Abstract
A comprehensive approach to 13C tracer studies, labeling measurements by gas chromatography-mass spectrometry, metabolite balancing, and isotopomer modeling, was applied for comparative metabolic network analysis of lysine-producing Corynebacterium glutamicum on glucose or fructose. Significantly reduced yields of lysine and biomass and enhanced formation of dihydroxyacetone, glycerol, and lactate in comparison to those for glucose resulted on fructose. Metabolic flux analysis revealed drastic differences in intracellular flux depending on the carbon source applied. On fructose, flux through the pentose phosphate pathway (PPP) was only 14.4% of the total substrate uptake flux and therefore markedly decreased compared to that for glucose (62.0%). This result is due mainly to (i) the predominance of phosphoenolpyruvate-dependent phosphotransferase systems for fructose uptake (PTSFructose) (92.3%), resulting in a major entry of fructose via fructose 1,6-bisphosphate, and (ii) the inactivity of fructose 1,6-bisphosphatase (0.0%). The uptake of fructose during flux via PTSMannose was only 7.7%. In glucose-grown cells, the flux through pyruvate dehydrogenase (70.9%) was much less than that in fructose-grown cells (95.2%). Accordingly, flux through the tricarboxylic acid cycle was decreased on glucose. Normalized to that for glucose uptake, the supply of NADPH during flux was only 112.4% on fructose compared to 176.9% on glucose, which might explain the substantially lower lysine yield of C. glutamicum on fructose. Balancing NADPH levels even revealed an apparent deficiency of NADPH on fructose, which is probably overcome by in vivo activity of malic enzyme. Based on these results, potential targets could be identified for optimization of lysine production by C. glutamicum on fructose, involving (i) modification of flux through the two PTS for fructose uptake, (ii) amplification of fructose 1,6-bisphosphatase to increase flux through the PPP, and (iii) knockout of a not-yet-annotated gene encoding dihydroxyacetone phosphatase or kinase activity to suppress overflow metabolism. Statistical evaluation revealed high precision of the estimates of flux, so the observed differences for metabolic flux are clearly substrate specific.
Corynebacterium glutamicum is widely used for industrial production of amino acids such as glutamate and lysine. Great effort has been undertaken in order to optimize producer strains. The powerful toolbox of genetic engineering allows the targeted modification of genes in C. glutamicum (10). Past experience clearly shows that detailed and quantitative knowledge of metabolic physiology is crucial as a basis for rational design and optimization of strains, and the metabolism of lysine-producing C. glutamicum has been the object of various studies in the last decade (15). Almost all of these studies were based on glucose, as were studies on metabolic flux, which were mainly carried out on glucose (3). In contrast, little attention was paid to other carbon sources, with the consequence that most of our knowledge about the physiology of C. glutamicum is based on the metabolism of glucose. Only selected studies have investigated the metabolism of C. glutamicum on lactate, acetate, and fructose (1, 5, 11, 18). Fructose, especially, is a relevant substrate for the industrial production of amino acids. Depending on quality and pretreatment, industrial molasses, one of the major raw materials for industrial amino acid production, usually contains between 10 to 20% of its carbon source in the form of fructose. Additionally, other fructose-containing raw materials, such as high-fructose corn syrup, could be of interest for industrial amino acid production under certain economic conditions. The first evidence that fructose results in significantly different performance of C. glutamicum in the production of aromatic amino acids than does glucose was obtained (16). In this study, a diminished yield of phenylalanine resulted when fructose was the carbon source. In a recent comparison of lysine-producing C. glutamicum grown on fructose or glucose, drastic differences in process stoichiometry were observed, involving a 30%-lower lysine yield and a 20%-lower biomass yield on fructose (7). The results of these studies pointed to a potentially lower activity of the pentose phosphate pathway (PPP), which is important for the production of phenylalanine via supply of the precursor erythrose 4-phosphate and for the production of lysine via the supply of NADPH. Recently, a study of metabolic flux using glucose and fructose as carbon sources was carried out with C. melassecola ATCC 17695, a strain which is related to C. glutamicum (5). In this study, first estimates of certain flux parameters were obtained during exponential growth. Compared to that for glucose, the activity of the PPP on fructose was about 30% less, with a significantly lower growth rate. A key reason for the observation of different phenotypes on fructose and glucose might be the entry points of the two substrates into the central metabolism. Glucose and fructose are taken up by C. glutamicum via phosphoenolpyruvate-dependent phosphotransferase systems (PTS), whereby glucose is phosphorylated into glucose 6-phosphate (8). Fructose is taken up simultaneously by a fructose and a mannose PTS (5) and enters glycolysis at two locations: (i) fructose 1,6-bisphosphate via fructose 1-phosphate and (ii) fructose 6-phosphate, respectively.
In the present work, metabolic flux of lysine-producing C. glutamicum was analyzed in comparative batch cultures on glucose or fructose. The production of lysine is known to pose specific flux burdens on the metabolism of C. glutamicum involving a high demand for NADPH, oxaloacetate, and pyruvate. Metabolic flux analysis was based on a straightforward and precise approach of metabolite balancing, 13C tracer studies with gas chromatography-mass spectrometry (GC-MS), and isotopomer modeling. By this approach, significant substrate-specific differences in intracellular pathway activities were identified, providing important knowledge on the metabolism of lysine-producing C. glutamicum.
MATERIALS AND METHODS
Microorganism and medium.
Corynebacterium glutamicum ATCC 21526 was obtained from the American Type Culture Collection (Manassas, Va.). This homoserine auxotrophic strain excretes l-lysine during l-threonine limitation due to the bypass of concerted aspartate kinase inhibition. Precultures were grown in complex medium containing 5 g liter−1 of either fructose or glucose. For agar plates, the complex medium was additionally amended with 12 g of agar liter−1. For the production of cells as inoculum for the tracer experiments and the tracer study itself, a minimal medium amended with 1 mg of calcium pantothenate-HCl ml−1 was used (20). In this medium, concentrations of the carbon source, glucose or fructose, of the essential amino acids threonine, methionine, and leucine, and of citrate were varied as specified below.
Cultivation.
Precultivation consisted of three steps involving (i) a starter cultivation in complex medium with cells from the agar plates as inoculum, (ii) a short cultivation for adaptation to the minimal medium, and (iii) a prolonged cultivation on minimal medium with elevated concentrations of essential amino acids. Precultures inoculated from agar plates were grown overnight in 100-ml baffled shake flasks on 10 ml of complex medium. Afterwards, cells were harvested by centrifugation (8800 × g, 30°C, 2 min), inoculated into minimal medium, and grown to an optical density of 2 to obtain exponentially growing cells adapted to minimal medium. Then the cells were harvested by centrifugation (8800 × g, 30°C, 2 min), including a washing step with sterile 0.9% NaCl. They were then inoculated into 6 ml of minimal medium in 50-ml baffled shake flasks with initial concentrations of 0.30 g of threonine liter−1, 0.08 g of methionine liter−1, 0.20 g of leucine liter−1, and 0.57 g of citrate liter−1. As the carbon source, 70 mM glucose or 80 mM fructose was added. The cells were grown until depletion of the essential amino acids, which was checked by analysis with high-pressure liquid chromatography (HPLC). At the end of the growth phase, the cells were harvested and washed with sterile NaCl (0.9%). Subsequently, they were transferred into 4 ml of minimal tracer medium in 25-ml baffled shake flasks for metabolic flux analysis under lysine-producing conditions. The tracer medium did not contain any threonine, methionine, leucine, or citrate. For each carbon source, two parallel flasks containing (i) 40 mM 1-13C-labeled substrate or (ii) 20 mM 13C6-labeled substrate plus 20 mM of naturally labeled substrate were incubated. All cultivations were carried out on a rotary shaker (Inova 4230; New Brunswick, Edison, N.J.) at 30°C and 150 rpm.
