Abstract
The prevalence among all Escherichia coli bacteria of the LTIIa toxin gene and STII toxin gene, both associated with enterotoxigenic E. coli, and of three genes (stxI, stxII, and eaeA) associated with enterohemorrhagic E. coli was determined in farm waste disposal systems seasonally for 1 year. Single- and nested-PCR results for the number of E. coli isolates carrying each toxin gene trait were compared with a five-replicate most-probable-number (MPN) method. The STII and LTIIa toxin genes were present continuously at all farms and downstream waters that were tested. Nested-MPN-PCR manifested sensitivity increased over that of single-MPN-PCR by a factor of 32 for LTIIa, 10 for STII, and 2 for the stxI, stxII, and eaeA genes. The geometric mean prevalence of each toxin gene within the E. coli community in waste disposal site waters after nested MPN-PCR was 1:8.5 E. coli isolates (1:8.5 E. coli) for the LTIIa toxin gene and 1:4 E. coli for the STII toxin gene. The geometric mean prevalence for the simultaneous occurrence of toxin genes stxI, stxII, and eaeA, was 1:182 E. coli. These findings based on total population analysis suggest that prevalence rates for these genes are higher than previously reported in studies based on surveys of single isolates. With a population-based approach, the frequency of each toxin gene at the corresponding disposal sites and the endemic nature of diseases on farms can be easily assessed, allowing farmers and public health officials to evaluate the risk of infection to animals or humans.
Enterotoxigenic Escherichia coli (ETEC) has been known to cause diarrheal diseases in both animals and humans. The LTIIa and STII toxin genes associated with ETEC are responsible for diarrheal diseases affecting calves and piglets, respectively (14, 30, 41, 51). Outbreaks, such as mastitis in cows or edema in piglets, caused by these agents have led to economic losses (14, 51). The bacteria harboring these toxin genes have the ability to colonize the small intestine of humans and animals, demonstrating host specificity through colonization factors. Two colonization factors, K88 and K99, have been found to be important in ETEC colonization of the intestinal epithelia of pigs and cows, respectively (14, 21). Detection and treatment or vaccination have been two directions used to reduce disease caused by these organisms in agriculture (52). However, most studies focus on individual animals, screening isolates from fecal swabs as opposed to screening populations of E. coli from a herd of animals (14, 29, 49). This could limit detection because relatively few clones can be screened, and perhaps biases result in a manner that underestimates the carrier rates within herds.
Contamination of drinking or recreational waters with enterohemorrhagic E. coli (EHEC) has resulted in an increase in the number of outbreaks and deaths from hemolytic-uremic syndrome in humans (11, 28). Found in the gastrointestinal tract of cows, EHEC, which can carry stxI, stxII, and eaeA virulence genes, is transmitted by food or water supplies polluted with cow fecal waste. An outbreak in Wyoming during the summer of 1998 resulted in 157 people becoming ill from the consumption of spring water contaminated with E. coli O157:H7, a strain carrying the stxI, stxII, and eaeA genes (28). Continuing outbreaks, due primarily to contamination of beef with O157:H7, have led to 4,528 cases of reported illness in the United States in 2002 (6), illustrating the endemic nature of this pathogen in cow-raising and dairy facilities (24). A variety of public health benefits could be derived from assessing the status of pathogen frequency in herds prior to shipment to slaughterhouses or the use of waste for manure spreading (4, 8, 17), which can place ground or surface waters at risk for introduction of this pathogen. Most studies to assess the occurrence of EHEC on farms screen individual animals using fecal swabs (12, 34).
A review of the agricultural and veterinary literature indicated that earlier work examined individual waste samples of pigs, comparing the prevalence of STII with three other enterotoxins within ETEC, and found that 74% of diarrheic pigs carried STII (26). Sampling of individual healthy animals has shown these traits to be less common, as indicated in studies by Celemin et al. (5), where STII occurred in 3% of E. coli isolates from healthy piglets, and by Osek (30), where E. coli carrying STII was not detected in healthy piglets. Both diseases appear to decrease with age, but little is known about the frequency of toxin gene occurrence in the waste disposal sites for these animals. A multiplex PCR study (12) of 17,050 single isolates from cow fecal samples found that 0.08% were E. coli O157:H7 and carried stxI, stxII, and eaeA. Also, using single isolates, those researchers reported no detection of O157:H7 genotypes in 312 cow waste lagoons tested. Other work that examined the occurrence of stxI, stxII, and eaeA genes in isolates of E. coli from cow rectal fecal swabs showed the presence of these genes and that prevalence was highest during the summer season (23, 31).
This investigation was initiated to determine the frequency of enterotoxigenic and enterohemorrhagic toxin-associated genotypes among E. coli from cow and pig waste disposal facilities. A major aim of this research was to determine the prevalence and whether nested PCR increased detection of these toxin genes within the E. coli populations of each waste treatment system. This research used a five-membrane filter most-probable-number (MPN) method coupled with single or nested PCR to determine the prevalence within E. coli populations from waste disposal sites of the two enterotoxin genes, LTIIa and STII, and the three genes (stxI and stxII [Shiga toxins] and eaeA) associated with EHEC, thus designating these toxin genes as biomarkers of disease potential.
MATERIALS AND METHODS
Sample collection.
Twenty (250-ml) samples from five cow waste lagoons, six (250-ml) samples from five pig waste lagoons, and two (6-liter) samples from a stream located downstream from a pig farm were collected. From each lagoon at five different cow farms throughout California, one sample was collected for each season over a 1-year period. The stxI and -II and eaeA genes were not screened seasonally. Herd size ranged from 300 to 3,000 on these farms. Pig waste lagoon water samples were from farms in North Carolina, Colorado, and Iowa, and herd size for these farms ranged from 80 to 8,200. Seasonal collection for swine farms was not possible due to inconsistencies of farmer participation, resulting in six fall samples and two summer samples screened for STII. Upon collection, all samples were stored on ice, shipped overnight, and processed within 8 h of arrival.
