Abstract
Arctic wintertime sea-ice cores, characterized by a temperature gradient of −2 to −20°C, were investigated to better understand constraints on bacterial abundance, activity, and diversity at subzero temperatures. With the fluorescent stains 4′,6′-diamidino-2-phenylindole 2HCl (DAPI) (for DNA) and 5-cyano-2,3-ditoyl tetrazolium chloride (CTC) (for O2-based respiration), the abundances of total, particle-associated (>3-μm), free-living, and actively respiring bacteria were determined for ice-core samples melted at their in situ temperatures (−2 to −20°C) and at the corresponding salinities of their brine inclusions (38 to 209 ppt). Fluorescence in situ hybridization was applied to determine the proportions of Bacteria, Cytophaga-Flavobacteria-Bacteroides (CFB), and Archaea. Microtome-prepared ice sections also were examined microscopically under in situ conditions to evaluate bacterial abundance (by DAPI staining) and particle associations within the brine-inclusion network of the ice. For both melted and intact ice sections, more than 50% of cells were found to be associated with particles or surfaces (sediment grains, detritus, and ice-crystal boundaries). CTC-active bacteria (0.5 to 4% of the total) and cells detectable by rRNA probes (18 to 86% of the total) were found in all ice samples, including the coldest (−20°C), where virtually all active cells were particle associated. The percentage of active bacteria associated with particles increased with decreasing temperature, as did the percentages of CFB (16 to 82% of Bacteria) and Archaea (0.0 to 3.4% of total cells). These results, combined with correlation analyses between bacterial variables and measures of particulate matter in the ice as well as the increase in CFB at lower temperatures, confirm the importance of particle or surface association to bacterial activity at subzero temperatures. Measuring activity down to −20°C adds to the concept that liquid inclusions in frozen environments provide an adequate habitat for active microbial populations on Earth and possibly elsewhere.
The constraints on and sustainability of life in frozen environments are of considerable importance in a number of contexts, from polar microbial ecology and astrobiology to cryopreservation and other industrial applications (42). For example, a number of subzero environments, such as Antarctic and Arctic lakes (23, 25, 38), snow (3), glacial ice (46), and permafrost soils (41), have been investigated as Earth analogs for potential extraterrestrial habitats also at subzero temperatures. To date, fundamental questions underlying the behavior of bacteria in any frozen environment have not been adequately addressed: how do bacteria manage to persist and possibly remain active? At the lowest temperatures observed on Earth, what environmental factors enable and control bacterial survival and even sustained activity?
This study focused on Arctic wintertime sea ice, the coldest marine habitat on Earth (temperature range of −2 to −35°C) (31) and an important component of polar climate and ecosystems. From bulk measurements made with melted sea-ice samples, extensive microbial communities are known to flourish within the polar ice covers during the sunlit season, contributing significantly to the polar ocean carbon budget (49). The few reported wintertime studies of sea ice, even though based on melted samples and incubations at nearly seawater (warmer than in situ) temperatures and salinities, nevertheless have suggested that activity may continue under the more extreme conditions of the dark season (19). Studies that document metabolically active bacteria in the extremely cold horizons of wintertime sea ice, however, are not available, leaving a significant gap in the understanding of bacterial survival in frozen environments.
To assess the potential for continuing microbial activity during winter, new analysis techniques had to be developed and applied to wintertime collections of sea ice. Most sea-ice bacterial studies not only have been limited to “warm” sunlit conditions but also have relied almost entirely on destructive treatment (melting) of the ice matrix. For this study, samples were collected near Barrow, Alaska, during the winters of 1999, 2000, and 2001 and kept close to in situ temperatures throughout treatment and analysis (17). In one approach, bacteria were stained and observed microscopically within the three-dimensional network of brine inclusions in intact ice sections (no melting) for the first time (21). In the second, ice samples were melted but under highly saline brine conditions that protected against osmotic (and thermal) shock and allowed for incubation at nearly in situ temperatures and brine salinities with the fluorescent dye 5-cyano-2,3-ditoyl tetrazolium chloride (CTC). The CTC method identifies respiratory activity in highly active bacterial cells specifically undergoing oxidative metabolism (44). For a different and more generalized measure of cellular activity (as well as community structure), fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes (30) also was performed with ice samples melted under nearly in situ brine conditions. Hybridizable or rRNA probe-detectable cells are interpreted as a very sensitive measure of active cells in a community (26), since the threshold signal of FISH depends on the cellular rRNA content.
