Abstract
Background
Trichinella spiralis is a parasite with unique features. It is a multicellular organism but with an intracellular parasitization and development stage. T. spiralis is the helminthic pathogen that causes zoonotic trichinellosis and afflicts more than 10 million people worldwide, whereas the parasite's biology, especially the developmental regulation is largely unknown. In other organisms, small non-coding RNAs, such as microRNAs (miRNA) and small interfering RNAs (siRNA) execute post-transcriptional regulation by translational repression or mRNA degradation, and a large number of miRNAs have been identified in diverse species. In T. spiralis, the profile of small non-coding RNAs and their function remains poorly understood.
Methodology and Principal Findings
Here, the transcriptional profiles of miRNA and siRNA in three developmental stages of T. spiralis in the rat host were investigated, and compared by high-throughput cDNA sequencing technique (“RNA-seq”). 5,443,641 unique sequence tags were obtained. Of these, 21 represented conserved miRNAs related to 13 previously identified metazoan miRNA families and 213 were novel miRNAs so far unique to T. spiralis. Some of these miRNAs exhibited stage-specific expression. Expression of miRNAs was confirmed in three stages of the life cycle by qRT-PCR and northern blot analysis. In addition, endogenous siRNAs (endo-siRNAs) were found mainly derived from natural antisense transcripts (NAT) and transposable elements (TE) in the parasite.
Conclusions and Significance
We provide evidence for the presence of miRNAs and endo-siRNAs in T. spiralis. The miRNAs accounted for the major proportion of the small regulatory RNA population of T. spiralis, while fewer endogenous siRNAs were found. The finding of stage-specific expression patterns of the miRNAs in different developmental stages of T. spiralis suggests that miRNAs may play important roles in parasite development. Our data provide a basis for further understanding of the molecular regulation and functional evolution of miRNAs in parasitic nematodes.
Introduction
Parasites of the genus Trichinella are a group of pathogens with diverse biological and pathological features. Phylogenetic analysis of the mitochondrial DNA of the species identified so far indicated that there are two genetic clades that form unique monophyletic lineages [1]. One clade is represented by T. pseudospiralis, which does not capsulate in the muscle cells, while the other clade is represented by T. spiralis while does encapsulate in muscle cells, named nurse cells, with a parasite inside the cell, surrounded by a sick layer of collagen. Parasites of Trichinella genus are unique intracellular pathogens. Interestingly, their entire life cycle can be completed within an animal, and trichinellosis in human and other mammals was caused through the ingestion of parasite-contaminated meat. This is a typical zoonotic disease that affects more than 10 million people worldwide [2].
After being ingested with the infected muscle tissue, L1 larvae are released and activated in the small intestine, enter the epithelial layer and undergo four times of moultings before maturation into adult worms. Mating is initiated on day 2 after infection and newborn larvae are released by the females into the mucosa as early as 4 days post infection (dpi) [3]. The larvae migrate through the lymphatic and blood vessels, invade striated muscle cells and develop into the infective Ll stage over a period of 2–3 weeks to the next host which complete the life cycle [4]. Thus, unlike other nematodes, T. spiralis does not have an embryonic developmental stage in the egg, which differs markedly in biological and molecular characteristics from other nematodes, especially the well-characterized free-living worm Caenorhabditis elegans. Intriguingly, compared to other nematodes, T. spiralis has a much smaller genome with 64 Mb in nuclear DNA, which contains ≈15,808 genes [5]. The availability of genome sequence information has made it possible to dissect parasite biology.
Small non-coding RNAs (sncRNAs) are a large group of small endogenous RNAs that have been widely identified in animals [6], plants [7], fungi [8], [9] and some viruses [10]. They are generally 21–23 nucleotides in length, which guide various processes involving sequence-specific silencing through chromatin modification, mRNA degradation, and translational repression [11]–[13]. Based on their origins, structures, associated proteins and biological roles, sncRNAs are divided into three general categories: microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), and piwi-interacting RNAs (piRNAs) [14]. MiRNA and endo-siRNAs have been discovered in diverse animals and plants and fungi [8] [12], while piRNAs are found only in animals [15]. miRNAs are generated from precursor transcripts by two RNase III-type enzymes, Drosha and Dicer. In animals, single-stranded miRNA is incorporated into the argonaute (Ago) protein complexes (Ago) known as RNA induced silencing complexes (RISC) and binds by partially or completely complementary to the 3′ untranslated region (3′UTR) of a target mRNA [6], this results negative control of gene expression by cleavage or inhibition of translation or other regulatory functions [16]. The biological function of miRNAs was first demonstrated in C. elegans, where two miRNAs (Let-7 and Lin-4) were shown to be regulators for stage-specific differentiation of the worm [17], [18]. Endogenous siRNAs are mainly derived from three sources: transposable elements (TEs), complementary annealed transcripts, also called natural antisense transcripts (NAT) and long ‘fold-back’ transcripts called hairpin RNAs (hpRNAs) [19]. Unlike miRNAs, endo-siRNA's function requires perfect match with the target mRNA [6], [20]. MiRNAs and endogenous siRNAs play important roles in the regulation of fundamental cellular processes, including cell differentiation, stress response, apoptosis, proliferation [12] and metabolism [21], [22]. Recent studies further differentiated the functions of endo-siRNAs based on the sources they were generated from [14]. The TE-derived siRNAs are likely functional in the germline cells by repression of transposon activity and consequently keep genome stability, while NAT-derived siRNAs are more functional in somatic cells [14].