Bacterial strains.
ETEC strains containing the pTC201 plasmid or the pRAS-1 plasmid (19, 20) were used as positive controls for the LTIIa and STII toxin genes, respectively. EHEC strain 4718 donated by V. K. Sharma was used as the positive control for the stxI, stxII, and eaeA genes. E. coli control strains were grown with agitation (150 rpm; model 420; Orbital Shaker, Marietta, Ohio) overnight in Luria-Bertani media; only ETEC strains were supplemented with 20 μg of ampicillin/ml. Each strain was stored at −70°C in 30% sterile glycerol. The negative control used for hybridization was Aeromonas hydrophila (ATCC 7966), which was grown on ampicillin dextrin agar overnight at 35°C. The DNA extract was stored at −70°C until confirmation by hybridization.
Membrane filtration.
Dilutions of 100 to 10−3 ml, except in the case of stream water where volumes of 0.1 to 1,000 ml were used, were filtered through a 0.45-μm-pore-size (47 mm in diameter) nylon membrane filter (Osmonics). Each filter was placed on mTEC agar (Difco), and contents were incubated at 35°C for 1.5 h, followed by 44.5°C for 20 ± 2 h (9). Yellow colonies were enumerated, followed by flooding with 1 ml of phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, and 1.4 mM KH2PO4 [pH 7.3]). All colonies on each filter were suspended in the buffer with a sterile rubber policeman and were pipetted into 1.5-ml tubes. Afterward, tubes were centrifuged for 10 min at 12,000 × g, the supernatant was discarded, and pelleted cells were kept for DNA extraction.
DNA extraction.
Two DNA extraction methods were used. The first extraction procedure was a modified version of the glass bead method (42). The modifications were the addition of 53 μl of 10% cetyltrimethylammonium bromide in 0.7 M NaCl and 8 μl of 0.5 M NaCl to the lysis buffer after 1 ml of guanidine thiocyanate (5.3 M guanidine thiocyanate, 10 mM dithiothreitol, 1% Tween 20, 0.3 M sodium acetate, and 50 mM sodium citrate [pH 7.0]) had been added.
The phenol-chloroform extraction method (45) used omitted the addition of Tris-HCl-saturated phenol to the DNA after the freeze-thaw cycles. An additional wash step was added after the isopropanol precipitation of the DNA. The DNA was washed in 200 μl of 75% ethanol (reagent grade), kept at −20°C for 1 h, and processed as described in the method. The extracted DNA without further purification was stored at −50°C until PCR analysis.
PCR amplification.
Outer primers for the LTIIa toxin gene (19) and inner primers for the STII (20) toxin gene, shown in Table 1, were obtained from previous research. The specificity of primers was determined in these earlier studies. A second set of primers for each toxin trait was developed for nested PCR (Table 1). These primers were tested for cross-reactivity by screening all sequences contained in The Institute for Genomic Research (http://www.tigr.org/tdb/) and GenBank (http://www.ncbi.nlm.nih.gov/) by using BLAST (1). Amplification was performed by using 1 to 10 μl of DNA sample extract for each 50-μl reaction as recommended by the manufacturer (Perkin-Elmer or Promega). Thirty PCR cycles (95°C for 30 s, 61°C [LTIIa] for 30 s or 57°C [STII] for 30 s, and 72°C for 30 s) were carried out in a Perkin-Elmer (model 9600, version 1.05; Boston, Mass.) DNA thermal cycler with an initial start at 95°C for 1 min and a final extension at 72°C for 6 min. Conditions for the second reaction were the same, except the LTIIa and STII annealing temperatures were 56 and 47°C, respectively. PCR primers for stxI, stxII, and eaeA and their amplification were as described by Sharma et al. (40).
TABLE 1.
PCR primers and probes
| Gene | Targeted regiona (bp) | Name | Sequence (5′ to 3′) | Annealing temp (°C) | Product size (bp) |
|---|---|---|---|---|---|
| LTIIa | 697-714 | LTIIa-F | GGTGTGCATTTCAGCGAC | 61 | 358 |
| 1035-1054 | LTIIa-R | TGGTATATTCCGGGTGGACG | |||
| 815-834 | IC-F | GCATGGAGAAAGAGATGAGC | 56 | 204 | |
| 999-1019 | IC-R | CTTACCACATAGATCCCACG | |||
| STII | 470-494 | STIIO-F | GCATCTATGTTCGTTTTTTCTATTG | 57 | 173 |
| 625-642 | STIIO-R | GCAACCATTATTTGGGCG | |||
| 502-521 | STII-F | TGCCTATGCATCTACACAAT | 47 | 113 | |
| 595-614 | STII-R | TAGAGATGGTACTGCTGGAG | |||
| LTIIa | 862-899 | prLTIIa | CTGATGACTGGCATTCTGTCTGGTCAGGTATATGCTGG | 58 | 38 |
| STII | 533-566 | prSTII | GATCTGTGTGAACATTATAGACAAATAGCCAAGG | 58 | 34 |
Product visualization.
PCR products were visualized on a 2% agarose gel containing 5 μg of ethidium bromide/ml by using a UV transilluminator (UVP, Inc., model TM-20, Upland, Calif.) and were documented by using ImageStore 5000 (version 2.03, Upland, Calif.). A 25-bp ladder (Gibco Life Technologies, Carlsbad, Calif.) was used to determine fragment size.
Controls.
