Q column step were separated on a 10% SDS-Laemmli gel and stained with silver. Sizes of the molecular mass markers are indicated in kD (lane M). The marker and the purified protein ran on the same gel, but an intervening lane was omitted.
The complete cDNA is 2123 nucleotides long and contains an open reading frame from nucleotide 134 to nucleotide 1805. Upstream and downstream of this open reading frame are multiple stop codons in all three frames. There are two small open readings frames upstream of the putative initiation codon, starting at nucleotides 7 and 38 of the cDNA. However, these ATG codons are unlikely to be used as initiation codons, as predicted by initiation codon-prediction programs (Genetics Computer Group), and both are followed in-frame by stop codons after three, and in the second case five, amino acids. If recognized as start codons, only short peptides would be expressed from these ATG codons. The gene encoding this cDNA was designated GMTRE1, for Glycine max trehalase 1 gene.
Protein Characteristics
The sequences identified from the major peptide fractions of the purified protein fraction are 100% homologous to sequences in GMTRE1 (underlined residues in the GMTRE1 sequence in Fig. 2). However, an Asn is encoded instead of a Ser at position 288 of the GMTRE1 protein. Since this position is a potential N-glycosylation site (Fitchette-Lainé et al., 1998), it is likely that this site is glycosylated in the mature protein, which resulted in a misinterpretation during the automated Edman degradation (see Methods).
Figure 2.
Comparison of GMTRE1 with other trehalases. Trehalases from several organisms were screened for homologies (see Methods). The trehalases used were: GMTRE1 (this publication); ATTRE, Arabidopsis trehalase isolog T19F06.15 (accession no. AC002343); Ecoptre: E. coli periplasmic trehalase precursor TREA ECOLI (accession no. P13428); Ecoctre: E. coli probable cytoplasmic trehalase TREF ECOLI (accession no. P37196); Humtre: trehalase [Homo sapiens] (accession no. AB000824); Rabtre: trehalase precursor from rabbit: TREA RABIT (accession no. P19813); Tentre: trehalase precursor Tenebrio molitor; TREA TENMO (accession no. P32359); Bomtre: trehalase precursor Bombyx mori TREA BOMMO) (accession no. P32358); and Celtre: trehalase isolog from C. elegans (accession no. AF039713). Four additional trehalase isologs from C. elegans also show strong homologies to GMTRE1 but are not shown here. The underlined residues in GMTRE1 indicate the identified peptide sequences from the purified soybean nodule trehalase.
The deduced protein sequence of GMTRE1 showed that the protein is closely related to other trehalases. The predicted GMTRE1 protein is 557 amino acids long and thus has the same length as the recently identified trehalase isolog T19F06.15 from Arabidopsis (accession no. AC002343). The predicted molecular mass of GMTRE1 is 56 kD.
GMTRE1 is most homologous to the Arabidopsis trehalase isolog, with 59% of the amino acids identical in total (Fig. 2). It is worth noting that soybean trehalase is more similar to trehalases in Escherichia coli, human, rabbit, insects, and Caenorhabditis elegans than to those from the yeast Saccharomyces cerevisiae and other fungi. Therefore, homologies are fewer and mainly restricted to four small blocks of conserved residues that start at position 195 (PGSRFREVYYWDSY), position 261 (RSQPP), and positions 586 (GGGGEY) and 596 (GFGW) of the comparison presented in Figure 2. Overall homologies are small, however, they extend along the entire length of the proteins with blocks of amino acids, as well as single amino acids (e.g. D at position 76, W at position 288, or G at position 411), shared among the various trehalases.
Southern Analysis of GMTRE1
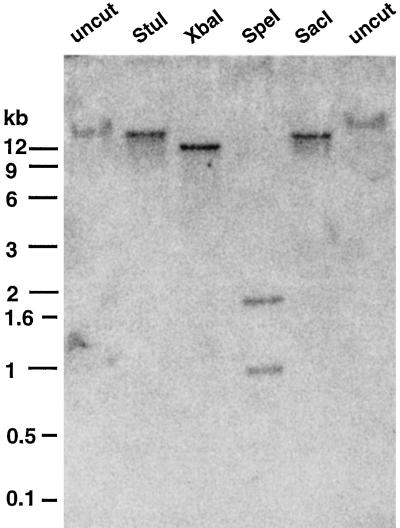
The genomic organization of GMTRE1 was analyzed by Southern hybridization. In undigested DNA from plant material grown under sterile conditions, the probe for GMTRE1 hybridized to the high Mr genomic DNA (Fig. 3). In DNA digested individually with the restriction enzymes StuI, SacI, and XbaI, single fragments of 20, 18, and 13 kb, respectively, were detected. Digestion with the restriction enzyme SpeI yielded two hybridizing fragments of 1.0 and 1.9 kb. This is most likely due to a restriction site for SpeI within an intron in GMTRE1. Since the hybridization was done under moderate-stringency conditions, we concluded that there are no genes closely related to GMTRE1 present in soybean and that GMTRE1 is a single-copy gene in soybean.
