Abstract
Particles are often regarded as microniches of enhanced microbial production and activities in the pelagic ocean and are vehicles of vertical material transport from the euphotic zone to the deep sea. Fluorescence in situ hybridization (FISH) can be a useful tool to study the microbial community structures associated with these particles, and thus their ecological significance, yet an appropriate protocol for processing deep-sea particle-rich water samples is lacking. Some sample processing considerations are discussed in the present study, and different combinations of existing procedures for preservation, size fractionation sequential filtration, and sonication were tested in conjunction with FISH. Results from this study show that water samples should be filtered and processed within no more than 10 to 12 h after collection, or else preservation is necessary. The commonly used prefiltration formaldehyde fixation was shown to be inadequate for the rRNA targeted by FISH. However, prefiltration formaldehyde fixation followed by immediate freezing and postfiltration paraformaldehyde fixation yielded highly consistent cell abundance estimates even after 96 days or potentially longer storage. Size fractionation sequential filtration and sonication together enhanced cell abundance estimates by severalfold. Size fractionation sequential filtration effectively separated particle-associated microbial communities from their free-living counterparts, while sonication detached cells from particles or aggregates for more-accurate cell counting using epifluorescence microscopy. Optimization in sonication time is recommended for different specific types of samples. These tested and optimized procedures can be incorporated into a FISH protocol for sampling in deep-sea particle-rich waters.
Particles play an important role in the biogeochemical cycling of global oceans. Ranging from submicrometer-diameter colloids to transparent exopolymer particles, large mineral flocs, and marine snow, particles serve as vehicles of material transport enhancing fluxes from the euphotic zone to the deep-sea (6, 23, 34). The general paradigm of marine nutrient cycling is that remineralization occurs during the sinking of particulate organic matter out of the euphotic zone through the water column and that the recycled nutrients will eventually recharge surface biological production via upwelling. Most of these remineralization processes are microbially mediated, yet little is known of the actual microbial communities and activities on particles in the deep-sea water column. In surface and coastal ocean environments, particles are considered the nutrient-rich microscale hot spots for microbial production compared to ambient seawater (2, 10, 27). Such enhanced microbial production on particles is more significant in the deep-sea water column, where primary production is even more limited.
Qualitative phylogenetic studies have revealed disparate microbial communities between particle-associated assemblages and their free-living counterparts in aquatic environments (1, 12, 16, 33). However, more quantitative measurements are required to elucidate the ecological significance of particle-associated microbes. Fluorescence in situ hybridization (FISH) has recently become a very useful tool to identify and enumerate microbes in mixed communities at the domain to species levels (3). FISH can reveal microbial community structures, and relate microbial communities to geochemical cycling, especially in the case of particular functional groups like nitrifiers (19, 28, 42) or methanotrophs (9, 30). However, a working protocol is lacking that differentiates particle-associated microbes from free-living microbes in particle-rich seawater.
Water from the deep-sea is commonly sampled using 10- to 30-liter Niskin bottles mounted on a conductivity-temperature-density (CTD) rosette package. Three major challenges have to be met for FISH subsampling from these Niskin bottles. First, the abundance of cells and particles in the deep-sea are relatively low, so that a large volume of water has to be concentrated to yield sufficient working materials. This is conventionally done either by centrifugation (4) or filtration onto filter membranes followed by cell transfers to gelatin-coated slides for FISH (24). Both methods cause severalfold greater cell losses than a protocol in which FISH is directly performed on the same filter membranes (26). Consequently, a direct filtration-to-FISH method (21) was chosen as the basic protocol in this study.
The second challenge concerns sample preservation. It usually takes at least an hour from water collection at depth to on-deck subsampling, and then up to another 24 h until the water is filtered for FISH. Therefore, it is essential to test for any storage effects both with and without preservatives. An efficient preservation method is imperative to ensure sample integrity until the actual hybridization procedure is performed in a shore-based laboratory. The two most commonly used preservation methods are postfiltration, 30-min 4% paraformaldehyde fixation (21, 32) and prefiltration formaldehyde (2% final concentration) overnight fixation (17, 25). Another option is to preserve the whole water sample with formaldehyde (2% final concentration) followed by immediate freezing (25). However, no systematic studies have been published to date that compare the effectiveness of these various preservation methods for FISH through time.
The third challenge is the differentiation between particle-associated and free-living microbial assemblages. Size fractionation sequential filtration approaches have been successfully applied in a few phylogenetic studies to separate these two microbial assemblages in aquatic environments (12, 33), and sonication has been used to detach cells from particles for FISH (7, 22, 50). The sonication times employed in published studies range from 2 s to 30 min. While too short a sonication time is obviously insufficient to detach cells from aggregates or particles, prolonged sonication likely disrupts the cells. Other available methods of microbe-particle separation include homogenization and chemical treatments with surfactants like tetrasodium pyrophosphate (41, 49), Tween 20 or 80 (11, 18, 51), and Triton X-100 (31, 36), among which only tetrasodium pyrophosphate has been used with FISH thus far (5, 22, 52). Only sonication was tested in the present study.
The objectives of this study are to evaluate the effects of storage and various preservation methods over time as well as the efficiency of combined size fractionation sequential filtration and sonication in differentiating and enumerating particle-associated and free-living microbes in marine samples. The ultimate goal is to establish a sampling protocol for FISH that is optimized for studying particle-associated versus free-living microbial assemblages in the deep sea.