Positive and negative controls were run with each PCR. Negative PCR results were further tested for interference by the addition of target DNA to the DNA extract from each negative sample, followed by PCR and visualization as described above.
Confirmation.
Results were confirmed by Southern hybridization with dot blot or gel transfer (3) to positively charged 0.45-mm-pore-size nylon filters (MSI). Filters were exposed to X-Omat film (Eastman Kodak, Rochester, N.Y.) at −80°C for 6 to 18 h depending on the activity of the probe. The probes were 5′ end labeled with [γ-32P]ATP by using T4 kinase as suggested by the manufacturer (Gibco Life Technologies, Carlsbad, Calif.).
Quantification of toxin gene occurrence.
Four dilutions with five replicates per dilution were made for each sample, but only three dilutions were used in computing the MPN (13). PCR analysis indicated a positive or negative result for each filter, and MPN values were calculated from these numbers. One toxin gene per E. coli was assumed for MPN calculations based on the pTC201 plasmid or the pRAS-1 plasmid and our limit of detection (19, 20). Each prevalence value was calculated by dividing the MPN value for toxin occurrence by the MPN value for total number of E. coli isolates per 100 ml with 95% confidence intervals calculated by Excel.
Statistical analysis.
The Friedman test was used to analyze the prevalence among seasons and farms. The percent change between single and nested PCRs was analyzed with the Wilcoxon signed-ranks test. All statistical analysis was performed with the SPSS statistical software system (version 9.0; SPSS, Inc., Chicago, Ill.).
RESULTS
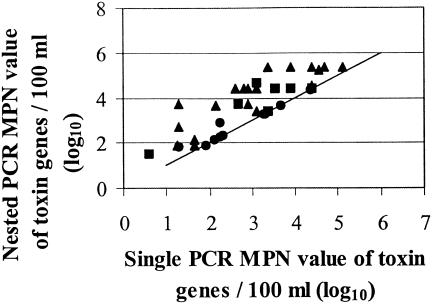
The occurrence of LTIIa and STII per 100 ml of waste lagoon water with single and nested PCRs is shown in Fig. 1. Interference with PCR from impurities extracted along with the DNA was not detected with our interference control test in single- or nested-PCR negative results.
FIG. 1.
Improvement in detection of LTIIa, STII and stxI, stxII, and eaeA toxin genes gained from using single- and nested-PCR MPN values. Symbols: LTIIa (▴), STII (▪), and stxI, stxII, and eaeA (•).
Overall, when single PCR was used, only 5% of all cow waste lagoon samples were negative for LTIIa, while no pig waste lagoon and 50% of stream water samples were negative for STII. Of the two single-PCR negative samples, one from a cow waste lagoon and one from a sampling point located downstream from a pig farm, both were positive after nested PCR. This resulted in a small improvement in detection of four and two times, respectively. These two data points are near the intercept of the x and y axes in Fig. 1.
Single- and nested-PCR results for LTIIa samples (Fig. 1) indicate that 50% of the samples increased by less than 1 log and 10% increased by 2 logs. The data from pig farms (Fig. 1) indicate that 67% of samples had a <1-log increase, while 33% showed an improvement of ≥1 log but ≤2 logs. The overall increase found with nested PCR in the detection frequencies across farms and seasons was 0.6 and 1 log, for the STII and LTIIa, respectively. A statistically significant improvement was shown in the detection of the LTIIa trait (P < 0.01) with the Wilcoxon test to compare single- and nested-PCR results, but no improvement was found for the STII trait (P ≥ 0.05).
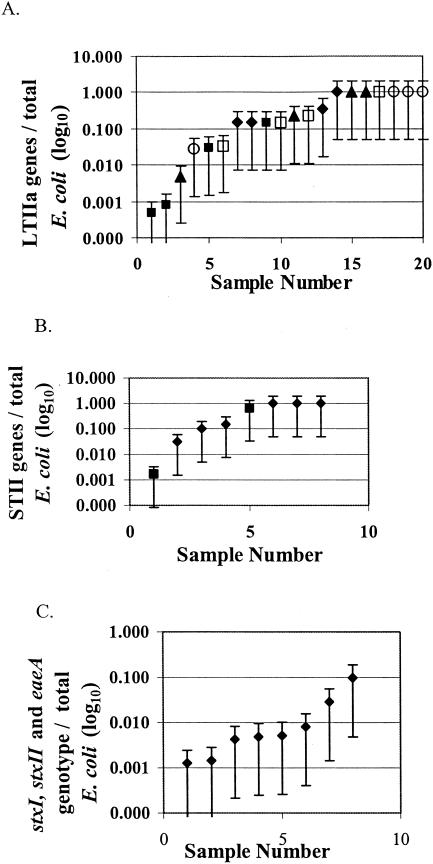
In Fig. 2A the prevalence of LTIIa by farm is shown. Farm 3 had the lowest prevalence values (1:>1,000 E. coli isolates [1,000 E. coli]), while farm 5 had the highest (1:1 E. coli). The 95% confidence intervals shown suggest that there is variability among samples. The prevalence for pig farm waste lagoons was never less than 1:33 E. coli, while the prevalence in the two stream samples below pig farms showed the greatest range from 1:2 to 1:600 E. coli.
FIG. 2.
Prevalence of toxin gene occurrence among all E. coli in waste lagoon waters with 95% confidence interval. Prevalence is displayed from lowest to highest frequency of occurrence. (A) LTIIa toxin gene. Symbols: ♦, farm 1; ▴, farm 2; ▪, farm 3; □, farm 4; and ○, farm 5. (B) STII toxin gene. Symbols: ♦, farms 1 to 5; and ▪, stream water samples. (C) stxI, stxII, and eaeA genotype. Each sample represents a different site.