Attachment to surfaces is a well-known microbial strategy for surviving a variety of conditions in marine and other systems (7, 14), even though underlying mechanisms remain poorly known (29). Despite high numbers of potential interior attachment sites (48), however, Arctic sea ice had not been investigated from this perspective. In order to evaluate attachment as an adaptive strategy for bacteria to maintain activity at extremely low temperatures, we used our new methods to test whether (i) bacteria were associated with particles or surfaces under in situ conditions in intact ice sections, (ii) particle-associated bacteria were more active with decreasing temperature, and (iii) bacterial types characterized by a surface-associated lifestyle—Cytophaga-Flavobacteria-Bacteroides (CFB) (27), already known to exist in springtime sea ice (24) and to predominate in Antarctic waters (45)—increased in abundance with decreasing temperature.
We further investigated whether Archaea, known to be abundant in Antarctic seawater during winter (35) and recently shown to be enriched in Arctic nepheloid (particle-rich) layers (51), also could be detected in wintertime sea-ice samples by FISH. This possibility was of particular interest, since Archaea could not be detected in 16S ribosomal DNA clone libraries prepared from Antarctic or Arctic sea ice (2) sampled during spring or summer.
MATERIALS AND METHODS
Sample collection and processing.
Sea-ice samples were collected during the coldest period of the Arctic winter, in March 1999, 2000, and 2001 (when air temperatures typically ranged between −35 and −40°C), from two sites readily accessed from Barrow, Alaska—one on the coastal fast ice of the Chukchi Sea (at 71.33°N, 156.68°W, in 1999; 71.33°N, 156.70°W, in 2000; and 71.33°N, 156.67°W, in 2001) and the other in nearby Elson Lagoon (at 71.35°N, 156.52°W, in all 3 years). Ice cores were taken by using a 10-cm-diameter ice auger. Samples were representative of first-year sea ice, with characteristic temperature and salinity profiles (17). While most of the physical properties of the Elson Lagoon ice were comparable to those of the Chukchi Sea ice, the particulate content in the upper sediment-rich layers of the shallow lagoon ice was markedly higher, generally exceeding 10 mg liter−1 (48).
The ice cores were placed in insulated containers that maintained the samples at or near the lowest in situ temperature (−20°C) (17) during sample transport to the nearby laboratory at Barrow to prepare immediately for in situ respiration studies. The cores collected last were shipped in these containers to the University of Alaska Fairbanks (UAF) for in situ microscopy work. Upon arrival at UAF, they were transferred to a −20°C freezer until processing. Sample processing for in situ microscopy, begun as soon as possible after return from Barrow, was performed in a temperature-controlled freezer room at UAF, which accommodated the equipment required for the preparation of ice sections as well as the microscope and image acquisition system.
Sterile conditions were maintained carefully during sampling and processing in the field and laboratory. In the field, ice temperatures in the ice-core interior were measured immediately after coring with a thermistor probe (precision, <0.1°C) (17). Ice horizons corresponding to in situ temperatures (and in situ brine salinities) of −2°C (38 ppt), −5°C (87 ppt), −10°C (143 ppt), −15°C (178 ppt), and −20°C (209 ppt) were cut from the cores by using a surface-sterilized saw immediately after coring and then transported in the insulated containers to the laboratory in Barrow. After being rinsed with sterile distilled water (melting away ∼2 mm of the exterior surfaces), the ice sections were placed in sterile plastic bags, weighed, crushed, and melted in measured volumes of prefiltered (0.2-μm-pore size), sterile, high-salinity brine solutions at the relevant in situ temperatures (−2, −5, −10, −15, and −20°C) and with final melt salinities of 65, 108, 150, 200, and 220 ppt for the temperatures, respectively (as described previously [21]). This isothermal-isohaline melting resulted in samples with salinities high enough to prevent freezing during subsequent incubation within the ice sheet but close enough to the in situ brine salinities to reduce the possibility of cell lysis due to sudden changes in osmotic pressure.
Microscopic analysis of ice sections.