MATERIALS AND METHODS
Water samples for the various experiments were taken from three saltwater aquaria—RS1, RS2, and WA—and a coastal site at Hawaii Kai on the island of Oahu, Hawaii (HK). Samples WA and HK were particularly loaded with visible particles. A direct-filtration-to-FISH method with postfiltration paraformaldehyde fixation (21) was used as the basic protocol. Briefly, water samples were filtered onto 0.2-μm-pore-size, 25-mm-diameter white polycarbonate membrane filters (Osmonics, Inc.), supported by 0.45-μm-pore-size nitrocellulose membrane filters (Whatman, Inc.) under a low vacuum pressure (5 in. of Hg). While still on the glass filtering tower with the vacuum broken, each sample filter was fixed with 3 ml of cold freshly prepared and filtered (pore size, 0.2 μm) 4% paraformaldehyde in phosphate-buffered saline (1×; pH 7.2). After a 30-min fixation, the fixative was drawn off by again applying low vacuum. Each filter was then rinsed once with 3 ml of phosphate-buffered saline (1×) and once with 3 ml of deionized water; both solutions were drawn off by vacuum. An additional 1-min cell permeabilization treatment was performed with 1 ml of 50% ethanol and 2% (wt/vol) sodium chloride. The filters were removed from the filtering towers, air dried in the dark, and stored in sterile petri dishes at −20°C until hybridization. Modifications made in the various experiments are described in the following sections.
Experiments on storage effects without preservatives.
Water from RS1 and RS2 was subsampled within 2 h after collection and processed according to the basic protocol. The remaining water samples were refrigerated at 4°C until the next subsampling time, when the same procedures of filtration and fixation were repeated. The subsampling time intervals were 2, 3, 4, 5, 18, 21, 24, and 27.5 h in series RS1 and were 1, 3, 7, 13.8, 21, 36, and 52 h in series RS2. Replicate samples were collected at each time interval.
Experiments on preservation methods.
Two preservation methods were tested using the WA sample: (i) half of the water sample was immediately filtered and preserved with 4% paraformaldehyde as in the basic protocol, and (ii) the remaining water sample was immediately preserved with formaldehyde (2% final concentration), and refrigerated (4°C) overnight (16 h) before filtering onto 0.2-μm-pore-size white polycarbonate membrane filters.
In the RS2 sample series, five preservation methods were tested: (i) postfiltration 4% paraformaldehyde fixation as in the basic protocol (the PFA method), (ii) prefiltration fixation with formaldehyde (2% final concentration) for at least 8 h (the F method), (iii) prefiltration formaldehyde fixation (2% final concentration, ≥8 h) followed by an additional postfiltration 4% paraformaldehyde fixation (the F+PFA method), (iv) prefiltration formaldehyde fixation (2% final concentration) and immediate freezing at −20°C (the FF method), and (v) prefiltration formaldehyde fixation with immediate freezing and postfiltration 4% paraformaldehyde fixation (the FF+PFA method). One large acid-washed bottle was used to collect the original aquarium water, which was then divided into 1-liter and 2-liter acid-washed polyethylene bottles for treatments i and treatments ii and iii, respectively. For treatments iv and v, 125-ml acid-washed bottles were filled with the original water sample, fixed with 2% formaldehyde (final concentration), and frozen immediately. At the designated time intervals, replicate bottles were taken out to thaw at room temperature 2 h prior to filtration. The subsamples for different time intervals of treatments iv and v were drawn from different smaller bottles instead of one single large bottle, in order to eliminate potential degradation effects of repeated freezing and thawing. Caution was taken to ensure complete thawing before filtration. Subsampling time intervals were approximately 1, 3, 7, 14, 21, 36, and 52 h and 4 days for treatments i and iii, while treatment ii was additionally subsampled after 37 days. Owing to the time required for freezing and thawing, and the limited availability of water samples, treatments iv and v targeted longer time intervals of ∼1, 4, 37, and 96 days. All resulting 0.2-μm-pore-size membrane filters were stored frozen until hybridization.
Experiments on particle separation and sonication.
The WA and HK samples were used in the particle separation and sonication experiments. The water samples were preserved with formaldehyde (2% final concentration) immediately upon sample collection. The purpose of this fixation procedure is to strengthen cellular structures and to help preserve cellular rRNAs targeted by FISH during sonication (e.g., see references 5, 8, 22, and 50). Three size fractions were investigated in this study: ≥10 μm, 5.0 to 10 μm, and 0.2 to 5.0 μm. The water samples were first gravity filtered through sterilized 10-μm-pore-size Nitex screens mounted on sterilized polypropylene beakers without bottoms and then were filtered through 5.0-μm-pore-size polycarbonate membrane filters in sterilized polycarbonate filtration units. The 5.0-μm-diameter filtrates were subsequently filtered onto 0.2-μm-pore-size polycarbonate membrane filters as the free-living (0.2- to 5.0-μm-diameter) fractions. The 10-μm-pore-size Nitex screens and the 5.0-μm-pore-size membrane filters were thoroughly rinsed with double-filtered (pore size, 0.2 μm) seawater into separate sterile graduated beakers. The rinsates, together with the corresponding Nitex screens or filters, were sonicated for 0 or 5 s in the WA series and were sonicated for 0, 5, 10, 15, 20, 25, and 30 s in the HK series. An ultrasonic bath with a 130-W output (Branson Ultrasonics Corp.) and 6- to 7-cm water depth was used. Then, the rinsates were filtered onto separate 0.2-μm-pore-size polycarbonate membrane filters, which represented the particle-associated fractions (pore sizes, >10 μm and 5.0 to 10 μm). All 0.2-μm-pore-size membrane filters were preserved with 4% paraformaldehyde and stored at −20°C until hybridization. Replicates were taken for each size fraction, and whole-water samples were also processed without size fractionation nor sonication.
FISH.