In this study, the transcriptional profiles of both miRNA and endo-siRNA in three developmental stages, namely adult (Ad), new born larvae (NBL) and muscle larvae (ML) of T. spiralis were systematically investigated and compared by high throughput cDNA sequencing technology (“RNA-seq”). We found that miRNAs were mainly expressed in the adult worm stage and endo-siRNAs were predominantly derived from transposable elements in the genome.
Results and Discussion
Summary of small RNA sequencing
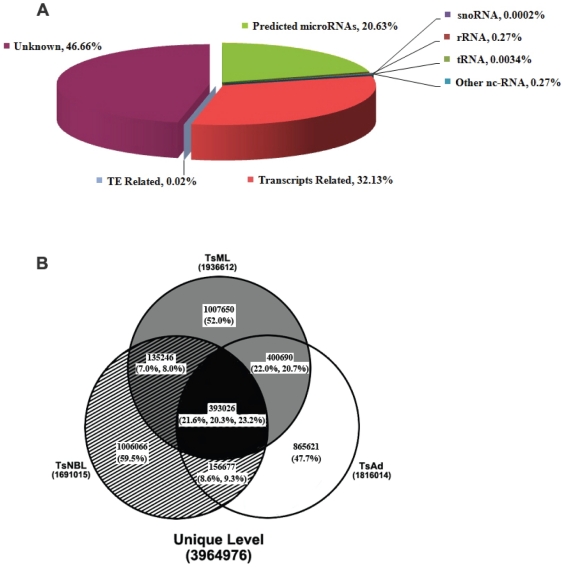
To identify miRNA and endo-siRNA involved in the development of T. spiralis, small RNA libraries were generated from three life cycle stages, the new-born larvae, musclar larvae and adult worms. The libraries were directly sequenced using Solexa sequencing technology. A total of about 40 million unfiltered sequence reads (12,368,833 reads from adult worms, 12,867,246 reads from newborn larvae and 14,495,649 reads from muscular larvae) with sizes between 18 and 30 nucleotides were obtained, respectively. After removal of low quality reads and adaptor sequences, the clean reads obtained in Ad, NBL and ML were 11,878,917 (96.0% of total reads), 12,357,960 (96.0% of total reads) and 14,078,375 (97.1% of total reads) respectively. Sequence characterization suggested that the small RNA pools contained miRNAs (20.63%), other non-coding RNAs (rRNA, tRNA, snoRNA) (0.55%), mRNA-related small RNAs (32.13%), TE-related small RNAs (0.02%) and unknown small RNA transcripts (46.66%) (Fig. 1A, Table S1). The GC content of the small RNAs was around 38.7% and the distribution of the small RNA populations in the libraries generally followed the patterns identified in other organisms, such as schistosomal parasites [23]–[29]. The libraries generated in this study have significantly high coverage than those in a recent report, which only showed a proportion of miRNAs in one single stage of the parasite [30].
Figure 1. Small non-coding RNA identified in three developmental stages of T. spiralis.
A The composition in percentages of clean reads of small RNAs. B Numbers and proportions (bracketed) of unique small RNAs identified in the three stages.
As shown in Fig. 1B, the unique clean reads obtained in Ad, NBL and ML were 1,816,014, 1,691,015 and 1,936,612, respectively (Table S2). The number of small RNAs existed in all three life cycle stages was 393,026 reads, which accounted for 21.6%, 20.3% and 23.2% of the unique clean reads of Ad, NBL and ML stage, respectively (Fig. 1B). The small RNAs commonly expressed in Ad stage and ML stage were 3 times more than that between Ad stage and NBL stage (Fig. 1B), indicating that many genes of Ad stage were activated already in ML and NBL had more stage-specific small RNAs.
Length variation of small RNAs in T. spiralis
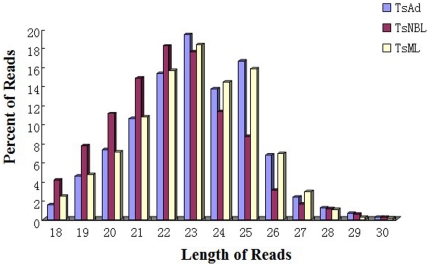
The length of predicted small RNAs varied from 18 to 30 nt in the three life stages, with most of them between from 18 to 27 nt. Here the size and number of sequences in Ad and ML almost followed the same pattern, whereas the number of sequences between 18 and 22 nt was enriched in NBL (Fig. 2 and Table S3). This implied that, apart from stage-specific activation in NBL, small RNAs may undergo size differentiation during early development of the parasite. A previous study indicated that the length of the non-conserved connector helix in Dicer was the main determinant of product size of the mature small RNAs [31], providing a possible explanation for the stage-specific variation we observed. The mechanism behind the stage-specific small RNA processing remains unknown. Since both piRNA and endo-siRNA were more common in the germline, it is likely that the small RNAs found in NBL were predominantly miRNAs, which may play fundamental roles in stage-specific differentiation. Longer RNAs may be more important for parasite reproduction [32]–[37].
Figure 2. Length distribution of small RNAs in different developmental stages.
Length of small RNAs is given on the x-axis in base pairs. And he left Y-axes indicate the percentage of small RNAs afer removal of low quality sequences but prior to selection for complete matches to the genome sequence.
Identification of miRNAs in T. spriralis
To identify candidate miRNAs of T. spiralis, the clean unique reads obtained were mapped to the draft T. spiralis genome sequences (http://genome.wustl.edu/pub/organism/Invertebrates/Trichinella_spiralis/assembly/Trichinella_spiralis-1.0/) [5] sing SOAP. A total of 72,110 (out of 2,099,966) unique small RNA reads that perfectly matched are referred to as miRNA candidates. After screening for secondary structure of the inverted repeats (found with Einverted of Emboss) with RNAfold and evaluation by MirCheck, a total of 240 T. spiralis predicted miRNAs were identified.