Procedures for direct microscopic observation of stained bacteria in Arctic sea-ice sections closely followed the protocol developed for in situ investigation of sea-ice samples by Junge et al. (21). Briefly, sample processing, including preparation of thin sections (sawing and cutting with a microtome), staining, and microscopic observations were performed in three stages at −5, −15, and −20°C (for the corresponding ice sections) in the freezer room at UAF, which accommodated the microscope and image acquisition system. Placement of a Hobo temperature logger (Onset Computers Corporation, Pocasset, Mass.) on the microscope stage verified that the examination unit maintained the desired temperature within 1°C during sample processing and analysis.
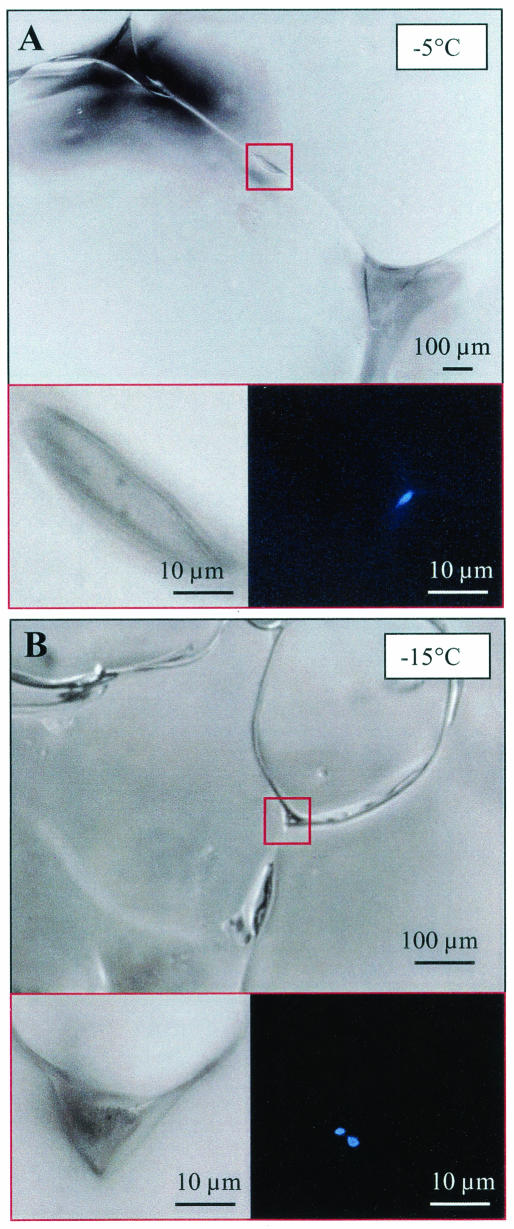
After preparation of a thin section, a temperature- and salt-specific solution of the DNA-specific fluorescent stain 4′,6′-diamidino-2-phenylindole 2HCl (DAPI) was added to the microtome-prepared surface. The staining solution was prepared to a final concentration of 20 μg of DAPI ml−1 in brine prepared from artificial sea salts (Instant Ocean) at final concentrations of 87 ppt (for −5°C), 178 ppt (for −15°C), and 209 ppt (for −20°C) (18), equilibrated to their respective temperatures, for a minimum of 2 days prior to use (see reference 21 for more details). After application to the microtome-prepared surface, the stain was allowed to diffuse into the sample for at least 1 h and up to 24 h in the dark before microscopic examination. Examination by epifluorescence microscopy with the optical filter set for DAPI then was begun at least 10 μm below the ice surface for a total examination thickness of 100 μm and a minimum of 500 microscopic fields examined (requiring many hours of observer time in the dark at freezing temperatures). When bacteria were encountered, images were recorded under both DAPI excitation and transmitted light. These images were later analyzed to enumerate bacteria and to determine their locations within the ice matrix and associations with ice features (e.g., brine channels, ice crystal walls, or particles) (Fig. 1). Bacteria were considered “attached” when an association with ice crystal walls or particles was clearly observed (by focusing in three dimensions); without such an association, bacteria were judged to be “free living” (Fig. 2). Microscopic observations also were made at decreasing magnifications with ×63, ×20, ×5, and ×2.5 objectives to combine observations of bacteria with those of other particles and with the morphology of the pore space.
FIG. 1.