In summary, the prevalence found by using single PCR ranged from 1 LTIIa:2 E. coli to 1 LTIIa:8,889 E. coli in cow waste lagoons. After nested PCR, the number of positives increased, changing the range of the prevalence to 1 LTIIa:1 E. coli to 1 LTIIa:2,051 E. coli. For pig waste lagoon waters, the range was 1 STII:1 E. coli to 1 STII:327 E. coli after single PCR and 1 STII:1 E. coli to 1 STII:33 E. coli after nested PCR. For the two stream water samples below the pig farm, the prevalence of the STII trait was 1 STII:>24 E. coli and 1 STII:13 E. coli with single PCR. After nested PCR, the prevalence increased to 1 STII:600 E. coli and 1 STII:2 E. coli.
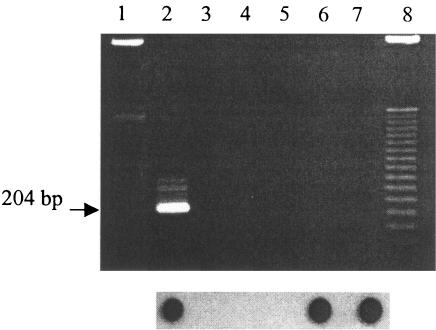
Positive PCR results were assessed with band visualization and/or Southern hybridization. Only one sample required Southern hybridization to detect the positive nested-PCR results (Fig. 3). The MPN positives changed from 200 to 400 for the winter season at cow farm 3, which increased the prevalence of the trait by 2.9-fold (from 1 LTIIa:356 E. coli to 1 LTIIa:123 E. coli). Southern hybridization did not change results for STII.
FIG. 3.
Electrophoretic analysis of nested-PCR products (LTIIa toxin gene), from cow waste lagoon sample farm 3 winter season, on an ethidium bromide-stained 2% agarose gel (top panel) and dot blot of corresponding samples (bottom panel). Lanes 1 and 8, 50-bp marker; lane 2, positive control; lane 3, negative control; lane 4, dilution 10−1 replicate A; lane 5, dilution 100 replicate D; lane 6, dilution 100 replicate C; and lane 7, dilution 100 replicate E.
The simultaneous occurrence of the three genes associated with the O157 genotype was analyzed by farm across seasons using single or nested PCR (Table 2; Fig. 1). The prevalence of the three genes, stxI, stxII, and eaeA, occurring simultaneously varied from 1:10 E. coli to 1:>889 E. coli with nested PCR (Table 2 and Fig. 2C), while individual toxin gene prevalence ranged from 1:1.4 to 1:800 E. coli for stxI, 1:6 to 1:889 E. coli for stxII, and 1:2 to 1:235 E. coli for eaeA. There was no statistically significant change in prevalence of these traits from single to nested PCR (P > 0.05) due to the small number of farms (n = 10).
TABLE 2.
MPN values and prevalence of all three genes (stxI, stxII, and eaeA) in cow waste lagoon waters using nested PCR
| Season | Farm | E. coli MPN/100 ml | MPN/100 ml for:
|
Data for simultaneous occurrence of stxI, stxII, and eaeA
|
|||
|---|---|---|---|---|---|---|---|
| stxI | stxII | eaeA | MPN/100 ml | Prevalence of stxI, stxII, and eaeA per no. of E. coli isolates | |||
| Fall | 2 | 1.6 × 105 | 1.7 × 103 | 1.8 × 102 | 1.4 × 103 | <1.8 × 102 | 1:>889 |
| 3 | 1.6 × 104 | 8.3 × 101 | 8.3 × 101 | 1.4 × 102 | 8.3 × 101 | 1:193 | |
| 4 | 1.6 × 105 | 2.8 × 104 | 7.0 × 103 | 4.6 × 103 | 4.6 × 103 | 1:35 | |
| 5 | 1.6 × 105 | 7.8 × 102 | 7.8 × 102 | 7.8 × 102 | 7.8 × 102 | 1:205 | |
| Spring | 1 | 1.6 × 104 | 1.7 × 102 | 1.3 × 102 | 2.1 × 102 | 1.3 × 102 | 1:123 |
| 3 | 1.6 × 105 | 4.9 × 102 | 3.3 × 102 | 5.4 × 103 | 2.3 × 102 | 1:696 | |
| 4 | 1.6 × 106 | 2.0 × 103 | 9.3 × 103 | 2.7 × 104 | 2.0 × 103 | 1:800 | |
| 5 | 2.4 × 105 | 1.8 × 103 | <1.8 × 103 | 6.8 × 103 | <1.8 × 103 | 1:>133 | |
| Summer | 3 | 1.6 × 104 | 9.3 × 101 | 9.3 × 101 | 6.8 × 101 | 6.8 × 101 | 1:235 |
| 4 | 2.4 × 105 | 1.7 × 105 | 4.3 × 104 | 1.4 × 105 | 2.3 × 104 | 1:10 | |
Variation in the prevalence of the LTIIa and STII traits among dairy and swine wastewaters can also be seen in Fig. 2A and B. When the prevalence of the LTIIa toxin gene was analyzed among farms and among seasons with the Friedman ranked-means test, no statistically significant differences were found (P > 0.05). The STII toxin gene was not analyzed for variance in prevalence among farms or seasons due to lack of complete seasonal data.
The stxI, stxII, and eaeA genes were screened in the fall, spring, and summer and were found to be present in all farms except at one farm at which stxII was absent in the spring sampling. Loss of participating farms over the course of the study prevented actual seasonal comparisons. Statistical analysis of the prevalence of the stxI, stxII, and eaeA genes among farms with the Friedman test showed no significant differences (P > 0.05).