To further characterize miRNAs in T. spiralis, all predicted miRNAs obtained above were compared against a miRNA database, miRBase (Release 15.0). In total, 21 miRNAs were found that had been identified in other species, and belong to 13 different miRNA families (Table 1, Table S4 and S5). Most miRNAs contain a 7 nt region (typically positioned at the 5′ side 2–8 nt) known as the “miRNA seed” sequence (Fig. 3). It has been suggested that the seed regions serv to anchor miRNAs to their mRNA targets [14].
Table 1. Conserved (common) miRNAs identified in different developmental stages.
| Name | Mature Arm | miR*a | Most abundant sequence | Length | Expression (TPMb) | ||
| Ad | NBL | ML | |||||
| tsp-miR-228 | 5′ | Y | AAUGGCACUGGAUGAAUUCACGG | 23 | 23725 | 19859 | 60017 |
| tsp-miR-100 | 5′ | Y | AACCCGUAGAUCCGAACUUGUGU | 23 | 3028 | 35995 | 541 |
| tsp-let-7 | 3′ | N | UGAGGUAGUAGGUUGUAUAGUU | 22 | 25573 | 1067 | 5554 |
| tsp-miR-1 | 3′ | Y | UGGAAUGUAAAGAAGUAUGUAG | 22 | 1515 | 9759 | 3351 |
| tsp-miR-31 | 5′ | N | AGGCAAGAUGUUGGCAUAGCUGA | 23 | 1004 | 7728 | 2025 |
| tsp-miR-125 | 5′ | Y | UCCCUGAGACCCAAACUUGUGA | 22 | 745 | 10 | 414 |
| tsp-miR-252 | 5′ | Y | CUAAGUAGUAGUGCCGCAGGUC | 22 | 193 | 279 | 290 |
| tsp-miR-9-1 | 5′ | Y | UCUUUGGUUAUCUAGCUGUAUGA | 23 | 239 | 203 | 179 |
| tsp-miR-87 | 3′ | N | GUGAGCAAAGUUUCAGGUGUGU | 22 | 78 | 160 | 120 |
| tsp-miR-9-2 | 3′ | Y | AUAAGCUAGUUGACCAAAGA | 20 | 47 | 25 | 97 |
| tsp-miR-29 | 3′ | Y | UAGCACCAUUUGAAUUCAGUG | 21 | 21 | 8 | 24 |
| tsp-miR-9-3 | 3′ | Y | UAAAGCUGGAUGACCAAAGU | 20 | 18 | 8 | 26 |
| tsp-miR-993 | 3′ | Y | GAAGCUCGUUUCUACAGG | 18 | 5 | 5 | 16 |
| tsp-miR-133 | 5′ | Y | ACUGGUUGAGGACGUACCAAAUUG | 24 | 1 | 3 | 1 |
| tsp-miR-34 | 5′ | Y | UGGCAGUGUAAUUAGCUGGUUGU | 23 | 1 | 1 | 2 |
Y indicates that sequences from both arms of a pre-miRNA species were found, while N means that only a sequence from one arm was identified.
The abundance value of each miRNA was normalized to “transcripts per million (TPM)”. If the value after normalization was less than 1, the normalized value was set as 1.
doi:10.1371/journal.pone.0026448.t001
Figure 3. Alignment of the tsp -mir-31 sequence with homologues from other organisms.
The seed sequences, typically positioned at the 5′ side 2–8 nucleotides, are shadowed in black. Abbreviations: B. floridae, bfl; C. familiaris, cfa; A.aegypti, aae; O.anatinus, oan; C. quinquefasciatus, cqu; D. grimshawi, dgr; D. ananassae, dan; D. erecta, der; D. melanogaster, dme; D. mojavensis, dmo; D. persimilis, dpe; D. pseudoobscura, dps; D. sechellia, dse; D. simulans, dsi; D. willistoni, dwi; D. virilis, dvi; D. yakuba, dya; B. malayi, bma; D. pulex, dpu; C. elegans, cel; S. mediterranea, sme.
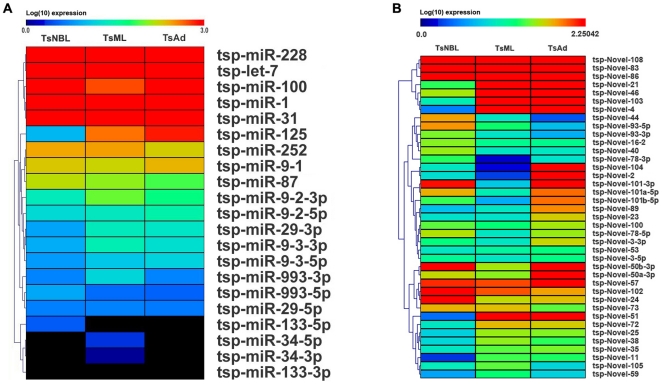
The sequencing data showed that, of the 21 conserved miRNAs, tsp-let-7 and tsp-miR-87 were found to locate only in the 3′ arm of their pre-miRNAs, and tsp-miR-31 was located only in the 5′ arm of the hairpin structures. The remaining 18 miRNAs were derived from both arms of the pre-miRNAs (Table S4 and S5). According to the current model of miRNA maturation, Dicer recognizes the double-strand of pre-miRNAs, cleaves away the loop structure and de-associates the duplex to generate antisense and sense strands, i.e. the mature miRNA and miRNA* [38]. Although the antisense and sense strands may have different thermostability, which also determined there relative abundance in different tissues, both strands could be functional in miRNA-mediated regulatory pathways [39]. For instance, tsp-miR-1-3p was detected at 1,515 TPM (transcripts per million) in the Ad stage, 9,759 in the NBL stage and 3,351 in the ML stage, respectively, whereas its counterpart tsp-miR-1-5p was much less abundant (Fig. 4, Table S5). The TPM of tsp-miR-100-5p and tsp-miR-100-3p, tsp-miR-125-5p and tsp-miR-125-3p, tsp-miR-9-1-5p and tsp-miR-9-1-3p, tsp-miR-9-2-3p and tsp-miR-9-2-5p behaved in similar fashion (Fig. 4, Fig. 5A and Table S5). Thus the anti-sense strands complementary to mRNA targets may play a main regulatory role. However, it cannot be ruled out that both strands generated from the same pre-miRNA execute similar functions. This is because, in some miRNAs, both strands were either similarly expressed or showed stage-specific expression patterns (Fig. 5 A and B; Table S5 and S6).