Chemicals.
Ninety-nine percent [1-13C]glucose, 99% [1-13C] fructose, 99% [13C6]glucose and 99% [13C6]fructose were purchased from Campro Scientific (Veenendaal, The Netherlands). Yeast extract and tryptone were obtained from Difco Laboratories (Detroit, Mich.). All other applied chemicals were from Sigma (St. Louis, Mo.), Merck (Darmstadt, Germany), or Fluka (Buchs, Switzerland) and were of analytical grade.
Substrate and product analysis.
The concentration of cells was determined by measurement of optical density at 660 nm (OD660 nm) using a photometer (Marsha Pharmacia Biotech, Freiburg, Germany) or by gravimetry. The latter was determined by harvesting 10 ml of cells from cultivation broth at room temperature by centrifugation for 10 min at 3700 × g, including a washing step with water. The washed cells were dried at 80°C until their weight became constant. The correlation factor (g of biomass to OD660 nm) between dry cell mass and OD660 nm was determined as 0.353.
The concentrations of extracellular substrates and products in the cultivation supernatants were determined via 3 min of centrifugation at 16000 × g. Fructose, glucose, sucrose, and trehalose were quantified by GC after derivatization into oxime trimethylsilyl derivatives. For this purpose, an HP 6890 gas chromatograph (Hewlett Packard, Palo Alto, Calif.) with an HP 5MS column (5% phenyl-methyl-siloxane-diphenyldimethylpolysiloxane, 30 m × 250 μm; Hewlett Packard) and a quadrupole mass selective detector with electron impact ionization at 70 eV (Agilent Technologies, Waldbronn, Germany) was applied. Sample preparation included lyophilization of the culture supernatant, dissolution in pyridine, and subsequent two-step derivatization of the sugars with hydroxylamine and (trimethylsilyl)trifluoroacetamide (BSTFA; Macherey & Nagel, Düren, Germany) (13, 14). β-d-ribose was used as the internal standard for quantification. The injected sample volume was 0.2 μl. The time program for GC analysis was as follows: 150°C (0 to 5 min), 8°C min−1 (5 to 25 min), 310°C (25 to 35 min). Helium was used as the carrier gas, with a flow of 1.5 liter min−1. The inlet temperature was 310°C, and the detector temperature was 320°C. Acetate, lactate, pyruvate, 2-oxoglutarate, and dihydroxyacetone levels were determined by HPLC, utilizing an Aminex-HPX-87H Bio-Rad Column (300 × 7.8 mm; Hercules, Calif.) with 4 mM sulfuric acid during the mobile phase at a flow rate of 0.8 ml min−1 and UV detection at 210 nm. Glycerol was quantified by enzymatic measurement (Boehringer, Mannheim, Germany). Amino acids were analyzed by HPLC (Agilent Technologies), utilizing a Zorbax Eclypse-AAA column (150 × 4.6 mm, 5 μm; Agilent Technologies) with automated online derivatization (o-phtaldialdehyde plus 3-mercaptopropionic acid) at a flow rate of 2 ml min−1 and fluorescence detection. Details are given in the instruction manual. α-Amino butyrate was used as the internal standard for quantification.
13C-labeling analysis.
The labeling patterns of lysine and trehalose in cultivation supernatants were quantified by GC-MS, and single mass isotopomer fractions were determined. In the present work, they are defined as M0 (relative amount of nonlabeled mass isotopomer fraction), M1 (relative amount of single-labeled mass isotopomer fraction), and corresponding terms for higher labeling. GC-MS analysis of lysine was performed after conversion into the t-butyl-dimethylsilyl derivate as described previously (14). Quantification of mass isotopomer distributions was performed in selective ion monitoring mode for the ion cluster m/z 431 to 437. This ion cluster corresponds to a fragment ion, which is formed by loss of a t-butyl group from the derivatization residue and thus includes the complete carbon skeleton of lysine (21). The labeling pattern of trehalose was determined from its trimethylsilyl derivate (22). The labeling pattern of trehalose was estimated via the ion cluster at m/z 361 to 367 corresponding to a fragment ion that contained an entire monomer unit of trehalose and thus a carbon skeleton equal to that of glucose 6-phosphate. All samples were measured first in scan mode, thus excluding isobaric interference between the analyzed products and other sample components. All measurements by selective ion monitoring were performed in duplicate. The experimental errors for single mass isotopomer fractions in the tracer experiments on fructose were 0.85% (M0), 0.16% (M1), 0.27% (M2), 0.35% (M3), and 0.45% (M4) for lysine on [1-13C]fructose; 0.87% (M0), 0.19% (M1), 0.44% (M2), 0.45% (M3), and 0.88% (M4) for trehalose on [1-13C] fructose; and 0.44% (M0), 0.54% (M1), 0.34% (M2), 0.34% (M3), 0.19% (M4), 0.14% (M5), and 0.52% (M6) for trehalose on 50% [13C6]fructose. The experimental errors for MS measurements in glucose tracer experiments were 0.47% (M0), 0.44% (M1), 0.21% (M2), 0.26% (M3), and 0.77% (M4) for lysine on [1-13C]glucose; 0.71% (M0), 0.85% (M1), 0.17% (M2), 0.32% (M3), and 0.46% (M4) for trehalose on [1-13C]glucose; and 1.29% (M0), 0.50% (M1), 0.83% (M2), 0.84% (M3), 1.71% (M4), 1.84% (M5), and 0.58% (M6) for trehalose on 50% [13C6]glucose.
Metabolic modeling and parameter estimation.
All metabolic simulations were carried out on a personal computer. The metabolic network of lysine-producing C. glutamicum was implemented in Matlab 6.1 and Simulink 3.0 (Mathworks Inc., Nattick, Mass.). The software implementation included an isotopomer model in Simulink to calculate the 13C-labeling distribution in the network. For parameter estimation, the isotopomer model was coupled with an iterative optimization algorithm in Matlab. Details of the applied computational tools are given by Wittmann and Heinzle (20).