The FISH procedures followed the basic protocol described in reference 21. Each membrane filter was cut into four sections for hybridization with different oligonucleotide probes. The filter sections were placed into a prewarmed sterile 24-well microtiter plate (Nunc), one filter section per well, and overlaid with 20 μl of prewarmed sterile hybridization solution. The hybridization solutions contained 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 0.01% sodium dodecyl sulfate, 2.5 ng of fluorescently labeled oligonucleotide probes μl−1, and 20 to 55% formamide according to the stringencies used with the corresponding probes (Table 1). The microtiter plate was placed in a prewarmed equilibrated chamber saturated with an atmosphere of hybridization solution minus the probes and incubated at 46°C for 2 h. Each filter section was next transferred to a prewarmed (48°C), sterile 20-ml glass vial filled with prewarmed sterile washing solution. These solutions contain 56 to 225 mM NaCl (Table 1), 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, and 0.01% sodium dodecyl sulfate. The filter sections were left freely floating within the vials without agitation at 48°C for 15 min, whereupon the washing solutions were replaced with fresh solutions. After another 15-min incubation, the filter sections were air dried in the dark. In case of a second hybridization, the dried filter sections were returned to the microtiter plate, and the same hybridization and washing procedures were repeated. Each filter section was then stained with 30 μl of DAPI (4′,6-diamidino-2-phenylindole) solution (50 μg ml−1) for 8 min in the dark, rinsed with double-deionized water, and air dried. Finally, the filter sections were mounted onto glass microscope slides with FluoroGuard Antifade Reagent (Bio-Rad Laboratories, Inc.) and stored at −20°C before viewing. Fixed samples from a pure culture of Nitrosomonas cryotolerans were used as positive controls during each run of FISH.
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this studya
| Probe name | Sequencec | Target organisms | Target site (E. coli positions) | % FA | NaCl concn (mM) | Reference(s) |
|---|---|---|---|---|---|---|
| UNI | 5′-GWA TTA CCG CGG CKG CTG-3′ | Universal | 519-536 | 20 | 70 | 20 |
| EUB338 | 5′-GCT GCC TCC CGT AGG AGT-3′ | Domain bacteria | 338-355 | 20 | 225 | 3, 42 |
| NON338 | 5′-ACT CCT ACG GGA GGC AGC-3′ | Negative control | NAb | 20 | 225 | 46 |
| NSO190 | 5′-CGA TCC CCT GCT TTT CTC C-3′ | β-AOB | 190-208 | 55 | 20 | 28, 42 |
| NSO1225 | 5′-CGC GAT TGT ATT ACG TGT GA-3′ | β-AOB | 1225-1244 | 35 | 191 | 28, 42 |
Probes are shown together with the target organisms, target sites on the 16S rRNA gene with respect to Escherichia coli positions, formamide concentrations (% FA) in hybridization solutions, and NaCl concentrations in the stringent washing solutions.
NA, not applicable.
W = A or T; K = G or T.
The slides were viewed under an Eclipse E400 microscope equipped with Y-FL Epi-fluorescence attachments (Nikon, Inc.) and filter sets specific for each fluorochrome used (Chroma Technology Co.). At least 20 fields or 1,000 cells were counted for each hybridization. The arithmetic means of all replicates counted were averaged and reported with standard errors (SE). All reported FISH cell counts have been corrected for any nonspecific binding to nontargeted cells or noncellular particles, and for any autofluorescence under the same wavelengths, by subtracting the counts of the negative control NON338 of the same fluorochrome used in the probes.
Oligonucleotide probes.
The sequences of the oligonucleotide probes employed in this study are listed in Table 1, together with the respective targeted microorganisms and the stringencies used in hybridization and washing solutions. A universal oligonucleotide probe (UNI) (20) was used in the sonication experiments, while EUB338, a probe specific for the domain Bacteria (referred to as eubacteria in the following sections) (3), and two probes specific for β-proteobacterial ammonia-oxidizing bacteria (β-AOB) (NSO190 and NSO1225) (28) were used in the storage effects and preservation experiments. A negative-control oligonucleotide probe, NON338, (46), which does not target any organisms, was used in all samples. All oligonucleotide probes were purchased custom made with the fluorochromes Cy3 or 6FAM attached at the 5′ ends of the oligonucleotide probes (Integrated DNA Technologies, Inc.).
Statistical analyses.
All statistical analyses were performed using Statistica 6.0 software (StatSoft). The normality of data sets was checked prior to any parametric tests; nonparametric tests were used for nonnormal data sets. The significance of all parameters in the regression analyses presented has been verified (P < 0.05) by one-way analysis of variance (ANOVA). The overall goodness of fit and significance of the regression analyses are indicated by the R2 and P values from one-way ANOVA. In each sample series, the first data point of any type of cell counts acquired with the basic protocol is taken as the reference value, which is assumed to be the closest to the in situ abundance.
RESULTS
Storage effects without preservatives.
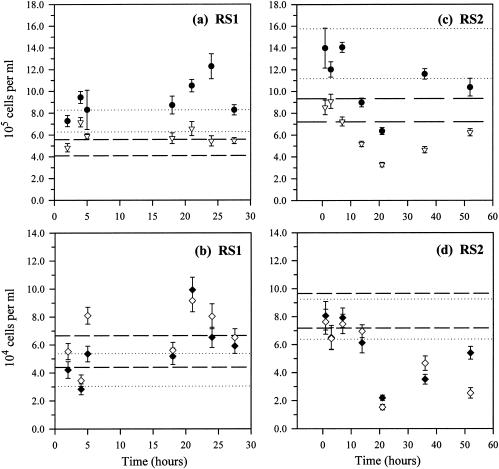
In sample RS1, although the sample means of DAPI and eubacterial cell counts were significantly different from their reference values (P < 0.05 and P < 0.01, respectively, t tests) (Fig. 1a), no systematic temporal trends are discernible by one-way ANOVA or any trend analyses (P > 0.05). β-AOB abundance, as enumerated with both NSO190 and NSO1225, were not significantly different from reference values (P > 0.05; t tests), and no temporal trends were evident either (P > 0.05; ANOVA) (Fig. 1b).