Figure 4. Sequence and predicted secondary stem-loop structure of tsp-mir-1 identified in T. spiralis.
Sequences and the number of sequencing reads matching the tsp-mir-1 hairpin were listed. The mature miRNA of tsp-mir-1 and the complementary miR* are represented in red and blue respectively. The predicted structure of the pre-miRNA is represented on the right side.
Figure 5. Clustering of expression level of conserved (A) and novel (B) miRNAs.
Log 10 (expression) is used instead of raw expression. The abundance of each miRNA was normalized to “transcripts per million (TPM)”. If the value after normalization was less than 1, the normalized value was set as 1 to avoid the negative values.
We found 213 novel miRNAs (Table 2, Table S7). Comparison of novel miRNAs expressed in the three developmental stages of T. spiralis revealed stage-specific expression patterns (Fig. 5). A majority of the novel miRNAs in NBL stage has a lower expression level than those in Ad stage and ML stage (Fig. 5B, Table S6 and S7). For instance, tsp-novel-21, tsp-novel-50a-3p, tsp-novel-50b-3p were relatively enriched in adult worm, tsp-novel-46 were abundant in muscular larvae, whereas tsp-novel-108 and tsp-novel-83 exhibited a high abundance in all the three stages (Table S7). Of the novel miRNAs, the expression patterns of tsp-novel-93-5p and tsp-novel-93-3p, tsp-novel-50a-3p and tsp-novel-50b-3p, tsp-novel-101-3p and tsp-novel-101a-5p and tsp-novel-101b-5p were interesting. Tsp-novel-93-5p and tsp-novel-93-3p were derived from the same pre-miRNA, but tsp-novel-93-5p was mainly expressed in the NBL stage, less in ML and least in Ad. While tsp-novel-93-3p was much less expressed than tsp-novel-93-5p in all three stages. (Fig. 5B, Table S7). Tsp-novel-50a-3p and tsp-novel-50b-3p were derived from the 3′ arm of two gene copies with a similar sequence. Tsp-novel-50a-3p was dominantly expressed in Ad, while tsp-novel-50b-3p was mainly expressed in NBL and Ad but less in ML. The genes coding for tsp-novel-101-3p and tsp-novel-101a-5p and tsp-novel-101b-5p and their expression patterns were even more complicated. They were encoded by three genes in the genome of T. spiralis. The sequences of tsp-novel-101-3p derived from the 3 genes were the same, while tsp-novel-101b-5p was derived from 1 gene which had a single nucleotide change from tsp-novel-101a-5p (Fig. 5B and Table S6 and S7). The later was derived from two gene copies. All these data suggested that there may be multiple layers of control for the stage-specific expression of miRNA genes, as well as their potential regulatory function in the development of the parasite [40]. Although the mRNA targets of these regulatory miRNAs have not been identified, with the availability of the genome sequence of the parasite, genome-wide association study can be pursued. For instance, miR-1 has been found in many species, from Drosophila to human [41]–[43], suggesting that they are evolutionary conserved, which has been characterized to play essential functions in regulating proliferation and differentiation of muscle cell [41],[44]. One homologous to mir-1, tsp-miR-1, was observed to express in three developmental stages of T. spiralis and may have similar functions. Further, MiR-100 and let-7, the two conserved miRNA in metazoa, play a role in regulation of developmental timing [18],[42],[45]. Their homologs, tsp-miRNA-100 and tsp-let-7, were found throughout the life cycle of T. spiralis. Tsp-let-7 showed very low expression in NBL stage, whereas tsp-miRNA-100 was detected in rather high abundance at the same development stage. The abundance of tsp-miRNA-100 was almost identical with that of tsp-let-7 in both Ad and ML stage, indicating that miRNA-100 may be more functional in NBL stage.
Table 2. Novel miRNAs identified in different developmental stages (only the most abundant are shown here).