The metabolic network was based on previous work and comprised glycolysis, the PPP, the tricarboxylic acid (TCA) cycle, anaplerotic carboxylation of pyruvate, biosynthesis of lysine and other secreted products (Tab. 1), and anabolic flux from intermediary precursors into biomass. In addition, uptake systems for glucose and fructose were alternatively implemented. Uptake of glucose involved phosphorylation to glucose 6-phosphate via a PTS (10). For fructose, two uptake systems were considered: (i) uptake by PTSFructose and conversion of fructose into fructose 1,6-bisphosphate via fructose 1-phosphate and (ii) uptake by PTSMannose, leading to fructose 6-phosphate (5). In addition, fructose 1,6-bisphosphatase was implemented into the model to allow carbon flux in both directions in upper glycolysis. Reactions regarded as reversible were those of transaldolase and transketolases in the PPP. Additionally, the reaction of glucose 6-phosphate isomerase was considered reversible for the experiments on glucose, whereby the trehalose labeling sensitively reflected the reversibility of this enzymatic reaction. In contrast, the reversibility of the reaction of glucose 6-phosphate isomerase could not be determined on fructose. In fructose-grown cells, glucose 6-phosphate is exclusively formed from fructose 6-phosphate, leading to identical labeling patterns for the two pools. Therefore, interconversion between glucose 6-phosphate and fructose 6-phosphate by a reversible glucose 6-phosphate isomerase reaction does not result in labeling differences that could be used for the estimation of the reversibility of the glucose 6-phosphate isomerase reaction. The measured labeling of lysine and trehalose was not sensitive toward (i) the reversibility of the flux between the lumped pools of phosphoenolpyruvate-pyruvate and malate-oxaloacetate and (ii) the reversibility of the reactions of malate dehydrogenase and fumarate hydratase in the TCA cycle. Accordingly, these reactions were regarded as irreversible. The labeling of alanine from a mixture of naturally labeled and 13C6-labeled substrate, which is sensitive for these flux parameters, was not available for this study. Based on previous results, the glyoxylate pathway was assumed to be inactive (20).
Stoichiometric data on the growth, product formation, and biomass composition of C. glutamicum together with MS labeling data on secreted lysine and trehalose were used to calculate metabolic flux distributions. The set of data that had the minimum deviation between experimental (Mi, exp) and simulated (Mi, calc) mass isotopomer fractions of lysine and trehalose from the two parallel experiments was taken as the best estimate for the intracellular flux distribution. As described in the appendix, the two networks of glucose-grown and fructose-grown cells were overdetermined. A least-squares approach was therefore possible. As the error criterion, a weighted sum of least-squares was used, where Si, exp is the standard deviation of the measurements (equation 1).
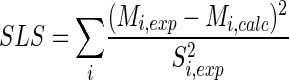 |
(1) |
Multiple parameter initializations were applied to investigate whether an obtained flux distribution represented a global optimum. For all strains, flux for glucose uptake during lysine production was set to 100% and the other fluxes in the network are given as relative molar flux normalized to flux for glucose uptake.
Statistical evaluation.
Statistical analysis of the results obtained for metabolic flux was carried out by a Monte-Carlo approach as described previously (20). For each strain, the statistical analysis was carried out by 100-parameter estimation runs, whereby the experimental data, comprising measurements of mass isotopomer ratios and flux, were varied statistically. From the obtained data, 90% confidence limits for the single parameters were calculated.
RESULTS
Lysine production by C. glutamicum on fructose or glucose.
In the present work, metabolic flux of lysine-producing C. glutamicum was analyzed in comparative batch cultures grown on glucose or fructose. For this purpose, pregrown cells were transferred into tracer medium and incubated for about 5 h. Analysis of the substrates and products at the beginning and end of the tracer experiment revealed drastic differences between results for the two carbon sources. Overall, 11.1 mM lysine was produced on glucose, whereas a lower concentration of only 8.6 mM was reached on fructose. During the 5-h incubation, the cell concentration increased from 3.9 g liter−1 to 6.0 g liter−1 (glucose) and from 3.5 g liter−1 to 4.4 g liter−1 (fructose). Because threonine and methionine were not present in the medium, internal sources were probably utilized by the cells for biomass synthesis. The mean specific sugar uptake rate was higher on fructose (1.93 mmol g−1 h−1) than on glucose (1.71 mmol g−1 h−1). As depicted in Table 1, the yields of C. glutamicum ATCC 21526 obtained differed drastically for fructose and glucose and involved the main product, lysine, and various by-products. The yield for lysine on fructose was 244 mmol mol−1, which was lower than the yield on glucose (281 mmol mol−1). Additionally, the carbon source had a drastic influence on the biomass yield, which was reduced by almost 50% on fructose in comparison to that on glucose. The most significant influence of the carbon source on by-product formation was observed for dihydroxyacetone, glycerol, and lactate. On fructose, accumulation of these by-products was strongly enhanced. The yield for glycerol was 10-fold higher, whereas dihydroxyacetone and lactate secretion were increased by a factor of six. Dihydroxyacetone was the dominating by-product on fructose. Due to the lower biomass yield, a significantly reduced demand for anabolic precursors resulted for fructose-grown cells (Table 2).
TABLE 1.
Yields of biomass and metabolites in the stages of lysine production by Corynebacterium glutamicum ATCC 21526 from glucose or fructosea
| Yield | Lysine production on:
|
|
|---|---|---|
| Glucose | Fructose | |
| Biomass | 54.1 ± 0.8 | 28.5 ± 0.0 |
| Lysine | 281.0 ± 2.0 | 244.4 ± 23.3 |
| Valine | 0.1 ± 0.0 | 0.0 ± 0.0 |
| Alanine | 0.1 ± 0.0 | 0.4 ± 0.1 |
| Glycine | 6.6 ± 0.0 | 7.1 ± 0.4 |
| Dihydroxyacetone | 26.3 ± 15.3 | 156.6 ± 25.8 |
| Glycerol | 3.8 ± 2.4 | 38.4 ± 3.9 |
| Trehalose | 3.3 ± 0.5 | 0.9 ± 0.1 |
| α-Ketoglutarate | 1.6 ± 0.4 | 6.5 ± 0.3 |
| Acetate | 45.1 ± 0.3 | 36.2 ± 5.7 |
| Pyruvate | 1.2 ± 0.4 | 2.1 ± 0.5 |
| Lactate | 7.1 ± 1.7 | 38.3 ± 3.5 |
The yields are the mean values of two parallel incubations on (i) 40 mM 1-13C-labeled substrate and (ii) 20 mM 13C6-labeled substrate plus 20 mM naturally labeled substrate with the corresponding deviations between the two incubations. All yields are given in mmol of product mol−1 except the yield for biomass, which is given in mg of dry biomass mmol−1.
TABLE 2.
Anabolic demand of Corynebacterium glutamicum ATCC 21526 for intracellular metabolites in the stages of lysine production from glucose or fructosea
| Precursor demandb, mmol mol of glucose−1 | Lysine production on:
|
|
|---|---|---|
| Glucose | Fructose | |
| Glucose 6-phosphate | 11.09 ± 0.16 | 5.84 ± 0.05 |
| Fructose 6-phosphate | 3.84 ± 0.06 | 2.02 ± 0.02 |
| Pentose 5-phosphate | 47.50 ± 0.70 | 25.05 ± 0.21 |
| Erythrose 4-phosphate | 14.50 ± 0.22 | 7.64 ± 0.06 |
| Glyceraldehyde 3-phosphate | 6.98 ± 0.10 | 3.68 ± 0.03 |
| 3-Phosphoglycerate | 59.95 ± 0.89 | 36.85 ± 0.31 |
| Pyruvate/phosphoenolpyruvate | 107.80 ± 1.60 | 56.80 ± 0.48 |
| α-Ketoglutarate | 92.51 ± 1.37 | 48.73 ± 0.41 |
| Oxaloacetate | 48.91 ± 0.72 | 45.76 ± 0.38 |
| Acetyl coenzyme A | 135.30 ± 2.00 | 71.25 ± 0.60 |
| Diaminopimelate+lysinec | 18.83 ± 0.28 | 9.92 ± 0.08 |
The data shown are the mean values of two parallel incubations on (i) 1-13C-labeled substrate and (ii) a 1:1 mixture of naturally labeled and 13C6-labeled substrate with the deviations between the two incubations.