FIG. 1.
Effects of storage without preservatives: temporal changes in total microbial abundance estimated by DAPI staining (•) and eubacterial abundance estimated with EUB338 (▿) in RS1 (a) and RS2 (c) and temporal changes in β-AOB abundance detected with NSO190 (♦) and with NSO1225 (⋄) in RS1 (b) and RS2 (d). (a and c) The dotted and dashed lines indicate the 95% confidence intervals (95% CI) of DAPI and EUB reference values, respectively. (b and d) The dotted and dashed lines are the 95% CI of the NSO190 and NSO1225 reference values, respectively. Please note the longer time scale used for RS2 in panels c and d. Error bars, standard deviations.
However, longer storage time in RS2 resulted in reduced microbial abundance for both DAPI and FISH (Fig. 1c and d). The DAPI cell counts decreased by 26% after 52 h, and the mean was significantly different from the reference value (P < 0.05; t test), but there was no statistically significant temporal trend. All FISH cell counts were significantly different from their respective reference values (P < 0.05, t tests). Eubacterial abundance decreased exponentially with time (R2 = 0.72; P < 0.05 [ANOVA]). β-AOB abundance enumerated with NSO1225 also decreased (log-linear: R2 = 0.53; P < 0.05 [ANOVA]), and so did that enumerated with NSO190, though not significantly (linear: R2 = 0.61; P = 0.07 [ANOVA]).
Comparison of preservation methods.
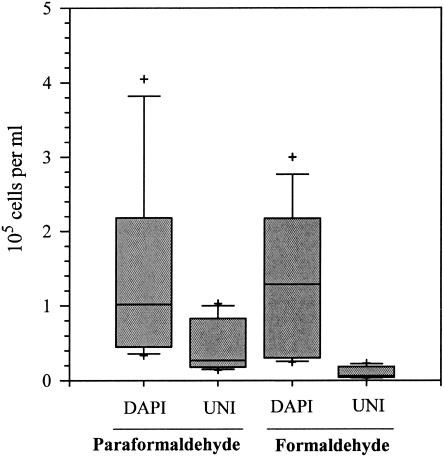
Overnight formaldehyde fixation in the WA sample yielded significantly lower UNI-hybridized cell counts than paraformaldehyde fixation (P < 0.01; Wilcoxon matched-pairs test), yet no significant difference could be observed in DAPI cell counts obtained from the two preservation methods (P > 0.05; Wilcoxon matched-pairs test) (Fig. 2). These results imply that formaldehyde is not as effective a fixative as paraformaldehyde for rRNAs which FISH targets, even though it may be an effective fixative for DNA to which DAPI binds.
FIG. 2.
Combined microbial abundance estimates of different size fractions with postfiltration paraformaldehyde and prefiltration overnight formaldehyde fixation in sample WA. Total microbial abundance was estimated by DAPI staining and FISH with the probe UNI. The mid-lines represent median values, the boxes and bars are the 50th and 75th percentiles, and plus signs indicate the outliers.
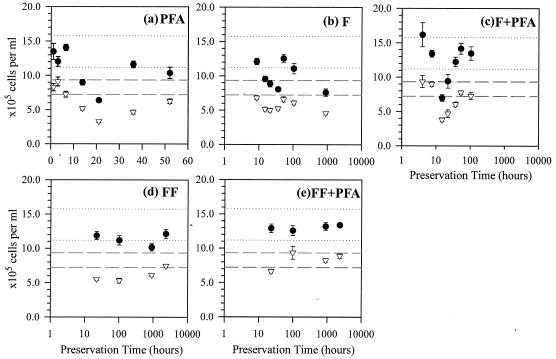
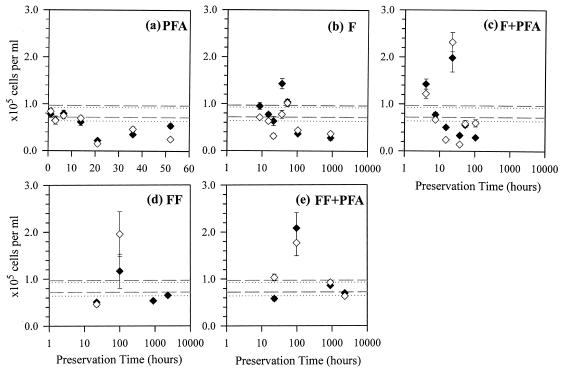
In the RS2 sample series, five preservation methods were tested. The results for the PFA method were the same as those for the RS2 storage effect experiment, in which all FISH cell counts showed significantly different means from reference values (P < 0.05; t tests) with systematic declining trends (Fig. 3a and 4a). Unlike the WA sample, the F method in the RS2 series did cause highly significantly lower cell counts in both DAPI and EUB with respect to reference values (P < 0.005 and P < 0.0005, respectively; t tests), probably due to the longer preservation time. There was a systematic decline in NSO190-detected β-AOB counts (natural logarithmic: R2 = 0.69; P < 0.05 [ANOVA]) (Fig. 4b), but not in other cell counts (P > 0.05; ANOVA) (Fig. 3b and 4b). In comparison, the F+PFA method resulted in no significant difference in any cell counts from the respective reference values (P > 0.05; t tests). However, if the anomalously high values at 22 h were excluded (Fig. 4c), the remaining data showed an exponential decrease in β-AOB abundance as detected with both NSO190 (R2 = 0.95; P < 0.05 [one-way ANOVA]) and NSO1225 (R2 = 0.76; P < 0.05 [ANOVA]). The anomalies at 22 h might be a result of heterogeneity introduced from particle associations common for β-AOB (33, 47). There were no noticeable temporal trends in DAPI and EUB, but the data variability was quite large (Fig. 3c).