| MicroRNA Name | Mature Arm | miR*a | Most abundant sequence | Length | Expression (TPMb) | ||
| Ad | NBL | ML | |||||
| tsp-novel-21 | 3′ | Y | UCACCGGGUAAUAAUUCACAGC | 22 | 4448 | 24 | 348 |
| tsp-novel-108 | 5′ | Y | CUUGGCACUGUAAGAAUUCACAGA | 24 | 2705 | 3722 | 4804 |
| tsp-novel-83 | 3′ | Y | UUGAGCAAUUUUGAUCGUAGC | 21 | 2024 | 1545 | 1143 |
| tsp-novel-50a | 3′ | Y | UCACCGGAUACUAAAACACGUU | 22 | 1022 | 47 | 36 |
| tsp-novel-50b | 3′ | Y | UCACCGGAUACUAAAACACGUGU | 23 | 854 | 288 | 40 |
| tsp-novel-103 | 5′ | Y | UUUUUAUGAAGUGGUAAGUAGG | 22 | 821 | 12 | 623 |
| tsp-novel-86 | 3′ | Y | UGAGAUCACCGUGAAAGCCUUU | 22 | 696 | 333 | 871 |
| tsp-novel-46 | 3′ | Y | UGGACGGCGAAUUAGUGGAAG | 21 | 539 | 40 | 1789 |
| tsp-novel-4 | 3′ | Y | UGGACGGAUGCUCAGUGGAUGU | 22 | 459 | 3 | 860 |
| tsp-novel-104 | 3′ | Y | UCACCGGGCACAAUUUGGCUGC | 22 | 270 | 1 | 2 |
| tsp-novel-2 | 3′ | Y | CACCCGGAUGCUAAAACACGUA | 22 | 194 | 9 | 2 |
| tsp-novel-51 | 5′ | Y | UCGAAUCGCCACAUCGGAAGGC | 22 | 182 | 3 | 171 |
| tsp-novel-101 | 3′ | Y | UCACCGGGCACUAAAUCACGUUU | 23 | 178 | 152 | 7 |
| tsp-novel-57 | 3′ | Y | UUGAGCAAUCACAGUCGUAG | 20 | 170 | 134 | 114 |
| tsp-novel-89 | 3′ | Y | CAUAGGAUUCUAAAACAUGCA | 21 | 80 | 6 | 1 |
| tsp-novel-102 | 5′ | Y | ACUGAAAGAGGGAAACGGUUAG | 22 | 69 | 251 | 107 |
| tsp-novel-24 | 3′ | Y | UGGCAUACUGGAAACGCUGUAGA | 23 | 56 | 189 | 46 |
| tsp-novel-72 | 3′ | Y | UAAUGAGCAUGUAGACCUGAGU | 22 | 52 | 1 | 60 |
| tsp-novel-100 | 3′ | Y | GACCAAUGCGUUGAUGUAGA | 20 | 40 | 29 | 19 |
| tsp-novel-73 | 5′ | Y | UGAAGUUGCACUGGGAUAUGGU | 22 | 29 | 63 | 58 |
Y indicates that the sequences from both strands of a pre-miRNA species were found, while N means that only the sequence from one arm was identified.
The abundance value of each miRNA was normalized to “transcripts per million (TPM)”. If the value after normalization was less than 1, the normalized value was set as 1.
doi:10.1371/journal.pone.0026448.t002
Characterization Of Stage-Associated Mirnas In T. Spiralis
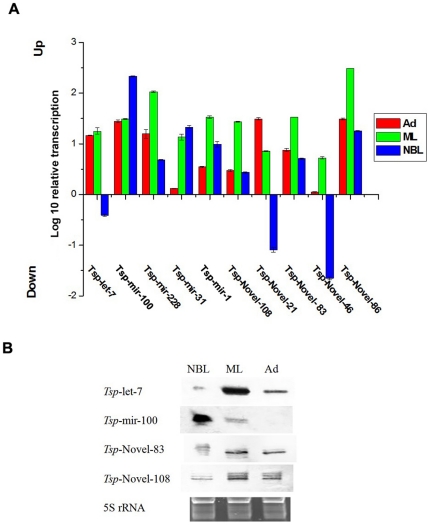
Quantitative real-time PCR and northern blots were performed to validate miRNAs identified in T. spiralis and their relative expression levels at different developmental stages. Five conserved miRNAs (tsp-miR-228, tsp-miR-100, tsp-let-7, tsp-miR-1 and tsp-miR-31) and five novel miRNAs (tsp-novel-108, tsp-novel-83, tsp-novel-46, tsp-novel-86 and tsp-novel-21) with relatively higher TPM values identified by sequencing were validated by qRT-PCR and Northern blot. First, the above miRNAs identified in sequencing were all amplified by qRT-PCR (Fig. 6A) and except the tsp-let-7 which was found less expressed in NBL, the qRT-PCR results were all consistent with the TPM values of sequencing results. The reason for tsp-let-7 being found less common in NBL is not known. This may be due to preferential amplification in the tag-generation step before RNA-seq or other unidentified factors. Further, all qPCR amplicons were cloned into the pMD-18T vector (Takara, Dalian, China) and sequenced. The sequencing results demonstrated that the amplicons were identical to the miRNAs sequences (data not shown). Four miRNAs were selected for characterization by northern blots. The results confirmed the qPCR data. However, the probe for tsp-mir-100 did not show any signal with total RNA purified from the Ad stage; this was likely due to probe modification or other unidentified factors that prevented annealing of the probes with the targets (Fig. 6B). Since this miRNA was found by both sequencing and qPCR in the Ad stage, we have no doubt that tsp-mir-100 was indeed expressed at this stage.
Figure 6. Confirmation of miRNA expression in three developmental stages of T. spiralis.
A The expression level of ten selected miRNAs was in 3 developmental stages of T. spiralis measured by qRT-PCR. GAPDH was chosen as an endogenous reference. B Identification of the miRNAs by northern-blot. Lanes from left to right: new born larvae (NBL), muscle larvae (ML) and adult (Ad). The 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls (bottom).
Identification Of Endogenous Sirna
Endogenous siRNA are extremely diverse and normally not conserved across species. Several types of endogenous siRNAs have been found in Drosopila melanogaster, C. elegans, Schistosoma japonicum, fungi, Arabidopsis thaliana and mice [8],[9],[27],[33],[36],[46]–[50]. Most of these endogenous siRNAs derive from transposable elements (TE), complementary annealed transcripts (also called natural antisense transcripts, NAT) and long ‘fold-back’ transcripts called hairpin RNAs (hpRNAs) [19]. TE and NAT are the main sources of endogenous siRNAs. Thus, the small RNA transcript reads perfectly matched to TE and NAT were regarded as endogenous siRNAs (endo-siRNAs) [47],[48].