The estimation of precursor demands was based on the biomass yield obtained for each strain (Table 1) and the biomass composition previously measured for C. glutamicum (9).
Diaminopimelate and lysine are regarded as separate anabolic precursors because in addition to the flux of lysine secretion, anabolic fluxes from pyruvate and oxaloacetate into diaminopimelate (cell wall) and lysine (protein) contribute to the overall flux through the lysine biosynthetic pathway.
Manual inspection of 13C-labeling patterns in tracer experiments.
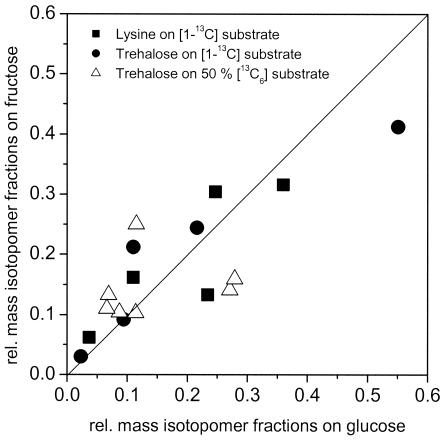
Relative mass isotopomer fractions of secreted lysine and trehalose were quantified with GC-MS. These mass isotopomer fractions are sensitive towards intracellular flux and therefore display fingerprints for the metabolic fluxes of the investigated biological system. As shown in Fig. 1, the labeling patterns of secreted lysine and trehalose differed significantly between glucose- and fructose-grown cells of C. glutamicum. The differences were found for both applied tracer labelings and for both measured products. This finding indicated substantial differences in the carbon flux pattern depending on the applied carbon source. Mass isotopomer fractions from two parallel cultivations of C. glutamicum on a mixture of [1-13C]glucose and [13C6]glucose were almost identical (22). Therefore, the differences observed here can be clearly related to substrate-specific differences in metabolic flux.
FIG. 1.
Comparison of relative mass isotopomer fractions of secreted lysine and trehalose measured by GC-MS in tracer experiments with Corynebacterium glutamicum ATCC 21526 during lysine production on glucose or fructose. Rel., relative.
Estimation of intracellular flux.
A central issue of the studies performed was the comparative investigation of the intracellular flux of C. glutamicum during lysine production on glucose or on fructose as the carbon source. For this purpose, the experimental data obtained from the tracer experiments were used to calculate metabolic flux distributions for each substrate, applying the flux estimation software as described above. The parameter estimation was carried out by minimizing the deviation between experimental and calculated mass isotopomer fractions. The approach performed utilized metabolite balancing during each step of the optimization. This included (i) stoichiometric data on product secretion (Table 1) and (ii) stoichiometric data on anabolic demand for biomass precursors (Table 2). The set of intracellular flux data that gave the minimum deviation between experimental and simulated labeling patterns was taken as the best estimate for intracellular flux distribution. For both scenarios, identical flux distributions were obtained with multiple initialization values, suggesting that global minima were identified. Obviously, good agreement between experimentally determined and calculated mass isotopomer ratios was achieved (Table 3).
TABLE 3.
Relative mass isotopomer fractions of secreted lysine and trehalose of lysine-producing Corynebacterium glutamicum ATCC 21526 cultivated on glucose or fructosea
| Substrate | Lysine on 1-13C-labeled substrate
|
Trehalose on 1-13C-labeled substrate
|
Trehalose on 50% 13C6-labeled substrate
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M0 | M1 | M2 | M3 | M4 | M0 | M1 | M2 | M3 | M4 | M0 | M1 | M2 | M3 | M4 | M5 | M6 | |
| Glucose | |||||||||||||||||
| Exp | 0.234 | 0.360 | 0.247 | 0.110 | 0.037 | 0.110 | 0.551 | 0.216 | 0.094 | 0.023 | 0.271 | 0.114 | 0.087 | 0.115 | 0.069 | 0.066 | 0.279 |
| Calc | 0.242 | 0.355 | 0.245 | 0.110 | 0.037 | 0.114 | 0.549 | 0.212 | 0.094 | 0.023 | 0.268 | 0.113 | 0.085 | 0.113 | 0.068 | 0.064 | 0.289 |
| Fructose | |||||||||||||||||
| Exp | 0.133 | 0.316 | 0.304 | 0.162 | 0.062 | 0.212 | 0.412 | 0.244 | 0.092 | 0.030 | 0.141 | 0.103 | 0.104 | 0.250 | 0.133 | 0.110 | 0.159 |
| Calc | 0.139 | 0.321 | 0.298 | 0.159 | 0.061 | 0.195 | 0.419 | 0.254 | 0.094 | 0.030 | 0.144 | 0.103 | 0.102 | 0.245 | 0.131 | 0.111 | 0.164 |
For both carbon sources, two parallel tracer experiments on (i) 1-13C-labeled and (ii) a 1:1 mixture of naturally 13C-labeled and 13C6-labeled tracer substrate were carried out. Exp, GC-MS data; Calc, values predicted by the solution of the mathematical model corresponding to the optimized set of fluxes. M0 denotes the relative amount of nonlabeled mass isotopomer fraction, M1 denotes the relative amount of single-labeled mass isotopomer fraction, and the corresponding terms stand for higher labeling.
Metabolic flux on fructose or glucose during lysine production.