FIG. 3.
Total microbial abundance estimated by DAPI staining (•) and eubacterial abundance by EUB338 in sample series RS2 preserved with paraformaldehyde (pfa) (a), formaldehyde (F) (b), combined formaldehyde-paraformaldehyde (F+pfa) (c), formaldehyde-freezing (FF) (d), and combined formaldehyde-freezing-paraformaldehyde (FF+pfa) (e). Dashed lines show the 95% CI of the EUB reference, and the dotted lines show the 95% CI of the DAPI reference. Note that the time scale is linear in panel a but logarithmic in panels b to e. Error bars, standard errors.
FIG. 4.
Abundance of β-AOB detected with NSO190 (♦) and with NSO1225 (⋄) in RS2 subsamples preserved by the PFA (a), F (b), F+PFA (c), FF (d), and FF+PFA (e) methods. Dashed lines show the 95% CI of the NSO1225 reference, and the dotted lines show the 95% CI of the NSO190 reference. Note that the time scale is linear in panel a but logarithmic in panels b to e. Error bars, standard errors.
The fourth preservation method, the FF method, resulted in DAPI and EUB cell counts significantly different from their reference values (P < 0.05; t tests), but no particular declining temporal trends were discernible in the data (Fig. 3d and 4d). Lastly, the combined FF+PFA method caused neither significantly different means from the respective references (P > 0.05; t tests) nor any declining temporal trends in any of the cell counts (P > 0.05; ANOVA), even after 96 days of storage. In fact, most of the measured values lie within the 95% confidence intervals (95% CI) of the respective original values (Fig. 3e and 4e). These results are summarized in Table 2.
TABLE 2.
Summary of results obtained by different preservation methods in saltwater aquarium RS2d
| Treatment | All microbes (DAPI)
|
Eubacteria (EUB338)
|
β-AOB (NSO190)
|
β-AOB (NSO1225)
|
||||
|---|---|---|---|---|---|---|---|---|
| Different meansa | Declining trendb | Different means | Declining trend | Different means | Declining trend | Different means | Declining trend | |
| PFA | Y | NS | Y | Exponential | Y | Linear | Y | Log-linear |
| F | Y | NS | Y | NS | N | Logarithmic | N | NS |
| F + PFA | N | NS | N | NS | N | Exponentialc | N | Exponentialc |
| FF | Y | NS | Y | NS | N | NS | N | NS |
| FF + PFA | N | NS | N | NS | N | NS | N | NS |
Significance of difference between results obtained with experimental treatments and the corresponding reference values was determined by t test (P < 0.05).
Significance of declining time trends was verified by one-way ANOVA (P < 0.05).
Exponential declining trend observed after excluding the data points at ≈22 h.
Abbreviations: Y, yes; N, no; NS, not significant.
Particle separation and sonication effects.
Substantial amounts of cell aggregates and exopolymeric substances were present in both the WA and HK whole-water samples, making cell counting under the microscope difficult. Size fractionation sequential filtration clearly facilitated cell counting. In the WA sample, the sums of all size fractions in DAPI- and UNI-hybridized cell counts yielded total microbial abundances that were 220 and 148%, respectively, of the nonfractionated whole sample (Table 3). Similarly, 239% of the total DAPI counts and 167% of the total UNI counts were obtained after size fractionation sequential filtration in the HK sample (Table 3).
TABLE 3.
Effects of size fractionation and sonication on total microbial abundance estimates by DAPI staining and FISH with UNI probesa
| Sample | Size class (μm) | WA samples
|
HK samples
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Nonsonicated
|
Sonicated
|
Nonsonicated
|
Sonicated
|
||||||
| DAPI | UNI | DAPI | UNI | DAPI | UNI | DAPI | UNI | ||
| Free living | 0.2-5.0 | 19.9 ± 1.8 | 10.8 ± 1.0 | 38.2 ± 2.3 | 19.5 ± 2.9 | 12.9 ± 1.2 | 8.3 ± 1.6 | ||
| Particle associated | 5.0-10 | 1.9 ± 0.7 | 0.5 ± 0.5 | 6.3 ± 0.5 | 3.1 ± 1.5 | 7.3 ± 0.5 | 7.0 ± 1.5 | 12.0 ± 0.6 | 9.6 ± 0.9 |
| >10 | 3.3 ± 0.3 | 1.5 ± 0.2 | 4.5 ± 0.2 | 5.4 ± 0.5 | 10.2 ± 0.7 | 7.1 ± 1.1 | 22.2 ± 0.9 | 18.7 ± 1.7 | |
| Σall fractions (% WS) | 25.1 ± 4.9 (220) | 12.7 ± 2.8 (148) | 33.2 ± 3.6 (291) | 28.0 ± 6.9 (326) | 30.3 ± 2.7 (239) | 22.5 ± 4.3 (167) | 47.0 ± 2.8 (370) | 36.7 ± 4.3 (272) | |
| Whole sample (% WS) | 11.4 ± 1.1 (100) | 8.6 ± 0.9 (100) | 12.7 ± 1.0 (100) | 13.5 ± 0.8 (100) | |||||
Cell abundances are expressed as 104 cells ml−1 (means ± SE) for the WA sample and 105 cells ml−1 (means ± SE) for the HK sample. Σall fractions represents the sum of results for all size fractions, and % WS indicates the percentage relative to the nonfractionated, nonsonicated whole sample. Since no sonicate data are available for the 0.2- to 5.0-μm-diameter size fraction in the HK sample, the nonsonicated cell counts were used to calculate the sonicated Σall fractions.