TEs are major components of the intergenic regions in the genomes of eukaryotic organisms [47],[48]. Based on their structures and modes of integration, TEs are comprised of two main classes [51]. One class includes retrotransposons and retrotransposon-like elements such as Long Interspersed Nuclear Elements (LINE), Long Terminal Repeat Elements (LTR), and Short Interspersed Elements (SINE). The other group includes DNA transposons. We predicted TE structures in the T. spiralis genome with RepeatMasker (http://www.repeatmasker.org/), and endo-siRNAs matched to TEs were further analyzed. As reported recently [5], the repetitive sequences in the T. spiralis genome estimated to be around 18%, which is much lower than in schistosomes and Drosophila melanogaster [52],[53]. Thus, TE-derived siRNAs accounted for a minor portion of the sncRNAs identified here (Fig. 1A).
The numbers for TE-derived siRNAs identified in the Ad, NBL and ML stages were 2,055, 2,234 and 2908 respectively. No obvious stage-related variations in endo-siRNA expression were found, suggesting that TE components in the genome might not be very active during embryogenesis or during other development stages. Further, most of these siRNAs were derived from transposons of Long Interspersed Nucleotide Elements (LINE) family and the transposon Charlie 24 accounted for a major portion (Table S8). Sequencing analysis showed that more siRNA are derived from the antisense strand than from the sense strands (data not shown). This suggested that antisense-derived siRNA may execute regulatory functions through hybridization with the mRNA template generated from the sense strand.
Since NAT-derived siRNAs were generated from double-stranded RNAs formed by complementary annealed transcripts (NAT) and long ‘fold-back’ transcripts (called hairpin RNAs, hpRNAs) [19], T. spiralis NAT-siRNAs were identified from predicted overlapping genes. The number of NAT-derived siRNAs identified in Ad, NBL and ML was 21,157, 16,243 and 21,251, respectively (Table S9). Thus the number of NAT-derived endo-siRNAs was much higher than that from TE, which suggests that these siRNAs played more regulatory roles in the development and parasitization of T. spiralis. Further, all NAT-derived siRNAs were trans-NAT siRNAs and no cis-NAT siRNAs were found (Fig. 7 and data not shown), which differs from observations with D. melanogaster and mice [33],[47]–[49]. This also suggests that there are fewer coding sequences with internal inverted repeat sequences in the genome of T. spiralis.
Figure 7. NAT-derived siRNA that matched to the sense and antisense strands.
The origin of the genomic sequence (genomic loci) is named on top. siRNAs matched to the sense strand are in blue and those matched to antisense strand are in red. The read numbers and reading direction of the siRNA are listed on the right side.
In summary, we identified and analyzed the expression of miRNAs and endo-siRNAs in three development stages of T. spiralis through high through-put RNA sequencing techniques. We found vastly more transcripts of miRNAs than that of endo-siRNAs. A total of 21 conserved miRNAs in 13 metazoan miRNA familis and 213 novel miRNAs were identified in the parasite. Some showed clear stage-specific expression patterns, suggesting a potential regulatory function in the corresponding developmental stages. Endo-siRNAs were mainly derived from natural antisense transcripts with TE-derived siRNAs accounting for only minor proportion of the small RNA population. Thus, the function of endo-siRNAs in T. spiralis is likely to regulate gene expression instead of maintaining genome stability. The data of this study provide insight information for further dissection of the parasite's biology.
Materials And Methods
Parasites
Muscle larvae (ML) of T. spiralis (strain ISS534) were obtained from rats 35 days post infection (dpi) by digestion of minced skeletal muscle in 1% pepsin, 1% HCl for 3 h at 37°C with agitation as previously described [54]. To purify adult worms and newborn larvae, Wistar rats at 6 weeks of age were orally inoculated with T. spiralis (strain ISS534) with a dose of 8000 larvae per rat. At 30 h or 6 dpi, all rats were killed and the entire intestines were removed, opened longitudinally and cut into small pieces (about 0.5–1 cm). The fragmented intestine was put on a layer of gauze which was immersed into 0.9% sodium chloride solution at 37°C and incubated for 3 h. Adult T. spiralis worms migrate into the liquid phase, which was harvested by centrifugation. To obtain new-born larvae, adult worms collected at 6 dpi were incubated in Iscove's Modified Dulbecco's Medium (IMDM) in 75-cm2 cell culture plates at 37°C, and the newborn larvae were harvested every 12 h. Our study was reviewed and approved by the Ethics Committee of Jilin University (Ethical clearance application number IZ-2009-III). All animal work was conducted according to Chinese and international guidelines.
Rna Isolation
Total RNA of T. spiralis (Ad, NBL and ML) was extracted using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer's instructions. RNAs were dissolved in diethylpyrocarbonate (DEPC)-treated water, aliquoted and stored at −80°C. RNA was quantified by measuring the absorbance at 260 nm with a Nanodrop 1000 machine (Thermo Scientific CA, USA).
Construction Of Small Rna Libraries And Sequencing
For small RNA library construction and deep sequencing, the 15–30 nt size range base-pair fraction of each RNA sample from the three life cycle stages was first enriched by 15% TBE urea polyacrylamide gel electrophoresis and the Illumina's proprietary adaptors (UCAGAGUUCUACAGUCCGACGAUC and UCGUAUGCCGUCUUCUGCUUGUidT) were ligated to the 5′ and 3′ termini of the purified small RNAs which were converted into single-stranded cDNA with Superscript II reverse transcriptase (Invitrogen, CA, USA) and Illumina's small RNA RT-Primer. The cDNAs were PCR-amplified with high fidelity Phusion DNApolymerase (Finnzymes Oy, Finland) in 18 PCR cycles using Illumina's small RNA primer sets. After purification using 6% TBE urea PAGE gels, the PCR products were sequenced by Solexa's sequencing-by-synthesis method (Fig. S1).