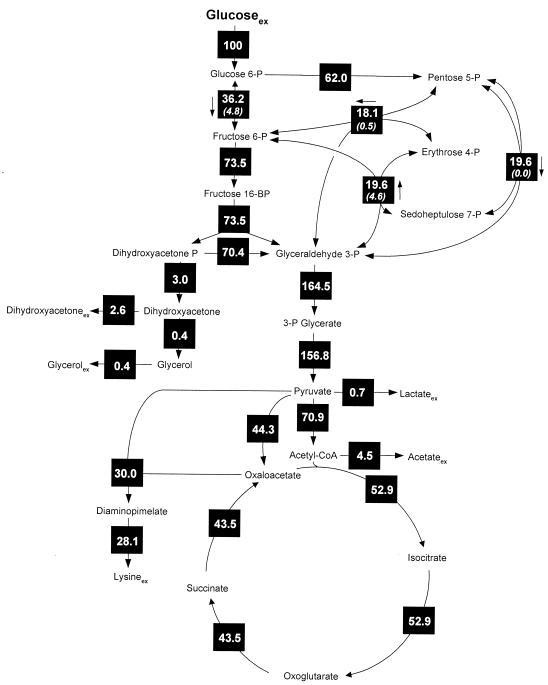
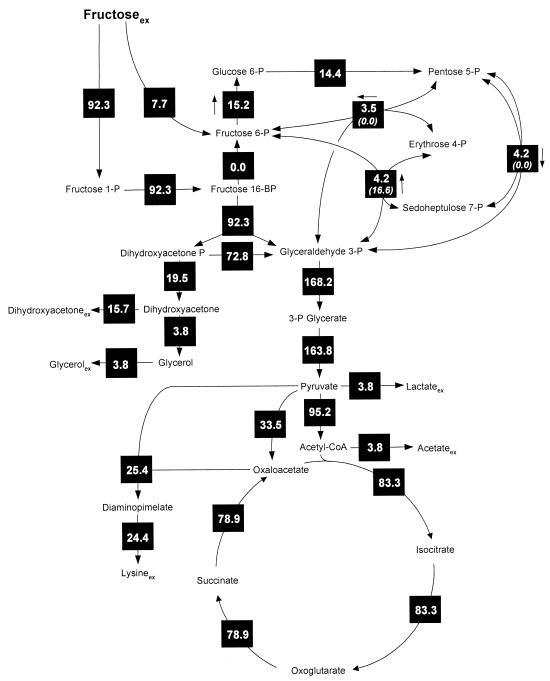
The intracellular flux distributions obtained for lysine-producing C. glutamicum on glucose or fructose are shown in Fig. 2 and 3. Obviously, the direction of intracellular flux differed tremendously depending on the carbon source applied. On glucose, 62% of the carbon flux was directed towards the PPP, whereas only 36% was channeled through the glycolytic chain (Fig. 2), which resulted in a relatively high amount (124%) of NADPH being generated by the PPP enzymes glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase. The intracellular flux distribution on fructose was completely different (Fig. 3). The flux analysis performed revealed the in vivo activity of two PTS for uptake of fructose, whereby 92.3% of the fructose was taken up by fructose-specific PTSFructose. A comparably small fraction, 7.7%, of fructose was taken up by PTSMannose. Thus, the majority of fructose entered glycolysis at the level of fructose 1,6-bisphosphate, whereas only a small fraction was channeled upstream at the level of fructose 6-phosphate into the glycolytic chain. The PPP of fructose-grown cells exhibited an activity of only 14.4%, dramatically reduced in comparison to that for glucose-grown cells. Glucose 6-phosphate isomerase operated in opposite directions on the two carbon sources. In glucose-grown cells, 36.2% of the net flux was directed from glucose 6-phosphate to fructose 6-phosphate, whereas a backward net flux of 15.2% was observed on fructose.
FIG. 2.
In vivo carbon flux distribution in the central metabolism of Corynebacterium glutamicum ATCC 21526 during lysine production on glucose estimated from the best fit to the experimental results using a comprehensive approach of combined metabolite balancing and isotopomer modeling for 13C tracer experiments with labeling measurements of secreted lysine and trehalose by GC-MS. Net fluxes are given in square symbols, and for reversible reactions the direction of the net flux is indicated by an arrow next to the corresponding black box. The numbers in parentheses below the fluxes of transaldolase, transketolase, and glucose 6-phosphate isomerase indicate flux reversibilities. All fluxes are expressed as molar percentages of the mean specific glucose uptake rate (1.77 mmol g−1 h−1). Acetyl-CoA, acetyl coenzyme A.
FIG. 3.
In vivo carbon flux distribution in the central metabolism of Corynebacterium glutamicum ATCC 21526 during lysine production on fructose estimated from the best fit to the experimental results using a comprehensive approach of combined metabolite balancing and isotopomer modeling for 13C tracer experiments with labeling measurement of secreted lysine and trehalose by GC-MS. Net fluxes are given in square symbols, and for reversible reactions the direction of the net flux is indicated by an arrow next to the corresponding black box. The numbers in brackets below the fluxes of transaldolase, transketolase, and glucose 6-phosphate isomerase indicate flux reversibilities. All fluxes are expressed as molar percentages of the mean specific fructose uptake rate (1.93 mmol g−1 h−1). Acetyl-CoA, acetyl coenzyme A.
On fructose flux through glucose 6-phosphate isomerase and PPP was about twice as high as that through the PTSMannose. However, this difference was not due to a gluconeogenetic flux of carbon from fructose 1,6-bisphosphate to fructose 6-phosphate, which could have supplied extra carbon flux towards the PPP. In fact, flux through fructose 1,6-bisphosphatase to catalyze this reaction was zero. The metabolic reactions responsible for the additional flux toward the PPP are those of the reversible enzymes transaldolase and transketolase in the PPP. About 3.5% of this additional flux was supplied by transketolase 2, which recycled carbon stemming from the PPP back into this pathway. Moreover, 4.2% of flux was directed towards fructose 6-phosphate and the PPP by the action of transaldolase.
Depending on the carbon source, completely different flux patterns in lysine-producing C. glutamicum were also observed around the pyruvate node (Fig. 2 and 3). On glucose flux into the lysine pathway was 30.0%, whereas a reduced flux (25.4%) was found on fructose. The elevated lysine yield on glucose compared to that for fructose is the major reason for this flux difference, but the higher biomass yield resulting in higher demands for diaminopimelate for cell wall synthesis and lysine for protein synthesis also contributes to it. Anaplerotic flux on glucose was 44.5% and was thus markedly higher than flux on fructose (33.5%). This difference is due mainly to the higher demand for oxaloacetate for lysine production but also to the higher anabolic demands for oxaloacetate and 2-oxoglutarate on glucose. On the other hand, flux through pyruvate dehydrogenase was substantially lower on glucose (70.9%) than on fructose (95.2%). This reduced carbon flux into the TCA cycle resulted in flux that was reduced more than 30% through TCA cycle enzymes on glucose (Fig. 2 and 3).
Statistical evaluation by a Monte-Carlo approach of the results obtained for flux was used to calculate 90% confidence intervals for the determined flux parameters. As shown for various key fluxes in Table 4, the confidence intervals were generally narrow. For example, the confidence interval for flux through glucose 6-phosphate dehydrogenase was only 1.2% for glucose-grown cells and 3.5% for fructose-grown cells. The chosen approach therefore allowed precise estimation of flux. We concluded that the differences in flux observed on glucose and fructose are clearly caused by the applied carbon source.
TABLE 4.
Statistical evaluation of metabolic fluxes of lysine-producing Corynebacterium glutamicum ATCC 21526 grown on fructose or glucosea
| Flux parameter | Glucose | Fructose |
|---|---|---|
| Net flux | ||
| Fructose uptake by PTSFructose | NAd | 90.0-96.1 |
| Fructose uptake by PTSMannose | NAd | 3.9-10.0 |
| Glucose 6-phosphate isomerase | 35.7-36.8 | 13.4-16.9 |
| Phosphofructokinaseb | 35.7-36.8 | NAd |
| Fructose 1,6-bisphosphatase | NAd | −2.1-3.4 |
| Fructose 1,6-bisphosphate aldolase | 73.7-73.8 | 91.7-92.9 |
| Glucose 6-phosphate dehydrogenase | 62.5-63.7 | 12.6-16.1 |
| Transaldolase | 19.4-19.8 | 3.6-4.1 |
| Transketolase 1 | 19.4-19.8 | 3.6-4.1 |
| Transketolase 2 | 17.9-18.3 | 2.9-4.0 |
| Glyceraldehyde 3-phosphate dehydrogenase | 158.1-164.5 | 163.3-174.6 |
| Pyruvate kinase | 156.2-167.4 | 158.9-168.2 |
| Pyruvate dehydrogenase | 69.5-72.5 | 87.1-102.3 |
| Pyruvate carboxylase | 43.7-44.8 | 29.9-37.3 |
| Citrate synthase | 51.2-54.8 | 76.5-91.5 |
| Isocitrate dehydrogenase | 51.2-54.8 | 76.5-91.5 |
| Oxoglutarate dehydrogenase | 41.6-45.6 | 70.9-86.0 |
| Aspartokinase | 29.6-30.3 | 21.8-29.2 |
| Flux reversibilityc | ||
| Glucose 6-phosphate isomerase | 4.5-5.1 | NAd |
| Transaldolase | 4.3-4.9 | 14.5-18.2 |
| Transketolase 1 | 0.0-0.0 | 0.0-0.1 |
| Transketolase 2 | 0.4-0.6 | 0.0-0.1 |
Values were determined by 13C tracer studies with MS and metabolite balancing. Ninety percent confidence intervals of key flux parameters were obtained by a Monte-Carlo approach, including 100 independent parameter estimation runs for each substrate with statistically varied experimental data.