Brief sonication (5 s) in the WA sample increased the DAPI cell counts detected in all individual size fractions by 0.3- to 2.2-fold (P < 0.05; one-tailed paired t test) and, similarly, increased the UNI cell counts by 1.5- to 10-fold (P = 0.09; one-tailed paired t test). Consequently, the sums of all sonicated size fractions yielded 291% of the DAPI and 326% of the UNI total microbial abundance relative to the nonsonicated whole sample (Table 3).
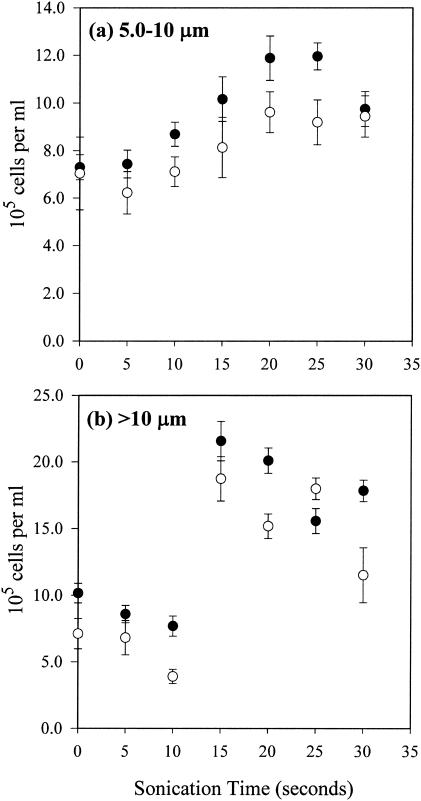
Longer sonication in the HK sample significantly increased both the DAPI and UNI cell counts, especially after >15 to 20 s in both 5.0- to 10-μm (P < 0.01; t tests for both DAPI and UNI)- and >10-μm-diameter fractions (DAPI, P < 0.005; UNI, P < 0.05 [t tests]) (Fig. 5). Trend analyses indicate a significant third-order relationship between cell counts and sonication time in the 5.0- to 10-μm-diameter fraction (R2 = 0.996 and P < 0.001 for DAPI; R2 = 0.945, P < 0.05 for UNI), but not in the >10-μm-diameter fraction (R2 = 0.842 and P = 0.35 for DAPI; R2 = 0.729 and P = 0.22 for UNI). The total microbial abundance as the sum of the two sonicated large size fractions and the nonsonicated 0.2- to 5.0-μm-diameter fraction (no sonication was performed in this size fraction) is equivalent to 370 and 272% of the nonsonicated whole-sample estimates by DAPI and UNI, respectively (Table 3). Further sonication (>20 to 25 s), however, started to reduce the cell counts in both the 5.0- to 10-μm- and >10-μm-diameter fractions. In general, more-even cell distribution was achieved on filters, and cell counting became much easier with less flocculent material present after sonication.
FIG. 5.
Increases in total microbial abundance estimated by DAPI staining (•) and UNI hybridization (○) after sonication, with an optimal sonication time at around 15 to 20 s in both 5.0- to 10-μm-diameter (a) and >10-μm-diameter (b) size fractions. Error bars, standard deviations.
DISCUSSION
Sample storage and preservation.
Prolonged storage of unpreserved water samples in a container may result in either increases or decreases in observed microbial abundance. Examples of these “bottle effects” include biofilm formation on the interior walls of bottles, cell aggregation, unusually fast microbial growth due to exclusion of some large natural predators during sampling, decreases due to cell deaths or substrate depletion, and changes in microbial community structure due to protistan predation. In the RS1 sample, the storage time of 27.5 h without preservation induced only minor positive changes to the microbial abundance estimates. However, after approximately 10 to 12 h, all abundance estimates in the RS2 sample decreased systematically, falling below the 95% CI. Although the exact temporal trends observed may not necessarily apply to all samples from various environments, these results strongly imply that unpreserved water samples can only be stored for a limited time without significant changes in microbial abundance—in this case no more than 10 to 12 h. If longer processing time is required, preservation is imperative.
Prefiltration 2% formaldehyde fixation is the most common alternative to postfiltration paraformaldehyde fixation (e.g., see references 17, 25, and 40) and is also a routine fixation method in DAPI staining for total microbial abundance (35). Good sample preservation for FISH should retain all cellular rRNA content, protect cell integrity and morphology, and allow good probe penetration during hybridization. Paraformaldehyde (O-CH2-O-CH2-O-CH2) is a trimer of formaldehyde (HCHO), and both are cross-linking fixatives which form DNA-protein cross-links within the cells. Due to its monomer structure, formaldehyde is the least cross-linking, takes longer to form sufficient irreversible cross-links within the cells, and is thus the least stabilizing among all aldehydes. Thus, the required fixation time was usually at least overnight or 16 h in the above-described studies. In both the WA and RS2 samples, DAPI and most FISH cell counts obtained using formaldehyde fixation alone were significantly lower than the reference values. In RS2, the 46% reduction in DAPI cell counts after 37 days of preservation in formaldehyde is similar to the 39% reduction reported in glutaraldehyde-fixed samples over 40 days in another study (48) but slower than the 30 to 40% reduction observed in viral abundance in formaldehyde-fixed samples after 7 days (14). Since RNA is less stable than DNA, such ineffectiveness in formaldehyde-fixation is even more pronounced in FISH cell counts. The EUB338-detected cell abundance in the same RS2 sample was reduced by 53% after 37 days, the NSO190-detected cell abundance in β-AOB was reduced by 71%, and the NSO1225-detected cell abundance in β-AOB was reduced by 48%. Although all cell counts at 2 and 4 days seemed to be close to the reference values, judging from the overall temporal trends these data points at 2 and 4 days appeared to be anomalies rather than representative measurements of the sample. The anomalies could have resulted from sample heterogeneity due to particle associations. On the whole, formaldehyde is an inadequate preservative for both rRNA in FISH and DNA in DAPI staining after prolonged storage.