Microscopic images of wintertime sea ice from the Chukchi sea near Barrow, Alaska, at −5 (A) and −15°C (B). Ice-grain boundaries and triple-point junctures (upper panels) and details of brine pockets (lower left panels, which are enlargements of the areas boxed in red in the upper panels) are visible by transmitted light. DAPI-stained bacteria (blue) attached to the wall of a brine pocket (A) or to particulate material within the pocket (B) are visible in the same fields as those shown in the lower left panels when examined by epifluorescence light (lower right panels).
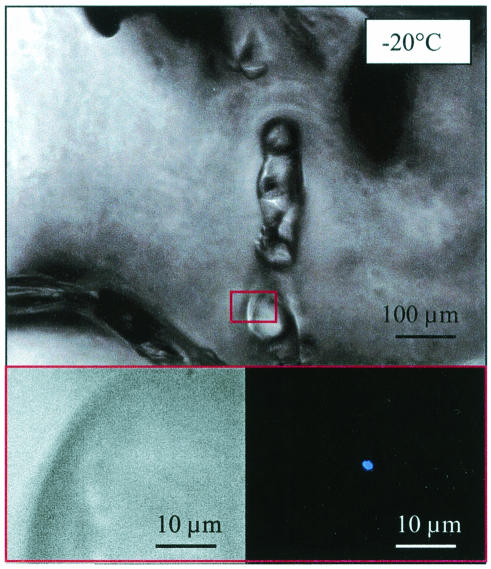
FIG. 2.
Microscopic images of wintertime sea ice at −20°C. The lower left panel is an enlargement of the area boxed in red in the upper panel. The images are similar to those in Fig. 1, except that a triple-point juncture is not obvious and the DAPI-stained bacterium (lower right panel) is not attached to a surface.
A Zeiss Axioskop 2 microscope, fitted with a set of ×2.5, ×5, ×20, and ×63 (immersion) objectives, an epifluorescence illumination system, and a 50-W mercury lamp, was used for transmission and epifluorescence microscopy of the ice sections. The microscope was modified in the factory for operation at subzero temperatures (−2 to −25°C) by exchanging all lubricants to maintain low viscosities at low temperatures. Nonfluorescent, filter-sterilized artificial brine prepared as described by Junge et al. (21) was used as the immersion fluid for the ×63 objective, which was a ceramic-tipped model. For epifluorescence work, the microscope was equipped with an optical filter set for DAPI (365-nm excitation, 395-nm beam splitter, and long-pass 420-nm emission).
Images of fluorescent cells and other features were captured by using an MTI DC330E3 charge-coupled device color camera and a Scion CG-7 RGB color PCI frame grabber. At the highest magnification, a measure of 1 μm on the sample slide corresponded to 10 linear pixel dimensions, with a total image size of 768 by 576 pixels. Pixels were digitized to 8 bits (512 gray levels) for each of the three RGB color channels. Image analysis was done on the host computer (G3 Macintosh with 128 MB). All images were acquired, calibrated, analyzed, and displayed by using a variant of NIH Image software, version 1.62a (39). The microscope features coupled with the video imaging system used facilitated visualization at a maximum magnification of ×3,230.
DAPI and CTC analyses of melted ice samples.
Concentrations of total, attached, and free-living bacteria were determined for triplicate 20-ml subsamples by using the DNA-specific fluorescent stain DAPI. Abundances of free-living bacteria were determined for sample filtrates obtained by gentle filtration through a 3-μm-pore-size polycarbonate TE membrane filter. Attached or particle-associated bacteria were defined as the total counts minus the free-living counts. The number of cells in each case was scaled to the volume of ice sampled as well as to the volume of brine within the ice sample (determined as described below).
Concentrations of actively respiring cells (ARC) were similarly determined for total, attached, and free-living cells by using the fluorescent electron transport system-specific reagent CTC (44) at a final concentration of 5 mM (24). Samples were equilibrated for several hours at or below their intended incubation temperatures (−2, −10, −15, and −20°C) prior to amendment with CTC. Twenty-four-hour incubations with CTC were performed within the ice sheet at the appropriate temperature horizon (−2, −5, −10, −15, and −20°C) by returning the contained samples, suitably spaced within a plastic ice-core sleeve, to an open ice-core hole (approach adapted from that described in reference 33). To account for false-positives and autofluorescence, formalin-killed controls were incubated along with live samples. After incubation, samples were preserved with formaldehyde (final concentration, 2%) and stored at −20°C until slide processing and counting as described by Junge et al. (24). No CTC-positive cell was found in any of the formalin-killed controls (up to 1,300 cells were checked per sample).