Since no statistical differences were found in occurrence across farms and seasons for each trait separately, an overall geometric mean was calculated for the prevalence of each toxin gene within the E. coli communities in the waters of waste disposal sites. After nested PCR, these were 1:8.5 E. coli for the LTIIa and 1:4 E. coli for the STII toxin genes. The geometric mean prevalence of all three stxI, stxII and eaeA genes occurring together was 1:182 E. coli with nested PCR. The geometric mean prevalence of stxI, stxII and eaeA genes occurring individually after nested PCR was 1:67 E. coli, 1:101 E. coli, and 1:43 E. coli, respectively. Thus, the relative frequency of each gene within the O157 genotype is eaeA > stxI > stxII. This finding also suggests that in this study the frequency of these three genes associated with the O157 genotype is significantly lower than that of the LTIIa or STII toxin genes associated with ETEC.
At the 95% confidence level (Fig. 2), MPN values used to calculate prevalence lead to broad confidence intervals. We estimated the prevalence of traits per filter to gain a perspective on our prevalence values derived from geometric MPN mean values. We screened all the colonies that grew on a filter for toxin gene occurrence in one PCR. For example, the ratio of PCR-positive results for LTIIa to the total number of E. coli isolates screened per filter for all samples ranged from 1:1 to ≥1:10,000. As only one positive is obtained per filter, it is impossible to ascertain whether only one colony was positive or a number of colonies were positive for LTIIa if the total number of E. coli isolates exceeds one. Hence, ≥1 is used for ratios based on filters. Filter analysis also showed that prevalence was highly variable among our samples, so direct enumeration, even if possible, may not have yielded narrower confidence intervals in this instance. If Southern hybridization probes had been used on our 47-mm filter results at countable dilutions, six samples would have been negative. The MPN method, in which dilutions yield E. coli numbers that are too numerous to count, allowed us to detect these traits at very low frequencies. The prevalence data for the enterotoxin and hemorrhagic genes shown in Fig. 2 indicate that broad 95% confidence intervals demonstrate that, while some of our data produced one-for-one occurrence of a trait among the E. coli population, it may be that occurrence is actually 1:10 to 30 E. coli.
Since there are reports in the literature of STII associated with E. coli isolated from diarrheic-cow fecal swabs (41, 48), we also screened for the presence of STII in cow waste lagoon waters. Single-PCR results were negative for all cow farms tested. However, when nested PCR was used, all farms screened were positive for STII. One of the farms screened (farm 2) raised swine and thus was eliminated from further analysis. Of the four remaining farms, the geometric mean occurrence of STII after nested PCR was 1 STII per 968 E. coli, compared to the geometric mean occurrence of 1 LTIIa:8 E. coli in these four farms. The LTIIa trait is 120 times more frequent in the E. coli population than is STII in the cow waste lagoon waters screened. These results suggest that the STII toxin gene trait is carried by cows but at very low frequencies. The design of the study precluded the determination of whether the trait was associated with healthy or diarrheic cows.
DISCUSSION
Nested PCR increased detection of the LTIIa and STII traits 1 to 2 orders of magnitude over the levels previously reported by Khatib et al. (19, 20), who used growth and single PCR. Khatib et al. (19, 20) examined approximately 33 dairy farms from four states with herd sizes of 145 to 5,000 for LTIIa and 33 pig farms from seven states with herd sizes from 900 to 18,000 for STII. These authors also found no difference in occurrence among farms but did find a statistically significant relationship between the number of E. coli isolates screened and PCR positives for STII or LTIIa. In their study, LTIIa was detected in 87% of the cow waste lagoon samples when ≤2.0 × 104 E. coli isolates were screened and in 100% of the samples when >105 E. coli isolates were screened. Khatib et al. (19, 20) hypothesized that increased sensitivity was necessary to detect toxin genes at farms at which these traits occurred at low frequencies. In this study, where nested replaced single PCR, all cow farms were positive for LTIIa at each sampling time.
The present study detected STII with nested PCR in all the pig waste lagoon samples, and the prevalence was an order of magnitude greater than found by Khatib et al. (20), who reported an estimated prevalence of one trait per 102 E. coli when using growth and single PCR. The use of growth does not always increase gene detection, because it also increases extraneous DNA, which in turn may decrease the efficiency of single PCR (43). Overall, nested PCR improved sensitivity in 76% of the samples examined in this study, regardless of the trait being determined.
Differences in the number of traits detected at different sampling times are not unexpected, as the incidence of animals positive for these traits is dependent on factors such as the age distribution of the herd and herd immunity (14). Since the copy number of pTC201 is estimated to be between 10 and 30 per E. coli isolate carrying this plasmid (19, 20), it may be that wild-type strains also carry this trait in multiple copies per cell. This could produce the 1:1 prevalence values seen in Fig. 2. Our prevalence data for LTIIa provide evidence additional to that of Tsai et al. (46) who, having used magnetic bead hybridization with LTIIa as the probe, also suggested that this gene may have a copy number greater than 1 in some wild-type E. coli isolates. It could also be that E. coli carrying these traits does not die off at the same rate as E. coli with other toxin genes. Although this has not been reported for any of the diarrheic E. coli strains, such resistance has been shown for E. coli strains and other bacterial genera carrying antibiotic resistance (2, 25).
Our nested-PCR results on the occurrence of stxI, stxII and eaeA in cow waste lagoons were comparable to those of Ibekwe et al. (18), who used real-time PCR, suggesting that nested PCR provides a similar level of detection. The drawback of nested PCR is the potential for contamination through carryover (32). We found no evidence of carryover in this study.