Mapping The Sequence Reads Onto The Reference Genome
The bioinformatic analysis and work flow is shown in Figure S1. After removing the low quality sequence reads and the adapter sequences according to the criteria of Illumina, all identical sequences were retained with associated count numbers as their expression abundances To determine whether these clean small RNA sequences were candidate miRNAs, the unique reads were mapped onto the T. spiralis genome of the Genome Sequencing Center (GSC) at Washington University St. Louis, (http://genome.wustl.edu/pub/organism/Invertebrates/Trichinella_spiralis/assembly/Trichinella_spiralis-1.0/) with SOAP [55] (http://soap.genomics.org.cn).
The perfectly matched reads were searched against the Metazoa mature miRNA of Sanger miRBase with Patscan [56]. Sequence tags of more than 5 reads that matched perfectly or near-perfectly (no more than 1 mismatch and the mismatch not positioned in the seed region) were regarded to be conserved miRNA candidates. For reads that did not match to the miRNA database, we used the software Einverted of Emboss [57] to find the inverted repeats (stem loops or hairpin structure). Each inverted repeat was extended 10 nt on each side, and the secondary structure of the inverted repeat was predicted by RNAfold [58]. Unique reads with a folding free energy of at least 25 kcal/mole( △G°folding≤−25 kcal/mol) were evaluated by MirCheck [59] with modified parameters. Finally, precursors (hairpins) of miRNA that passed MirCheck were inspected manually in order to remove false predictions. The reads passing the inspections were regarded as novel miRNAs.
Similar to credibility interval approaches reported for the analysis of SAGE data [60], we employed IDEG6 [61] to identify miRNAs showing statistically significant differences in relative abundance (as reflected by the total count of individual sequence reads) between the three libraries (corresponding to the three developmental stages of the parasite). We used the general Chi-square method for comparison analysis, which has been commonly applied by others [27],[36]. Finally, miRNA with a P value≤0.01 were deemed to be significantly different between the samples of the three developmental stages of the parasite.
The repeated sequences, i.e. potential transposable elements, in the T. spiralis genome were predicted by using RepeatMasker (http://www.repeatmasker.org/) and the sequences of typical transposons were annotated (data not shown). The sequencing reads that perfectly matched the T. spiralis genome were aligned to repeats (TE) using SOAP. The reads that perfectly matched TEs were considered TE-derived siRNAs.
Natural antisense transcripts (NATs) were detected by aligning the T. spiralis predicted genes to each other. If a pair of overlapping genes were matched on opposite strands with an E value of ≤1e-9 [62], this pair of overlapping genes was defined as a NAT pair. The reads that perfectly matched the T. spiralis genome were aligned to overlapped sequences of NAT pairs with SOAP. The reads that perfectly matched the overlapped regions were considered NAT-derived siRNAs.
Mirnas Quantification By Real-Time Pcr
Total RNA purified from the three stages were polyadenylated with E. coli poly(A) polymerase (E-PAP) following the manufacturer's protocol of the poly(A)-tailing kit (Ambion, CA, USA). The RNA samples were purified separately from the parasites of the same developmental stages for sequencing purpose. After phenol-chloroform extraction and ethanol precipitation, the polyadenylated products were dissolved in DEPC-treated water and reverse-transcribed with 200 U of SuperScript™ III Reverse Transcriptase (Invitrogen, CA) and 3′ RACE Adapter (5′-GCGAGCACAGAATTAATACGACTCACTATAGGT12VN-3′) in the FirstChoice RLM-RACE Kit (Ambion) according to the manufacturer's protocol. The 20 µl RT reaction contained 1 µg total RNA, 2 µl 3′ RACE Adapter, 0.5 mM dNTP mix (Takara, Dalian, China), 10 U RNase inhibitor and 200 U SuperScript™ III Reverse Transcriptase.
In quantitative RT-PCR reactions, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was chosen as an endogenous reference. The forward and reverse primers for GAPDH were 5′-GTGCTGATTACGCTGTTG-3′ and 5′-CTAAGCCATTGGTAGTGC-3′. PCR was done on an Applied Biosystems 7500 system. The following forward primers were designed to confirm the sequencing results of miRNAs that showed differential expression patterns: tsp-miR-100 5′-AAC CCG TAG ATC CGA ACT TGT GT-3′; tsp-let-7 5′-TGA GGT AGT AGG TTG TAT AGT T-3′; tsp-miR-228 5′-AAT GGC ACT GGA TGA ATT CAC GG-3′; tsp-miR-1 5′-TGG AAT GTA AAG AAG TAT GTA G-3′; tsp-miR-31 5′-AGG CAA GAT GTT GGC ATA GCT GA-3′; tsp-novel-108 5′-CTT GGC ACT GTA AGA ATT CAC AGA-3′; tsp-novel-83 5′-TTG AGC AAT TTT GAT CGT AGC-3′; tsp-novel-46 5′-TGG ACG GCG AAT TAG TGG AAG-3′; tsp-novel-86 5′-TGA GAT CAC CGT GAA AGC CTT T-3′; tsp-novel-21 5′-TCA CCG GGT AAT AAT TCA CAG C-3′. The sequence 5′-GCG AGC ACA GAA TTA ATA CGA CT-3′ (complementary to the adaptor) was used as a common reverse primer. Relative expression was calculated by the comparative Ct method [63]. ANOVA and Tukey's HSD post-hoc test were used to analyze significant differences among three stage; P<0.05 was considered significant.