In some studies, formaldehyde has also been used in conjunction with another aldehyde or precipitating preservatives like methanol or ethanol (e.g., see references 5 and 15). The addition of 4% paraformaldehyde after filtration (the F+PFA method) in sample series RS2 appeared to have stabilized the nucleic acids in the first few hours. Most DAPI and EUB cell estimates in F+PFA-treated samples lay within the 95% CI, yet they also showed the largest variability among all tested methods (standard deviation = 22 and 23% of the means). This preservation method may have been adequate only for the first 10 h, after which both β-AOB estimates decreased exponentially, falling well below the 95% CI.
For long-term storage, the FF method, or even better the combined FF+PFA method, seems to be the most effective preservation method. Both methods resulted in the smallest variability among the five methods tested (e.g., standard deviation = 16 and 14% of the means of EUB), except for an outlier at 4 days for β-AOB. In fact, most abundance estimates for FF+PFA-treated samples lay within the 95% CI of the reference values even after 96 days. Early cryopreservation evidently helped the retention of cellular rRNA. The FF and especially FF+PFA preservation methods are particularly useful for deep-sea sampling, when time is often limited.
Particle separation and sonication.
Quantifying microbial abundance in particle-rich waters using epifluorescence microscopy can be challenging. For example, flocculent materials often introduce background fluorescence, and the three-dimensional structures of cell aggregates hinder accurate cell counting. In addition, some mineral particles autofluoresce in the same emission wavelengths. Separation of microbes from particles should result in more-accurate abundance estimates. In this study, size fractionation sequential filtration removed the large particles (diameter, >10 μm) from smaller particles (diameter, <10 μm) or free-living cells, which helped reduce clogging in subsequent filtrations. The division into various size fractions also provides an opportunity to examine microbial association with various particle sizes, while revealing the characteristics of the particles. In the WA and HK samples, significantly higher microbial abundance estimates from DAPI and FISH were obtained after size fractionation, and there seemed to be effective separation between the particle-associated and free-living microbial assemblages. Size fractionation sequential filtration has been commonly applied in various aquatic ecological studies (12, 29, 39, 45).
Sonication has been widely applied to separate microbes from particles in soil (37, 43), sediments (13, 38), biofilms (5), water samples (49), limnetic snow (22, 50), and a hydrothermal vent chimney (11). Sonication invariably increased microbial abundance estimates. In this study, very few aggregates remained after sonication, and cells could be counted with greater ease on one plane of view under the microscope. The optimal sonication time in the HK sample was determined to be 15 to 20 s for both particle size fractions, resulting in more than 270% of the reference cell counts when combined with the use of size fractionation sequential filtration. The optimal sonication time likely depends on the surface characteristics of cells and associated particles or the presence of any chemical surfactants. For instance, out of the few reported studies, the optimum sonication times were 60 s for bacteria on leaf litter (44) and 3 min for viruses in marine sediments (14). Moreover, since further sonication after the optimal time started to reduce cell counts, optimization tests for sonication time are highly recommended for specific applications.
Conclusions.
This study illustrates the importance of validating different combinations of existing procedures for FISH sampling in specific environments. In this study, the commonly used prefiltration formaldehyde fixation was inferior to postfiltration paraformaldehyde fixation for FISH processing and was inadequate for prolonged storage. If long-term storage is required, the prefiltration formaldehyde fixation followed by immediate freezing and postfiltration paraformaldehyde fixation is the optimal preservation method, and this method is particularly useful for intensive deep-sea sampling. Size fractionation sequential filtration is effective in separating particles into various size classes, and sonication is effective in detaching microbes from particles or aggregates. It is necessary to optimize the sonication time for specific sample types. Together, size fractionation sequential filtration and sonication can significantly increase cell abundance estimates compared to those obtained in nonfractionated and nonsonicated whole samples of particle-rich waters. These optimized procedures can be incorporated into one effective FISH sampling protocol to study particle-associated microbes versus free-living microbes in the deep-seawater column.
Acknowledgments
This work was supported by the National Science Foundation (OCE-0095297 and OCE-9618243).
We sincerely thank the Waikiki Aquarium and Rachel Shackelford for their kind permission to sample their aquaria, and Edward Laws for generously providing the preserved culture sample of N. cryotolerans. We are also grateful to Kimberly Shaner and Donald McGee for their conscientious technical assistance.
Footnotes
SOEST contribution number 6301.