Figure 3.
Southern analysis of GMTRE1. The sizes of DNA molecular size markers are indicated. uncut, Undigested genomic DNA. Lanes labeled with StuI, XbaI, SpeI, and SacI indicate the complete digests of genomic DNA using the corresponding restriction enzymes.
Expression of Trehalase
By RNA-blot analysis, we were unable to detect any signals from GMTRE1 transcripts in total RNA prepared from roots, leaves, flowers, or nodules, indicating that GMTRE1 is expressed at a low level in soybean. We therefore analyzed the expression by RT-PCR. To design primers for RT-PCR reactions, we amplified genomic DNA using sequence-specific primers of GMTRE1. In that way we were able to identify a 144-bp-long intron that maps to position 136/137 of the cDNA, immediately following the start codon. For the RT-PCR experiments, we used primers encompassing this intron to be able to distinguish the amplified cDNA from any potential genomic DNA contamination.
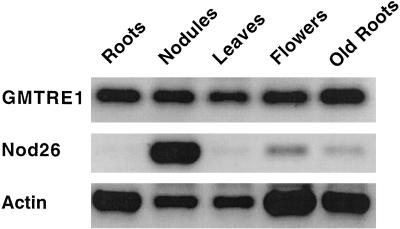
Figure 4 shows that GMTRE1 is expressed in several tissues at a similar level. Expression could be detected in nodules, leaves, flowers, and roots. However, the transcript of GMTRE1 did not appear to be induced in nodules. Nod26, which was used as a control for nodulation, was strongly induced in nodules. Actin was used as a constitutive control and showed a similar expression in all of the tissues tested, with perhaps a smaller level of expression in nodules. We therefore conclude that GMTRE1 is expressed in multiple tissues at a low but constant level.
Figure 4.
Expression analysis of GMTRE1 in different soybean tissues. The following tissues of soybean were used: roots from 4-d-old plants (Roots), mature nodules (Nodules), mature leaves (Leaves), flowers (Flowers), and roots from 3-month-old plants (Old Roots). cDNA fragments of GMTRE1, Nod26, and Actin were amplified by RT-PCR, as described in Methods. Ten microliters of the amplified reactions were resolved by agarose gel electrophoresis and analyzed under UV light. The sizes of the amplified PCR correspond to the expected size of amplified cDNA products.
DISCUSSION
Purification of the Soybean Nodule Trehalase and Cloning of Its cDNA
To date, trehalases in soybean have been identified in nodules, calli, cell cultures, roots, leaves, and flowers (Müller et al., 1995a). This parallels trehalase activity in other leguminosae and in other higher plants, in which trehalase in nonsymbiotic organs was found in pollen, cell cultures, roots, and leaves (Müller et al., 1992). Here we show that ineffective pseudonodules also express trehalase at a level similar to nodule tissue. This clearly demonstrates that the expressed trehalase is of soybean origin.
We have purified a soybean nodule trehalase 2600-fold to apparent homogeneity, using an improved purification scheme including a hydroxyapatite-affinity column and a Hi-Trap Q ion-exchange chromatography step. Microsequence analysis of the purified protein resulted in several peptide sequences derived from the nodule trehalase. Based on these sequences, we designed degenerate primers and were able to clone the cDNA encoding the GMTRE1 gene by performing RT-PCR experiments. GMTRE1 is a single-copy gene as shown by the Southern analysis, which did not reveal any closely homologous sequences.
GMTRE1 Encodes the Soybean Nodule Trehalase
The analysis of the cDNA sequence shows that the ATG at position 134 is the initiation codon for the open reading frame. The two ATGs present at positions 7 and 38 terminate in multiple stop codons and therefore would give rise to only short open reading frames (if any). Short open reading frames in the 5′-untranslated sequence of plant genes are well known in plants (Aeschbacher et al., 1991, 1995), but their function is unclear. GMTRE1 has the same predicted number of amino acids as the trehalase isolog T19F06.15 from Arabidopsis and both show homologies in the N terminus, again indicating that the ATG134 is used as start codon.
GMTRE1 is most likely glycosylated. This is indicated by the identification of a Ser at a potential gylcosylation site at position 288 during microsequencing, instead of the Asn predicted by the cDNA sequence. This is likely to be an artifact of microsequencing, in which the glycosyl residue attached to the Asn resulted in a misinterpretation of the signal obtained in this cycle. Glycosylation of GMTRE1 is also highly likely because we were able to purify this protein with a concanavalin A column, a selective adsorbent for glycoproteins.