FISH analyses of melted ice samples.
Samples were obtained from the same ice horizons and prepared in the same fashion as for CTC incubations, except that 2% formaldehyde was added to each brine solution prior to sample melting. DAPI and FISH counts of total and free-living cells were determined for filtered (3-μm-pore size) subsamples. Triplicate 80-ml subsamples of the whole sample and the filtrate were filtered onto a 0.2-μm-pore-size polycarbonate TE black membrane filter. Cy-3-conjugated probes specific for Bacteria (EUB338), the Cytophaga-Flavobacteria cluster of the CFB phylum (CF319a), and Archaea (ARCH915) were applied for hybridization by following standard filter hybridization protocols essentially as described by Wells and Deming (51; see also references 30 and 45). Pure cultures of Bacteria, CFB, and Archaea were used to optimize the hybridization signal and probe specificity.
Filters for each sample were cut aseptically into four equal sections. After dehydration by sequential 2-min washes in 50, 80, and 96% ethanol, each section was mounted on a coverslip. A 65-μl volume of hybridization buffer [0.9 M NaCl, 20 mM Tris-HCl (pH 8.0), 0.01% sodium dodecyl sulfate, 0.1 mg of poly(A) ml−1, 0.2 mg of bovine serum albumin ml−1, 20 to 40% formamide] was added, covering the entire filter, and prehybridization was carried out for 30 min at 46°C. After prehybridization, 250 ng of Cy-3-conjugated probe was added to the buffer, and hybridization was carried out for 6 h at 46°C. To account for autofluorescence, no probe was added to one of the sections.
After hybridization, the filter sections were washed for 25 min at 48°C in wash buffer (20 mM Tris-HCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate, 56 to 225 mM NaCl [depending on the formamide concentration]), dried, and mounted on slides with Vectashield mounting medium containing 1.5 μg of DAPI ml−1. Each hybridization reaction was accompanied by positive and negative controls from laboratory cultures. The filter sections were examined at a magnification of ×1,563 by using a Zeiss universal microscope with filter sets for DAPI- and Cy-3-labeled cells (365 nm excitation, 395 nm beamsplitter, 420 nm emission, and 545 nm excitation, 565 nm beamsplitter, 610 nm emission, respectively). Bacteria, CFB, and Archaea were enumerated in the whole sample and the free-living fraction, accounting for attached bacterial populations by subtraction. Total and rRNA probe-detectable cells were counted in at least 20 randomly selected fields (minimum count of 200 cells).
Determination of chemical variables.
All bulk chemical variables, including salinity, were measured in a standard fashion (15) for melted samples in parallel ice cores from the same ice horizons as those used for bacterial counts. Values for bulk salinity were used with field measurements of ice-core temperatures and the equations of Cox and Weeks (9) to determine the volume of brine within a given ice section. For analysis of dissolved organic carbon (DOC), ice samples were rinsed with sterile distilled water to remove potential contaminating traces of organic carbon before being melted in muffled glass beakers at room temperature. Six-milliliter samples were prefiltered with precombusted GF/F filters to remove particulate organic carbon (POC) at >0.7 μm. DOC concentrations in the filtrates were measured after acidification with HCl by the high-temperature catalytic oxidation method with a Shimadzu total organic carbon analyzer. For POC and particulate organic nitrogen (PON) contents, samples were filtered with precombusted GF/F filters, dried for ∼24 h at 60°C, and fumed with HCl (using the vapor method) to remove the carbonate fraction for 24 h. POC and PON quantification was done with a Leeman Labs model CEC440 elemental analyzer. Total particulate inorganic matter (PIM) and particulate organic matter (POM) contents were determined by filtering up to 1,000 ml of melted sample with precombusted and preweighed GF/F filters. POM and PIM contents were determined from dry weights before and after combustion. Concentrations of DOC, POC, PON, PIM, and POM were scaled to volume of ice melted as well as to volume of brine within a given ice section.