REFERENCES
- 1.Acinas, S. G., J. Anton, and F. Rodriguez-Valera. 1999. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 65:514-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alldredge, A. L., and M. Silver. 1988. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20:41-82. [Google Scholar]
- 3.Amann, R., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berelson, W. M. 2002. Particle settling rates increase with depth in the ocean. Deep-Sea Res. II 49:237-251. [Google Scholar]
- 7.Böckelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2002. Investigation of lotic microbial aggregates by a combined technique of fluorescent in situ hybridization and lectin-binding-analysis. J. Microbiol. Methods 49:75-87. [DOI] [PubMed] [Google Scholar]
- 8.Boetius, A., K. Ravenschlag, C. J. Schubert, R. D., F. Widdel, A. Gieseke, R. Amann, B. B. Jøgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apprently mediating anaerobic oxidation of methane. Nature 407:623-636. [DOI] [PubMed] [Google Scholar]
- 9.Bourne, D. G., A. J. Holmes, N. Iversen, and J. C. Murrell. 2000. Fluorescent oligonucleotide rDNA probes for specific detection methane oxidising bacteria. FEMS Microbiol. Ecol. 31:29-38. [DOI] [PubMed] [Google Scholar]
- 10.Caron, D. A., P. G. Davis, L. P. Madin, and J. M. Sieburth. 1986. Enrichment of microbial populations in macroaggregates (marine snow) from surface waters of the North Atlantic. J. Mar. Res. 44:543-565. [Google Scholar]
- 11.Chevaldonne, P., and A. Godfroy. 1997. Enumeration of microorganisms from deep-sea hydrothermal chimney samples. FEMS Microbiol. Lett. 146:211-216. [Google Scholar]
- 12.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danovaro, R., A. Dell'Anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 16.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanog. 38:924-934. [Google Scholar]
- 17.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.dos Santos Furtado, A. L., and P. Casper. 2000. Different methods for extracting bacteria from freshwater sediment and a simple method to measure bacterial production in sediment samples. J. Microbiol. Methods 41:249-257. [DOI] [PubMed] [Google Scholar]
- 19.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligonucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glöckner, F. O., R. Amann, A. Alreider, J. Pernthaler, R. Psenner, K. Trebesius, and K.-H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 22.Grossart, H.-P., and M. Simon. 1998. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat. Microb. Ecol. 15:127-140. [Google Scholar]
- 23.Jackson, G. A., and A. Burd. 2002. A model for the distrubution of particle flux in the mid-water column controlled by subsurface biotic interactions. Deep-Sea Res. II 49:193-217. [Google Scholar]
- 24.Karner, M. B., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archael dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 26.Lemke, M. J., C. J. McNamara, and L. G. Leff. 1997. Comparison of methods for the concentration of bacterioplankton for in situ hybridization. J. Microbiol. Methods 29:23-29. [Google Scholar]
- 27.Long, R. A., and F. Azam. 2001. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat. Microb. Ecol. 26:103-113. [Google Scholar]
- 28.Mobarry, B., M. Wagner, V. Urbain, B. Rittmann, and D. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. (Erratum, 63:815, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mousseau, L., L. Legendre, and L. Fortier. 1996. Dynamics of size-fractionated phytoplankton and trophic pathways on the Scotian Shelf and at the shelf break, Northwest Atlantic. Aquat. Microb. Ecol. 10:149-163. [Google Scholar]
- 30.Murrell, J. C., I. R. McDonald, and D. G. Bourne. 1998. Molecular methods for the study of methanotroph ecology. FEMS Microbiol. Ecol. 27:103-114. [Google Scholar]
- 31.Nebe-von-Caron, G., P. J. Stephens, C. J. Hewitt, J. R. Powell, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 32.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, C. J., Z. Smith, T. M. Embley, and J. I. Prosser. 1999. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilskaln, C. H., C. Lehmann, J. B. Paduan, and M. W. Silver. 1995. Spatial and temporal dynamics in marine aggregate abundance, sinking rate and flux: Monterey Bay, central California. Deep-Sea Res. II 45:1803-1837. [Google Scholar]
- 35.Porter, K., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanog. 25:943-948. [Google Scholar]
- 36.Proctor, L. M., and A. C. Souza. 2001. Method for enumeration of 5-cyano-2,3-ditoyl tetrazolium chloride (CTC)-active cells and cell-specific CTC activity of benthic bacteria in riverine, estuarine and coastal sediments. J. Microbiol. Methods 43:213-222. [DOI] [PubMed] [Google Scholar]
- 37.Ramsay, A. J. 1984. Extraction of bacteria from soil: efficiency of shaking or ultrasonication as indicated by direct counts and autoradiography. Soil Biol. Biochem. 16:475-481. [Google Scholar]
- 38.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph, C., G. Wanner, and R. Huber. 2001. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67:2336-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schallenberg, M., J. Kalff, and J. B. Rasmussen. 1989. Solutions to problems in enumerating sediment bacteria by direct counts. Appl. Environ. Microbiol. 55:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelley, B. C. L., and J. A. Perry. 2000. Evaluation of bacterial recovery efficiency and counting precision from decaying leaf litter in Little Rock Lake, Wisconsin, USA. J. Fresh. Ecol. 15:157-169. [Google Scholar]
- 45.Simek, K., J. Pernthaler, M. G. Weinbauer, K. Hornak, J. R. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 47.Stehr, G., S. Zörner, B. Böttcher, and H.-P. Koops. 1995. Exopolymers: an ecological characteristic of a floc-attached, ammonia-oxidizing bacterium. Microb. Ecol. 30:115-126. [DOI] [PubMed] [Google Scholar]
- 48.Turley, C. M., and D. J. Hughes. 1992. Effects of storage on direct estimates of bacterial numbers of preserved seawater samples. Deep-Sea Res. 39:375-394. [Google Scholar]
- 49.Velji, M. I., and L. J. Albright. 1986. Microscopic enumeration of attached marine bacteria of seawater, marine sediment, fecal matter, and kelp blade samples following pyrophosphate and ultrasound treatments. Can. J. Microbiol. 32:121-126. [Google Scholar]
- 50.Weiss, P., B. Schweitzer, R. Amann, and M. Simon. 1996. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl. Environ. Microbiol. 62:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon, W. B., and R. A. Rosson. 1990. Improved method of enumeration of attached bacteria for study of fluctuation in the abundance of attached and free-living bacteria in response to diel variation in seawater turbidity. Appl. Environ. Microbiol. 56:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zarda, B., D. Hahn, A. Chatzinotas, W. Schonhuber, A. Neef, R. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]