In clinical microbiology, single isolates are often used to study pathogenicity (37, 50), as well as to understand the genetic diversity within a species. However, a population approach may be more useful, when the frequency of the target trait is low (<1:50 E. coli). A population approach is applicable to screen herds for pathogens or to determine the effectiveness of a pathogen eradication program. One reason that direct extraction procedures gained widespread acceptance in microbial ecology is the inefficiency of screening single isolates, the inability to culture many species, and the bias introduced from studying small sample sizes (7). For example, Moon et al. (26) found 1 STII trait per 20 E. coli isolates from fecal samples of healthy pigs. After the usage of a population approach, our data from waste lagoon ponds showed a higher frequency of occurrence for STII among E. coli (1 STII:4 E. coli), which seems reasonable since Fairbrother (10) reported that 75% of E. coli isolates from diarrheic piglets harbored STII.
Although STII was detected in cow waste lagoon waters screened in this study, its occurrence is low (1 STII:968 E. coli or 0.1% of E. coli) and below those reported for STII toxin genes associated with E. coli from diarrheic animals other than pigs (15). While the published literature is limited, the frequency of STII in diarrheic hosts was 0.75% (27), 1.1% (40), and 4.4% (15) of humans, cows, and dogs, respectively. An explanation for the occurrence of STII in cattle waste was reported by Shin et al. (41), who isolated E. coli with the colonization factor, K99, from diarrheic piglets. Furthermore, shared homology among the subunits of adhesion factors may result in nontargeted hosts being colonized by ETEC (41).
Outside Thailand the LTIIa toxin gene has not been reported in hosts other than cattle (33). Individual isolate studies on the occurrence of the stxI, stxII and eaeA genes in E. coli reported this genotype in diarrheic hosts other than cattle. For example, 1.1% of horse fecal samples (16), 3.1% of dog fecal samples (16), and 1.7% of sheep rectal fecal swabs (31) have tested positive for this genotype.
In this study the simultaneous prevalence of the three traits associated with the O157 genotype is 1:182 E. coli (0.55% of E. coli), similar to reports in the literature (38, 47). Our study examined occurrence among total E. coli while most examined occurrence in fecal samples. The rate of infection in cows is estimated to be 1 to 2 percent (38) and is hence difficult to detect on a farm through single-isolate screening of fecal samples. Our data suggest that screening large numbers of isolates provides an easy approach for detecting this genotype in farm waste ponds.
Quantitation using either the three-tube (22, 36) or five-tube MPN method should be acceptable when a Poisson distribution (far more negatives than positives) is expected (13) as in this study, for which we used five-tube MPN in combination with single and nested PCR to determine the frequency of occurrence of toxin genes. All MPN methods (13) have large confidence intervals around the predicted MPN value per volume or weight. Thus, the prevalence rates presented in this paper should be considered in the context of the broad confidence intervals displayed in Fig. 2. The data do suggest that population studies provide unique insights into the level of trait occurrence in herds, even considering these confidence intervals. The agreement between the values for the frequency of stxI, stxII, and eaeA traits obtained by Ibekwe et al. (18) by using real-time PCR and our nested-PCR MPN values indicates the validity of an MPN approach.
Single-PCR data (19, 20) and the literature on LTIIa and STII occurrence (26, 30) suggest that the frequency of the traits after dilution in waste lagoon waters would be below detection if a direct assessment technique such as colony blots of E. coli grown on membranes was used (35, 39). For example, STII has been reported in 5.4% of individual isolates from healthy pigs (26). E. coli carrying the trait will be diluted by other E. coli, so the frequency of STII occurrence would be far below the 1:20 expected prevalence. Given the high frequencies of STII in this study, membrane filtration coupled with Southern hybridization, as reported by Todd et al. (44), could be the preferable method for computing prevalence rates. Narrower confidence limits should result if both total population and specific gene traits are enumerated at countable dilutions. However, in this study, prevalence rates based on LTIIa trait occurrence showed the number of E. coli colonies screened per filter yielding a positive ranged from 1 to 10,000, due to farm 3 (Fig. 2A). If colony blots had been used in this study, some positive samples would not have been detected and confidence intervals would still have been large based on sample-to-sample variability. Also, when frequency is highly variable as we found, the MPN test provides a relatively rapid and reliable method to ascertain trait frequency, because too-numerous-to-count samples are screened.
The stxI, stxII, and eaeA occurrence data compiled by Ibekwe et al. (18) from dairy waste lagoons and our data agreed for the same region in California. Importantly, his work showed that, after treatment in a facultative pond and a wetland, the effluent from these showed little if any reduction in the frequency of stxI, stxII, and eaeA. This suggests that E. coli is surviving and perhaps growing, introducing the potential for these pathogens to enter environmental waters. For example, LTIIa was used as a biomarker to track fecal contamination from a dairy farm in New York State. The farm's manure pit tested positive for the presence of LTIIa, as did the well water being used for human consumption below a dairy. It was hypothesized that E. coli was being transported by an underground stream to the user's well (unpublished data from 2002). These findings emphasize the importance of federal regulations requiring disinfection of well waters and treatment for groundwater under the influence of surface waters (http://www.epa.gov/safewater/therule.html#Surface).
Detection of toxin occurrence at the herd level could easily be accomplished through screening E. coli populations from composite fecal or waste lagoon samples. In turn this would increase our understanding of the occurrence of these traits and their endemicity within animal herds. This information could lead to better management practices on farms, preventing further disease spread with corresponding economic benefits, and could decrease the risk of disease to the public.
Acknowledgments
This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grant 99-25102-8596.