Glycosylation of GMTRE1 might explain the fact that we observed three isoforms of nodule trehalase in IEF gels (data not shown) and a molecular mass of GMTRE1 in SDS-PAGE gels (66 kD) that is bigger than the predicted mass of 56 kD. In soybean suspension-cultured cells, we found >80% of the total trehalase activity in the medium, suggesting that the trehalase activity is secreted (data not shown).
GMTRE1 is strongly homologous to trehalases from diverse organisms, including bacteria, insects, and mammals. These proteins have only a 30% to 40% identity, but they share conserved blocks dispersed over the entire protein. These blocks are also found in GMTRE1, and since we have cloned its cDNA based on a highly purified trehalase activity, we conclude that the cloned GMTRE1 cDNA is indeed encoding the purified functional nodule trehalase.
Expression of the GMTRE1 Gene
GMTRE1 is expressed at a low constitutive level in several tissues, including roots and nodules, as well as phototrophic tissue such as leaves and flowers. Although trehalase is about 10-fold more active in nodules than in roots (Müller et al., 1992), induction of GMTRE1 expression in nodules compared with roots was not observed at the RNA level. Since the actin probe used as a constitutive control showed a lower expression in nodules, a low level of induction might still exist. However, such fine levels of induction would have to be tested using a quantitative RT-PCR method. Whether GMTRE1 expression might be posttranscriptionally regulated awaits further examination.
The GMTRE1 expression pattern parallels the expression of the recently identified Arabidopsis trehalase isolog T19F06.15 (accession no. AC002343), in which we have found, using RT-PCR, a low but constitutive level of expression in various tissues as well (data not shown).
Trehalose Metabolism and Sugar Sensing
Production of trehalose in plants is of interest since trehalose might be exploited as a stress-protective agent in vivo (Goddijn et al., 1995). Therefore, attempts have been made to produce trehalose in higher plants. Tobacco and potato were transformed with trehalose-6-P synthase (otsA) and trehalose-6-P phosphatase (otsB) genes from E. coli (Goddijn et al., 1997). However, the transgenic plants accumulated trehalose only in very low amounts, if at all, and transgenic potatoes accumulated low amounts of trehalose in microtubers only when cultured on validamycin A, a strong inhibitor of trehalases. On the other hand, plants transformed with trehalose-6-P synthase exhibited strong alterations in growth (Goddijn et al., 1997). Strong pleiotropic growth phenotypes were also observed in tobacco plants transformed with trehalose-6-P synthase from yeast (Romero et al., 1997).
The causes of these growth alterations are unclear but the results point toward a regulatory role for trehalose or its metabolic intermediates in plant sugar sensing and/or in plant development (Goodijn and Smeekens, 1998). Exogenous application of trehalose to soybean roots affects Suc synthase and invertase activities (Müller et al., 1998). Whether microorganisms might produce and secrete trehalose to alter the carbohydrate allocation of the plant to their favor is, however, still unknown. Such a mechanism could supersede the regulation of the plant's endogenous carbohydrate partitioning and make it independent of environmental conditions such as light intensity and nutrient status. Since the soybean trehalase is expressed in multiple tissues at a low level and is secreted, it might have the function to protect the plant from exposure to exogenous microbial trehalose.
Thus far, we have been unable to identify endogenous trehalose production in soybean plants. However, the recent findings that higher plants may potentially produce trehalose and that even minute amounts of endogenously produced trehalose may have drastic effects on the long-term development of the plant (Goddijn and Smeekens, 1998) indicate a possible endogenous function of trehalose and its degrading enzyme, trehalase. Whether trehalose or its metabolic intermediates are regulators involved in sugar sensing, or whether they are simply toxic to plants and thus have to be removed by trehalases, awaits further analysis.
ACKNOWLEDGMENTS
We thank our colleagues Dr. Z.-P. Xie, Dr. C. Staehelin, Dr. K.-H. Bortlik, Dr. N. Sprenger, and Monica Alt (Botanisches Institut, Universität Basel, Switzerland) for their help with nodule harvest. We also thank the following members of the institute: Dr. G. Vogel for collaboration, Dr. M. Lüscher for helpful discussions concerning protein purification, N. Bürckert for initial help with sequencing, and Dr. J. Oetiker and Dr. I. Sanders for critical reading of the manuscript.
Abbreviations:
- RACE
rapid amplification of cDNA ends
- RT
reverse transcriptase
Note Added in Proof
A protein sequence from potato exhibiting a high homology to GMTRE1 was published in the EMBL database while this manuscript was in preparation (accession no. A52426).
Footnotes
This work was supported by grants from the Swiss National Science Foundation (no. 3100-042535.94 to A.W. and no. 3100-040837.94 to T.B.).