The negative flux for the lower confidence boundary is equal to a positive flux in the reverse direction (through phosphofructokinase).
Flux reversibility is defined as the ratio of back flux to net flux.
NA, not applicable.
Note that the mean specific substrate uptake of 1.93 mmol g−1 h−1 on fructose was slightly higher than that of 1.77 mmol g−1 h−1 found on glucose. Due to this difference, absolute intracellular flux expressed in mmol g−1 h−1 is slightly increased in relation to that for glucose compared to the relative flux values discussed above. The flux distributions of lysine-producing C. glutamicum on fructose and glucose are, however, so completely different that all comparisons drawn above also hold for absolute carbon flux.
DISCUSSION
Substrate-specific culture characteristics.
Cultivation of lysine-producing C. glutamicum on fructose and on glucose revealed that growth and product formation strongly depend on the carbon source applied. Significantly reduced yields of lysine and biomass on fructose were reported previously for another strain of C. glutamicum, where lysine and biomass yield were 30 and 20% less, respectively, than those for glucose (7). Cultivation of C. glutamicum and C. melassecola on fructose is linked to higher carbon dioxide production rates than those for glucose (5, 7), which coincides with the elevated level of flux through the TCA cycle observed in the present work for this carbon source. Substrate-specific differences were also observed for by-products. The formation of trehalose was lower on fructose than on glucose and might be related to different entry points into glycolysis for glucose and fructose (7). Because of the uptake systems in C. glutamicum, utilization of glucose leads to the formation of the trehalose precursor glucose 6-phosphate, whereas fructose is converted into fructose 1,6-bisphosphate and thus enters the central metabolism downstream from glucose 6-phosphate (5). The levels of other by-products, such as dihydroxyacetone, glycerol, and lactate, were strongly increased when fructose was applied as the carbon source. From the viewpoint of lysine production, this result is not desired, because a substantial fraction of carbon is withdrawn from the central metabolism into the formed by-products. The specific-substrate uptake on fructose (1.93 mmol g−1 h−1) was higher than on glucose (1.77 mmol g−1 h−1). This result differs from that of a previous study on exponentially growing C. melassecola ATCC 17965 (5), where similar specific uptake rates on fructose and glucose were observed. The higher uptake rate for fructose observed in our study might be due to the fact that the studied strains are different. C. melassecola and C. glutamicum are related species but might differ in certain metabolic properties. The strain studied in the present work was previously derived by classical strain optimization, which may have introduced mutations influencing substrate uptake. Another explanation is the difference in cultivation conditions. Fructose might be more effectively utilized under conditions of limited growth and lysine production.
Metabolic flux distributions.
The intracellular flux distributions obtained for lysine-producing C. glutamicum on glucose and fructose revealed tremendous differences. Statistical evaluation of the flux distributions obtained revealed narrow 90% confidence intervals, so that the observed flux differences can be clearly attributed to the applied carbon sources. One of the most remarkable differences concerns flux partitioning between glycolysis and PPP. On glucose, 62.3% of carbon was channeled through the PPP. The predominance of the PPP of lysine-producing C. glutamicum grown on this substrate has been previously observed in different studies (9, 19, 20). On fructose the flux into the PPP was reduced to 14.4%. As identified by the metabolic flux analysis performed, this reduction was due mainly to the unfavorable combination of the entry of fructose at the level of fructose 1,6-bisphospate and the inactivity of fructose 1,6-bisphosphatase. The observed inactivity of fructose 1,6-bisphosphatase agrees well with enzymatic measurements of C. melassecola ATCC 17965 during exponential growth on fructose and on glucose (5).
Surprisingly, flux through glucose 6-phosphate isomerase and PPP was about twice as high as flux through the PTSMannose when C. glutamicum was cultivated on fructose. Due to the inactivity of fructose 1,6-bisphosphatase, this difference was not caused by a gluconeogenetic flux. In fact, C. glutamicum possesses an operating metabolic cycle via fructose 6-phosphate, glucose 6-phosphate, and ribose 5-phosphate. Additional flux into the PPP was supplied by transketolase 2, which recycled carbon stemming from the PPP back into this pathway, and by the action of transaldolase, which redirected glyceraldehyde 3-phosphate back into the PPP, thus bypassing gluconeogenesis. This cycling activity may help the cell to overcome the limitation of NADPH on fructose. The drastically reduced flux at glucose 6-phosphate for fructose-grown C. glutamicum might also explain the reduced formation of trehalose on this substrate (7). Glucose 6-phosphate isomerase operated in opposite directions depending on the carbon source. In glucose-grown cells, net flux was directed from glucose 6-phosphate to fructose 6-phosphate, whereas an inverse net flux was observed in fructose-grown cells. This finding underlines the importance of the reversibility of this enzyme for metabolic flexibility in C. glutamicum.
NADPH metabolism.
The following calculations provide a comparison of the NADPH metabolism of lysine-producing C. glutamicum on fructose and glucose. The overall supply of NADPH was calculated from the estimated level of flux through glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and isocitrate dehydrogenase. On glucose, the PPP enzymes glucose 6-phosphate dehydrogenase (62.0%) and glucose 6-phosphate dehydrogenase (62.0%) supplied the major fraction of NADPH. Isocitrate dehydrogenase (52.9%) contributed to only a small extent. A completely different contribution of the PPP and TCA cycle to the NADPH supply was observed on fructose, where isocitrate dehydrogenase (83.3%) was the major source of NADPH. Glucose 6-phosphate dehydrogenase (14.4%) produced much less NADPH on fructose. NADPH is required for growth and formation of lysine. The NADPH requirement for growth was calculated from a stoichiometric demand of 11.51 mmol of NAPDH (g of biomass−1), which was assumed to be identical for glucose and fructose (5), and the experimental biomass yield of the present work (Table 1). C. glutamicum consumed 62.3% of NADPH for biomass production on glucose, which was much higher than that consumed on fructose as the carbon source (32.8%). The amount of NADPH required for product synthesis was determined from the estimated level of flux into lysine (Table 1) and the corresponding stoichiometric NADPH demand of 4 mol (mol of lysine−1) and was 112.4% for lysine production from glucose and 97.6% for lysine production from fructose. The overall NADPH supply on glucose was significantly higher (176.9%) than that for fructose (112.1%), which can be attributed mainly to the increased PPP flux on glucose. The NADPH balance was almost closed on glucose. In contrast, a significant apparent deficiency of NADPH (18.3%) was observed on fructose. This finding raises the question of what enzymes, in addition to the above-mentioned enzymes, glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and isocitrate dehydrogenase, might catalyze metabolic reactions that may supply NADPH. A likely candidate seems to be NADPH-dependent malic enzyme. Previously, an increased specific activity of this enzyme was detected on fructose-grown C. melassecola in comparison to that for glucose-grown cells (5). However, the flux through this particular enzyme could not be resolved by the experimental setup in the present work. Assuming malic enzyme is the missing NADPH-generating enzyme, a flux level of 18.3% would be sufficient to supply the apparently missing NADPH. Detailed flux studies of C. glutamicum with glucose as the carbon source revealed no significant activity of malic enzyme (12). The results for fructose might, however, be coupled to elevated in vivo activity of this enzyme.