Northern Blot Analysis Of Mirna Expression
Total RNA of T. spiralis (muscle larvae, adult worms and newborn larvae) were separated by electrophoresis on a 12.5% polyacrylamide gel under denaturing (8 M urea) conditions and transferred to Hybond-N+ nylon membranes (GE Healthsystems, Uppsala, Sweden). The membranes were crossslinked in a UV crosslinker and baked for 1 h at 80°C. Probes complementary to small RNA sequences were end-labeled with DIG at 5′ termini (Takara, Dalian, China). Prehybridization and hybridization were both performed overnight at 53°C in Northernmax Hybridization buffer (Ambion, CA, USA). The blots were washed four times for 30 min in 2×SSC, 0.05% SDS and twice for 15 min in 0.1×SSC, 0.1% SDS at room temperature. The hybridization signal was detected using a DIG Detection Kit (Roche) following manufacturer's instructions. The oligonucleotide probes used for hybridization are as follows:
tsp-mir-100 probes: 5′ACACAA*GTTC*GGATCT*AC*GGGTT3′
tsp-let-7 probes: 5′AACTAT*ACA*ACCT*ACT*ACCTCA3′
tsp-novel-108 probes: 5′TCT*GT*GAATTCTT*ACA*GTGCCAAG3′
tsp-novel-83 probes: 5′GCTAC*GATC*AA*AATT*GCTCAA3′
(LNA (Locked nucleic acid) substitutions are indicated by a “*”).
Supporting Information
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We very much appreciate the effort of scientists at BGI Genomics for their kind assistance with the sequencing of small RNA libraries and Dr. Haipo Sun for assistance with bioinformatics.
References
- 1.Zarlenga DS, Rosenthal BM, La Rosa G, Pozio E, Hoberg EP. Post-Miocene expansion, colonization, and host switching drove speciation among extant nematodes of the archaic genus Trichinella. Proc Natl Acad Sci U S A. 2006;103:7354–7359. doi: 10.1073/pnas.0602466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupouy-Camet J. Trichinellosis: a worldwide zoonosis. Vet Parasitol. 2000;93:191–200. doi: 10.1016/s0304-4017(00)00341-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu MY, Zhu XP, Xu KC, Lu Q, Boireau P. Biological and genetic characteristics of two Trichinella isolates in China; comparison with European species. Parasite. 2001;8:S34–38. doi: 10.1051/parasite/200108s2034. [DOI] [PubMed] [Google Scholar]
- 4.Wakelin D, Goyal PK. Trichinella isolates: parasite variability and host responses. Int J Parasitol. 1996;26:471–481. doi: 10.1016/0020-7519(96)89377-5. [DOI] [PubMed] [Google Scholar]
- 5.Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 10.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 11.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Pan X, Anderson TA. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 16.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 19.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 22.Gauthier BR, Wollheim CB. MicroRNAs: ‘ribo-regulators’ of glucose homeostasis. Nat Med. 2006;12:36–38. doi: 10.1038/nm0106-36. [DOI] [PubMed] [Google Scholar]
- 23.Xu MJ, Liu Q, Nisbet AJ, Cai XQ, Yan C, et al. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;11:521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedlander MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole CB, Davis PJ, Jin J, McReynolds LA. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol Biochem Parasitol. 2010;169:87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Cucher M, Prada L, Mourglia-Ettlin G, Dematteis S, Camicia F, et al. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. Int J Parasitol. 2011;41:439–448. doi: 10.1016/j.ijpara.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Hao L, Cai P, Jiang N, Wang H, Chen Q. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics. 2010;11:55. doi: 10.1186/1471-2164-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simoes MC, Lee J, Djikeng A, Cerqueira GC, Zerlotini A, et al. Identification of Schistosoma mansoni microRNAs. BMC Genomics. 2011;12:47. doi: 10.1186/1471-2164-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen MX, Ai L, Xu MJ, Zhang RL, Chen SH, et al. Angiostrongylus cantonensis: Identification and characterization of microRNAs in male and female adults. Exp Parasitol. 2011;128:116–120. doi: 10.1016/j.exppara.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 30.Chen MX, Ai L, Xu MJ, Chen SH, Zhang YN, et al. Identification and characterization of microRNAs in Trichinella spiralis by comparison with Brugia malayi and Caenorhabditis elegans. Parasitol Res. 2011 doi: 10.1007/s00436-011-2283-x. [DOI] [PubMed] [Google Scholar]
- 31.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 32.Saito K, Siomi MC. Small RNA-Mediated Quiescence of Transposable Elements in Animals. Dev Cell. 2010;19:687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 37.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 44.Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- 47.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Gogvadze E, Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci. 2009;66:3727–3742. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biemont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 54.Despommier DD, Campbell WC, Blair LS. The in vivo and in vitro analysis of immunity to Trichinella spiralis in mice and rats. Parasitology. 1977;74:109–119. doi: 10.1017/s0031182000047570. [DOI] [PubMed] [Google Scholar]
- 55.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 56.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 57.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 58.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast Folding and Comparison of RNA Secondary Structures. Monatshefte fur Chem Chem Month. 1994;125:167–188. [Google Scholar]
- 59.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 60.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 61.Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- 62.Li YY, Qin L, Guo ZM, Liu L, Xu H, et al. In silico discovery of human natural antisense transcripts. BMC Bioinformatics. 2006;7:18. doi: 10.1186/1471-2105-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)