We gratefully acknowledge dairy and swine farmers, N. Hall at the University of Iowa, T. Shultz of UC Cooperative Extension, and R. Sizemore and his graduate students at the University of North Carolina for their support with sample collection. We also thank V. K. Sharma for donating EHEC strain 4718, J. Wang for her help with DNA extractions, and Y. L. Tsai and L. Khatib for their assistance with the techniques used in this research.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zheng, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. L., J. J. Calomiris, and R. J. Seidler. 1982. Selection of antibiotic-resistant standard plate count bacteria during water treatment. Appl. Environ. Microbiol. 44:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 4.Avery, S. M., A. Small, C. A. Reid, and S. Buncic. 2002. Pulsed-field gel electrophoresis characterization of Shiga toxin-producing Escherichia coli O157 from hides of cattle at slaughter. J. Food Prot. 65:1172-1176. [DOI] [PubMed] [Google Scholar]
- 5.Celemin, C., P. Rubio, P. Echeverria, and S. Suarez. 1995. Gene toxin patterns of Escherichia coli isolated from diseased and healthy piglets. Vet. Microbiol. 45:121-127. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention 2002. Summary of notifiable diseases—United States, 2000. Morb. Mortal. Wkly. Rep. 49:1-102. [PubMed] [Google Scholar]
- 7.Cho, J. C., and S. J. Kim. 2000. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl. Environ. Microbiol. 66:956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, A., S. Morton, P. Wright, J. Corkish, F. J. Bolton, and J. Russell. 1997. A community outbreak of Vero cytotoxin producing Escherichia coli O157 infection linked to a small farm dairy. Commun. Dis. Rep. CDR Rev. 7:R206-R211. [PubMed] [Google Scholar]
- 9.Dufour, A. P., E. R. Strickland, and V. J. Cabelli. 1981. Membrane filter method for enumerating Escherichia coli. Appl. Environ. Microbiol. 41:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairbrother, J. M. 1992. Enteric colibacillosis, p. 489-497. In A. Leman (ed.), Diseases of swine, 7th ed. Iowa State University Press, Ames.
- 11.Feldman, K. A., J. C. Mohle-Boetani, J. Ward, K. Furst, S. L. Abbott, D. V. Ferrero, A. Olsen, and S. B. Werner. 2002. A cluster of Escherichia coli O157: nonmotile infections associated with recreational exposure to lake water. Public Health Rep. 117:380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galland, J. C., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton (ed.). 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 14.Gyles, C. L. (ed.). 1994. Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 15.Hammermueller, J., S. Kruth, J. Prescott, and C. Gyles. 1995. Detection of toxin genes in Escherichia coli isolated from normal dogs and dogs with diarrhea. Can. J. Vet. Res. 59:265-270. [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, D. D., T. E. Besser, D. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 17.Heuvelink, A. E., F. L. van den Biggelaar, E. de Boer, R. G. Herbes, W. J. Melchers, J. H. Huis in't Veld, and L. A. Monnens. 1998. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2002. A biomarker for the identification of cattle fecal pollution in water using the LTIIa toxin gene from enterotoxigenic E. coli. Appl. Microbiol. Biotechnol. 59:97-104. [DOI] [PubMed] [Google Scholar]
- 20.Khatib, L. A., Y. L. Tsai, and B. H. Olson. A biomarker for the identification of swine fecal pollution in water using the STII toxin gene from enterotoxigenic E. coli. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 21.Klemm, P. 1994. Fimbriae: adhesion, genetics, biogenesis and vaccines. CRC Press, Boca Raton, Fla.
- 22.Kowalchuk, G., A. Stienstra, G. Heilig, J. Stephen, and J. Woldendorp. 2000. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands). FEMS Microbiol. Ecol. 31:207-215. [DOI] [PubMed] [Google Scholar]
- 23.Lahti, E., M. Keskimaki, L. Rantala, P. Hyvonen, A. Siitonen, and T. Honkanen-Buzalski. 2001. Occurrence of Escherichia coli O157:H7 in Finnish cattle. Vet. Microbiol. 79:239-251. [DOI] [PubMed] [Google Scholar]
- 24.LeJeune, J. T., T. E. Besser, and D. D. Hancock. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl. Environ. Microbiol. 67:3053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leong, L. Y. C., D. Osaka, H. F. Ridgway, and B. H. Olson. 1982. Chlorine-resistance of coliform-tested bacteria isolated from raw and treated sewage effluent. Wat. Sci. Technol. 14:127-132. [Google Scholar]
- 26.Moon, H. W., R. A. Schneider, and S. L. Moseley. 1986. Comparative prevalence of four enterotoxin genes among Escherichia coli isolated from swine. Am. J. Vet. Res. 47:210-212. [PubMed] [Google Scholar]
- 27.Okamoto, K., Y. Fujii, N. Akashi, S. Hitotsubashi, H. Kurazono, T. Karasawa, and Y. Takeda. 1993. Identification and characterization of heat-stable enterotoxin II-producing Escherichia coli from patients with diarrhea. Microbiol. Immunol. 37:411-414. [DOI] [PubMed] [Google Scholar]
- 28.Olsen, S. J., G. Miller, T. Breuer, M. Kennedy, C. Higgins, J. Walford, G. McKee, K. Fox, W. Bibb, and P. Mead. 2002. A waterborne outbreak of Escherichia coli O157:H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg. Infect. Dis. 8:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osek, J. 1998. Application of polymerase chain reaction for determination of toxins in Escherichia coli strains isolated from pigs with diarrhea. Acta Microbiol. Pol. 47:409-413. [PubMed] [Google Scholar]