LITERATURE CITED
- Aeschbacher RA, Hauser MT, Feldman K, Benfey PN. The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 1995;9:330–340. doi: 10.1101/gad.9.3.330. [DOI] [PubMed] [Google Scholar]
- Aeschbacher RA, Schrott M, Potrykus I, Saul MW. Isolation and molecular characterization of PosF21, an Arabidopsis thaliana gene which shows characteristics of a b-Zip class transcription factor. Plant J. 1991;1:303–316. [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1992) Short Protocols in Molecular Biology. Greene Publishing Associates and John Wiley & Sons, New York
- Blázquez MA, Santos E, Flores C-L, Martínez-Zapater JM, Salinas J, Gancedo C. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 1998;13:685–690. doi: 10.1046/j.1365-313x.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM. The role of vitrification in anhydrobiosis. Annu Rev Physiol. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms. The role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- Elbein AD. The metabolism of α,α-trehalose. Adv Carbohydr Chem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- Fitchette-Lainé A-C, Denmai L-A, Lerouge P, Faye L (1998) Analysis of N- and O-glycosylation of plant proteins. In C Cunningham, AJR Porter, eds, Recombinant Proteins from Plants: Production and Isolation of Clinically Useful Compounds. Humana Press, Totowa, NJ, pp 271–290
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJM, Smeekens S. Mini-review: sensing trehalose biosynthesis in plants. Plant J. 1998;14:143–146. doi: 10.1046/j.1365-313x.1998.00140.x. [DOI] [PubMed] [Google Scholar]
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen R, de Graaf P, van Dun K, De Laat A, Van den Elzen P, Damm B, Pen J. Transgenic tobacco plants as a model system for the production of trehalose. Plant Physiol. 1995;99:1443–1448. [Google Scholar]
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen RWHH, de Graaf PTHM, Poels J, van Dun K, Ponstein AS, Damm BJP. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol. 1997;113:181–190. doi: 10.1104/pp.113.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger T, De Virgilio C, Hall MN, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur J Biochem. 1994;219:187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- Iwahashi H, Obuchi K, Fujii S, Komatsu Y. The correlative evidence suggesting that trehalose stabilizes membrane structure in the yeast Saccharomyces cerevisiae. Cell Mol Biol. 1995;41:763–769. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Müller J, Boller T, Wiemken A. Trehalose and trehalase in plants: recent developments. Plant Sci. 1995a;112:1–9. [Google Scholar]
- Müller J, Boller T, Wiemken A. Effects of validamycin A, a potent trehalase inhibitor, and phytohormones on trehalose metabolism in roots and nodules of soybean and cowpea. Planta. 1995b;197:362–368. [Google Scholar]
- Müller J, Boller T, Wiemken A. Trehalose affects sucrose synthase and invertase activities in soybean (Glycine max [L.] Merr.) roots. J Plant Physiol. 1998;153:255–257. [Google Scholar]
- Müller J, Staehelin C, Mellor RB, Boller T, Wiemken A. Partial purification and characterization of trehalase from soybean nodules. J Plant Physiol. 1992;140:8–13. [Google Scholar]
- Müller J, Xie Z-P, Staehelin C, Mellor RB, Boller T, Wiemken A. Trehalose and trehalase in root nodules from various legumes. Physiol Plant. 1994;90:86–92. [Google Scholar]
- Ribeiro MJS, Reinders A, Boller T, Wiemken A, De Virgilio C. Mol Microbiol. 1997;25:571–581. doi: 10.1046/j.1365-2958.1997.4961856.x. [DOI] [PubMed] [Google Scholar]
- Romero C, Bellés JM, Vayá JL, Serrano R, Culiáñez-Macià FA. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta. 1997;201:293–297. doi: 10.1007/s004250050069. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sprenger N, Bortlik K, Brandt A, Boller T, Wiemken A. Purification, cloning, and functional expression of sucrose:fructan 6-fructosyltransferase, a key enzyme of fructan synthesis in barley. Proc Natl Acad Sci USA. 1995;92:11652–11656. doi: 10.1073/pnas.92.25.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C, Granado J, Müller J, Wiemken A, Mellor RB, Felix G, Regenass M, Broughton WJ, Boller T. Perception of Rhizobium nodulation factors by tomato cells and inactivation by root chitinases. Proc Natl Acad Sci USA. 1994;91:2196–2200. doi: 10.1073/pnas.91.6.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Aeschbacher RA, Müller J, Boller T, Wiemken A. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J. 1998;13:673–683. doi: 10.1046/j.1365-313x.1998.00064.x. [DOI] [PubMed] [Google Scholar]
- Wiemken A. Trehalose in yeast, stress protectant rather than reserve carbohydrate. J Gen Mol Microbiol. 1990;58:209–217. doi: 10.1007/BF00548935. [DOI] [PubMed] [Google Scholar]