NADH metabolism.
On fructose, C. glutamicum revealed increased activity of NADH-forming enzymes. Glyceraldehyde 3-phosphate dehydrogenase, pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase, and malate dehydrogenase formed 421.2% NADH on fructose. On glucose the NADH production was only 322.4%. Additionally, the anabolic NADH demand was significantly lower on fructose than on glucose. The significantly enhanced NADH production coupled to a reduced metabolic demand could lead to an increased NADH/NAD ratio. For C. melassecola, it was previously shown that fructose leads to an increased NADH/NAD ratio compared to that for glucose (5). This finding raises a question about NADH-regenerating mechanisms during lysine production on fructose. Fructose-grown cells exhibited an enhanced secretion of dihydroxyacetone, glycerol, and lactate. The increased formation of dihydroxyacetone and glycerol could be due to a higher NADH/NAD ratio. NADH was previously shown to inhibit glyceraldehyde dehydrogenase, so overflow of dihydroxyacetone and glycerol might be related to a reduction of the flux capacity of this enzyme. The reduction of dihydroxyacetone to glycerol could additionally be favored by the high NADH/NAD ratio and thus contribute to regeneration of excess NADH. The NADH-demanding lactate formation from pyruvate could have a background similar to that for the production of glycerol. In comparison to that for exponential growth, the excess of NADH under lysine-producing conditions characterized by relatively high TCA cycle activity and reduced biomass yield might be increased.
Potential targets for optimization of lysine-producing C. glutamicum on fructose.
Based on the flux patterns obtained, several potential targets for the optimization of lysine production by C. glutamicum on fructose can be formulated. A central point surely is the supply of NADPH. Fructose 1,6-bisphosphatase displays an interesting target in order to increase the supply of NADPH. Amplification of its activity might lead to a higher level of flux through the PPP, resulting in increased NADPH generation and increased lysine yield. An increase of the level of flux through the PPP via amplification of fructose 1,6-bisphosphatase might also be beneficial for aromatic amino acid production (6). The inactivity of fructose 1,6-bisphosphatase during growth on fructose is surely bad from the viewpoint of lysine production but not too surprising, because this gluconeogenetic enzyme is not required during growth on sugars and is probably suppressed. In prokaryotes this enzyme is under efficient metabolic control by, e.g., fructose 1,6-bisphosphate, fructose-2,6 bisphosphate, metal ions, and AMP (17). It is known that C. glutamicum can grow on acetate (18), where this enzyme is essential to maintain gluconeogenesis. Another potential target to increase the level of flux through the PPP is the PTS for fructose uptake. Modification of flux partitioning between PTSFructose and PTSMannose could yield a higher proportion of fructose, which enters at the level of fructose 6-phosphate and thus also leads to an increased level of flux through the PPP. Additionally, amplification of malic enzyme, which probably contributes significantly to the NADPH supply on fructose, could be an interesting target.
Another bottleneck is the high secretion levels of dihydroxyacetone, glycerol, and lactate. The formation of dihydroxyacetone and glycerol could be blocked by deletion of the corresponding enzymes. The conversion of dihydroxyacetone phosphate to dihydroxyacetone could be catalyzed by a corresponding phosphatase. A dihydroxyacetone phosphatase has, however, not yet been annotated in C. glutamicum (http://www3.ncbi.nlm.nih.gov/Taxonomy/). Theoretically, this reaction could also be catalyzed by a kinase. Presently, two entries in the genome database of C. glutamicum relate to dihydroxyacetone kinase (http://www3.ncbi.nlm.nih.gov/Taxonomy/). Lactate secretion could be avoided by knockout of lactate dehydrogenase. Since glycerol and lactate formation could be important for NADH regeneration, negative effects on the overall performance of the organism can, however, not be excluded. In case carbon flux through the lower glycolytic chain is limited by the capacity of glyceraldehyde 3-phosphate dehydrogenase as previously speculated (5), the suppression of dihydroxyacetone and glycerol production could eventually lead to an activation of fructose 1,6-bisphosphatase and a redirection of carbon flux through the PPP. Note that dihydroxyacetone is not reutilized during the cultivation of C. glutamicum and thus displays wasted carbon with respect to product synthesis, whereas this is not the case for lactate (2).
The results obtained in this work for fructose also have some relevance for sucrose as the carbon source for lysine production by C. glutamicum. Sucrose is the major carbon source in molasses. As shown previously, the fructose unit of sucrose enters glycolysis at the level of fructose 1,6-bisphosphate (4). Therefore, this part of the sucrose molecule—assuming an inactive fructose 1,6-bisphosphatase as found in the present study—probably does not enter into the PPP, so that the supply of NADPH in lysine-producing strains may be limited.
APPENDIX
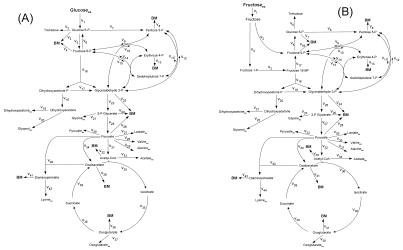
The following calculations show that the two networks of glucose-grown and fructose-grown cells were overdetermined, so that a least-squares approach was possible in the parameter estimation.
Metabolic network of glucose-grown cells. The network for the central metabolism of C. glutamicum on glucose is shown in Fig. A1A. In total it comprises 42 fluxes. Of these, 12 are directly accessible via quantification of the secretion of 11 products (v2, v19, v20, v21, v27, v28, v29, v30, v33, v37, v42) and the uptake of glucose (v1). Taking biomass composition and measured biomass yield into account, a further 11 fluxes from anabolic precursors into biomass (v3, v4, v8, v13, v22, v24, v26, v32, v36, v39, v41) can be estimated. In addition, the following 14 metabolite balances can be formulated (equations 2 through 15):
 |
(2) |
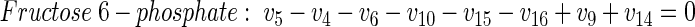 |
(3) |
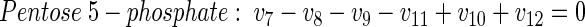 |
(4) |
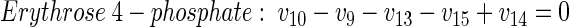 |
(5) |
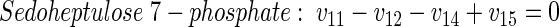 |
(6) |
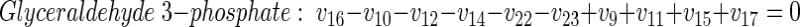 |
(7) |
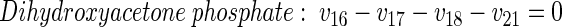 |
(8) |
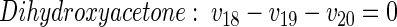 |
(9) |
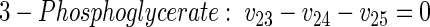 |
(10) |
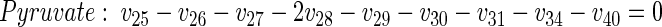 |
(11) |
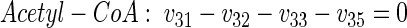 |
(12) |
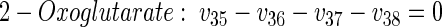 |
(13) |
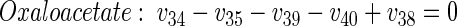 |
(14) |
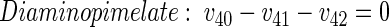 |
(15) |
FIG.A1.