- 30.Osek, J. 1999. Prevalence of virulence factors of Escherichia coli strains isolated from diarrheic and healthy piglets after weaning. Vet. Microbiol. 68:209-217. [DOI] [PubMed] [Google Scholar]
- 31.Paiba, G. A., J. C. Gibbens, S. J. Pascoe, J. W. Wilesmith, S. A. Kidd, C. Byrne, J. B. Ryan, R. P. Smith, M. McLaren, R. J. Futter, A. C. Kay, Y. E. Jones, S. A. Chappell, G. A. Willshaw, and T. Cheasty. 2002. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Vet. Rec. 150:593-598. [DOI] [PubMed] [Google Scholar]
- 32.Picken, M. M., R. N. Picken, D. Han, Y. Cheng, and F. Strle. 1996. Single-tube nested polymerase chain reaction assay based on flagellin gene sequences for detection of Borrelia burgdorferi sensu lato. Eur. J. Clin. Microbiol. Infect. Dis. 15:489-498. [DOI] [PubMed] [Google Scholar]
- 33.Pickett, C. L., D. L. Weinstein, and R. K. Holmes. 1987. Genetics of type IIa heat-labile enterotoxin of Escherichia coli: operon fusions, nucleotide sequence, and hybridization studies. J. Bacteriol. 168:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pradel, N., V. Livrelli, C. De Champs, J.-B. Palcoux, A. Reynaud, F. Scheutz, J. Sirot, B. Joly, and C. Forestier. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramotar, K., B. Waldhart, D. Church, R. Szumski, and T. J. Louie. 1995. Direct detection of verotoxin-producing Escherichia coli in stool samples by PCR. J. Clin. Microbiol. 33:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosado, A., L. Seldin, A. C. Wolters, and J. D. van Elsas. 1996. Quantitative 16S rDNA-targeted polymerase chain reaction and oligonucleotide hybridization for the detection of Paenibacillus azotofixans in soil and the wheat rhizosphere. FEMS Microbiol. Ecol. 19:153-164. [Google Scholar]
- 37.Sanchez-Carlo, V., J. S. McDonald, and R. A. Packer. 1984. Virulence factors of Escherichia coli isolated from cows with acute mastitis. Am. J. Vet. Res. 45:1775-1777. [PubMed] [Google Scholar]
- 38.Sargeant, J. M., J. R. Gillespie, R. D. Oberst, R. K. Phebus, D. R. Hyatt, L. K. Bohra, and J. C. Galland. 2000. Results of a longitudinal study of the prevalence of Escherichia coli O157:H7 on cow-calf farms. Am. J. Vet. Res. 61:1375-1379. [DOI] [PubMed] [Google Scholar]
- 39.Schultsz, C., G. J. Pool, R. van Ketel, B. de Wever, P. Speelman, and J. Dankert. 1994. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. J. Clin. Microbiol. 32:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13:291-302. [DOI] [PubMed] [Google Scholar]
- 41.Shin, S. J., Y. F. Chang, M. Timour, T. L. Lauderdale and D. H. Lein. 1994. Hybridization of clinical Escherichia coli isolates from calves and piglets in New York State with gene probes for enterotoxins (STaP, STb, LT), Shiga-like toxins (SLT-1, SLT-II) and adhesion factors (K88, K99, F41, 987P). Vet. Microbiol. 38:217-225. [DOI] [PubMed] [Google Scholar]
- 42.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theron, J., D. Morar, M. Preez, V. S. Brozel, and S. N. Venter. 2001. A sensitive seminested PCR method for the detection of Shigella in spiked environmental water samples. Water Res. 35:869-874. [DOI] [PubMed] [Google Scholar]
- 44.Todd, E. C., R. A. Szabo, J. M. MacKenzie, A. Martin, K. Rahn, C. Gyles, A. Gao, D. Alves, and A. J. Yee. 1999. Application of a DNA hybridization-hydrophobic-grid membrane filter method for detection and isolation of verotoxigenic Escherichia coli. Appl. Environ. Microbiol. 65:4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai, Y. L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai, Y. L., J. Y. Le, and B. H. Olson. 2003. Magnetic bead hybridization to detect enterotoxigenic Escherichia coli strains associated with cattle in environmental water sources. Can. J. Microbiol. 49:391-398. [DOI] [PubMed] [Google Scholar]
- 47.Tutenel, A. V., D. Pierard, J. Uradzinski, E. Jozwik, M. Pastuszczak, J. Van Hende, M. Uyttendaele, J. Debevere, T. Cheasty, J. Van Hoof, and L. De Zutter. 2002. Isolation and characterization of enterohaemorrhagic Escherichia coli O157:H7 from cattle in Belgium and Poland. Epidemiol. Infect. 129:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodward, M. J., R. Kearsley, C. Wray, and P. L. Roeder. 1990. DNA probes for the detection of toxin genes in Escherichia coli isolated from diarrhoeal disease in cattle and pigs. Vet. Microbiol. 22:277-290. [DOI] [PubMed] [Google Scholar]
- 49.Woodward, M. J., and C. Wray. 1990. Nine DNA probes for detection of toxin and adhesin genes in Escherichia coli isolated from diarrhoeal disease in animals. Vet. Microbiol. 25:55-65. [DOI] [PubMed] [Google Scholar]
- 50.Wooley, R. E., K. R. Spears, J. Brown, L. K. Nolan, and O. J. Fletcher. 1992. Relationship of complement resistance and selected virulence factors in pathogenic avian Escherichia coli. Avian Dis. 36:679-684. [PubMed] [Google Scholar]
- 51.Wray, C., and M. J. Woodward. 1997. Escherichia coli infections in farm animals, p. 49-84. In M. Sussman (ed.), Escherichia coli: mechanisms of virulence. Cambridge University Press, Cambridge, England.
- 52.Yamasaki, S. 2002. Development of vaccine for enterohemorrhagic Escherichia coli infection. Nippon Rinsho 60:1083-1088. (In Japanese.) [PubMed] [Google Scholar]