Metabolic network of the central metabolism of glucose-grown (A) and fructose-grown (B) lysine-producing Corynebacterium glutamicum, including transport fluxes, anabolic fluxes, and fluxes between intermediary metabolite pools.
The rank of the corresponding stoichiometric matrix was calculated as 14 using the software Matlab, showing that all metabolite balances were linearly independent. Together with 14 sets of labeling data contained in the 17 measured mass isotopomer fractions of trehalose and lysine from the parallel tracer experiments, 51 determinations were available. Thus, the network was overdetermined, and the calculation of values for the entire set of 42 fluxes could be realized by a least-squares approach.
Metabolic network of fructose-grown cells. The operation of the network of central metabolism of C. glutamicum on fructose is rather similar to that on glucose. In total, it comprises 44 fluxes (Fig. A1B). Of these, 12 could be directly measured via the secretion of products (v5, v21, v22, v23, v29, v30, v31, v33, v34, v38, v43) and the uptake of fructose (v1). Eleven anabolic fluxes were accessible (v7, v8, v13, v16, v24, v25, v28, v35, v37, v41, v43). In addition, the following 16 metabolite balances can be formulated for this network (equations 16 through 31):
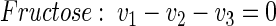 |
(16) |
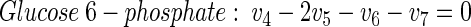 |
(17) |
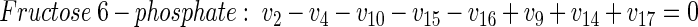 |
(18) |
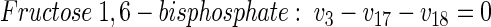 |
(19) |
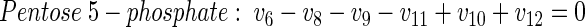 |
(20) |
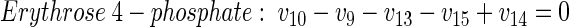 |
(21) |
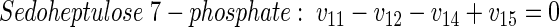 |
(22) |
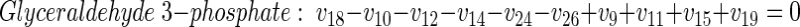 |
(23) |
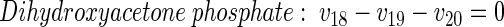 |
(24) |
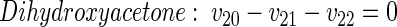 |
(25) |
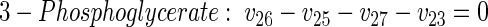 |
(26) |
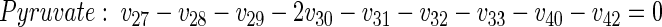 |
(27) |
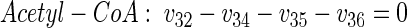 |
(28) |
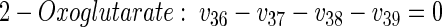 |
(29) |
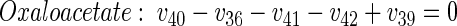 |
(30) |
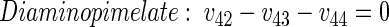 |
(31) |
The rank of the corresponding stoichiometric matrix was calculated as 16 using Matlab, showing that all metabolite balances were linearly independent. Together with 14 labeling data, 53 sets of data were available, resulting in an overdetermined network.
Acknowledgments
We appreciate the valuable assistance of Michel Fritz with mass spectrometric analysis.
This work was supported by BASF AG (Ludwigshafen, Germany).
REFERENCES
- 1.Cocaign, M., C. Monnet, and N. D. Lindley. 1993. Batch kinetics in Corynebacterium glutamicum during growth on various carbon substrates: use of substrate mixtures to localize metabolic bottlenecks. Appl. Microbiol. Biotechnol. 40:526-530. [Google Scholar]
- 2.Cocaign-Bousquet, M., and N. D. Lindley. 1995. Pyruvate overflow and carbon flux within the central metabolic pathways of Corynebacterium glutamicum during growth on lactate. Enz. Microbiol. Technol. 17:260-267. [Google Scholar]
- 3.de Graaf, A. A. 2000. Metabolic flux analysis of Corynebacterium glutamicum, p. 506-555. In: K. Schügerl and K. H. Bellgardt (ed.), Bioreaction engineering. Springer Verlag, Berlin, Germany.
- 4.Dominguez, H., and N. D. Lindley. 1996. Complete sucrose metabolism requires fructose phosphotransferase activity in Corynebacterium glutamicum to ensure phosphorylation of liberated fructose. Appl. Environ. Microbiol. 62:3878-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez, H., C. Rollin, A. Guyonvarch, J. L. Guerquin-Kern, M. Cocaign-Bousquet, and N. D. Lindley. 1998. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur. J. Biochem. 254:96-102. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda, M. 2003. Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 79:1-36. [DOI] [PubMed] [Google Scholar]
- 7.Kiefer, P., E. Heinzle, and C. Wittmann. 2002. Influence of glucose, fructose and sucrose on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 28:338-343. [DOI] [PubMed] [Google Scholar]
- 8.Malin, G. M., and G. I. Bourd. 1991. Phosphotransferase-dependent glucose transport in Corynebacterium glutamicum. J. Appl. Bacteriol. 71:517-523. [Google Scholar]
- 9.Marx, A., A. A. de Graaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi, J., S. Mitsuhashi, M. Hayashi, S. Ando, H. Yokoi, K. Ochiai, and M. A. Ikeda. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine producing mutant. Appl. Microbiol. Biotechnol. 58:217-223. [DOI] [PubMed] [Google Scholar]
- 11.Pelechova, J., J. Smekal, V. Koura, J. Plachy, and V. Krumphanzl. 1980. Biosynthesis of l-lysine in Corynebacterium glutamicum on sucrose, ethanol and acetic acid. Folia Microbiol. 25:341-346. [DOI] [PubMed] [Google Scholar]
- 12.Petersen, S., A. A. de Graaf, L. Eggeling, M. Möllney, W. Wiechert, and H. Sahm. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 75:35932-35941. [DOI] [PubMed] [Google Scholar]
- 13.Petersson, G. 1974. Gas-chromatographic analysis of sugars and related hydroxyacids as acyclic oxime and ester trimethylsilyl derivates. Carbohydr. Res. 33:47-61. [Google Scholar]
- 14.Rubino, F. M. 1989. Silylaldonitrile derivates for the determination of sugars by gas chromatography-mass spectrometry. J. Chromatogr. 473:125-133. [Google Scholar]
- 15.Sahm, H., L. Eggeling, and A. A. de Graaf. 2000. Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381:899-910. [DOI] [PubMed] [Google Scholar]
- 16.Shiio, I., S. Sugimoto, and K. Kawamura. 1990. Effects of carbon source sugars on the yield of amino acid production and sucrose metabolism in Brevibacterium flavum. Agric. Biol. Chem. 54:1513-1519. [Google Scholar]
- 17.Skrypal, I. G., and O. V. Iastrebova. 2002. Fructose-bisphosphatase of microorganisms. Mikrobiol. ZH. 64:82-94. [PubMed] [Google Scholar]
- 18.Wendisch, V. F., A. A. de Graaf, H. Sahm, and B. Eikmans. 2000. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 182:3088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittmann, C., and E. Heinzle. 2001. Application of MALDI-TOF MS to lysine-producing Corynebacterium glutamicum: a novel approach for metabolic flux analysis. Eur. J. Biochem. 268:2441-2455. [DOI] [PubMed] [Google Scholar]
- 20.Wittmann, C., and E. Heinzle. 2002. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl. Environ. Microbiol. 68:5843-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittmann, C., M. Hans, and E. Heinzle. 2002. In vivo analysis of intracellular amino acid labelings by GC/MS. Anal. Biochem. 307:379-382. [DOI] [PubMed] [Google Scholar]
- 22.Wittmann, C., H. M. Kim, and E. Heinzle. Metabolic network analysis of lysine-producing Corynebacterium glutamicum at miniaturized scale. Biotechnol. Bioeng., in press. [DOI] [PubMed]