RESULTS AND DISCUSSION
For the entire range of temperatures examined, including the coldest (−20°C), microscopic observations of intact ice sections revealed numerous liquid brine inclusions that were inhabited by bacteria (Fig. 1 and 2). On the scale of a bacterium, a substantial volume of habitable brine-filled pore space has been shown to exist within the ice matrix even at −20°C, with both isolated and fully connected brine tubes, veins, and junctures occurring at densities exceeding 150 mm−3 (17); these densities are almost 2 orders of magnitude higher than those reported in previous studies at lower magnifications. This reservoir of unfrozen water, a prerequisite for microbial activity, allows for fluid flow induced by local thermomolecular pressure gradients or larger-scale temperature gradients (10, 52) as well as for possible movement of bacteria within the ice (22, 37).
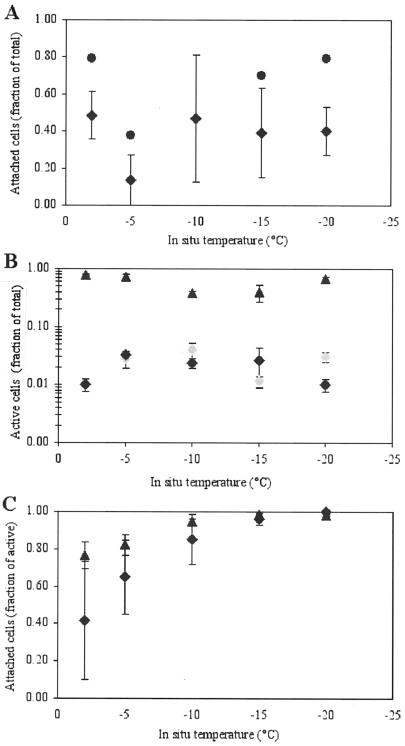
Most bacteria, however, were observed to be associated with a variety of surfaces (sediment grains, detritus, and ice-crystal boundaries). Higher proportions of attached bacteria were observed in intact ice sections (up to 79%) (Fig. 1; see also images shown in reference 21) than in parallel samples that had been melted and size fractionated (up to ∼50%) (Fig. 3A), a result which we attribute to the fact that ice-wall associations cannot be determined with melted samples. At ∼50%, the mean fraction of attached cells in the Chukchi Sea ice (with types and concentrations of particles typical of ordinary coastal sea ice) (40) is high compared to fractions of 1 to 20% in Arctic seawater (20) or temperate marine environments (4). In ice horizons with entrained seafloor sediments, obtained (only) from shallow Elson Lagoon, over 95% of the bacteria were found associated with particles, in keeping with the similarly high fraction of attached bacteria found in seafloor sediments (43).
FIG. 3.
Fractions of total bacteria that were attached (A) or active (B) and fractions of active cells that were attached (C) across the temperature gradient in wintertime sea ice. Circles indicate data from intact ice sections examined microscopically. Diamonds indicate mean values from isothermal-isohaline-melted ice samples used for CTC incubations: black diamonds indicate samples from the Chukchi Sea, and gray diamonds indicate samples from Elson Lagoon. Triangles indicate mean values from isothermal-isohaline-melted ice samples used for rRNA probing. Error bars indicate the SEM (n = 3).
The proportion of total attached cells in the Chukchi Sea ice samples did not change over the temperature range studied (Fig. 3A), arguing against preferential growth or physical accumulation of bacteria on particles as a function of temperature. This scenario changes, however, when the active part of the sea-ice bacterial population is examined (Fig. 3B and C). ARC were documented at all incubation temperatures (Fig. 3B), including −20°C; this temperature is 18°C lower than the temperature at which bacterial activity in sea ice was reported previously (19), 5°C colder than the temperature at which thymidine and leucine incorporation by bacterial isolates from glacial ice occurred (5), 3°C colder than the temperature at which thymidine uptake activity in Antarctic snow occurred (3), and on a par with the temperature at which rates of acetate metabolism in permafrost were near the detection limit (41). The mean percentages of active cells detected with CTC in this study ranged from 0.5 to 4% (Fig. 3B). These percentages were detected readily in melted ice samples of sufficient filtration volume but not in intact ice sections (the search time required to visualize a significant number of CTC-stained cells within microscopic brine pores was prohibitive, both for the observer, working at freezing temperatures, and for the ice section, given the onset of sublimation). The observed percentages of CTC-stained cells are similar to those found for summertime seawater bacteria in the Chukchi Sea (mean and standard error of the mean [SEM], 3.3% ± 1.3%) (24, 44), suggesting that bacterial populations in Arctic wintertime sea ice are proportionately as active as their summertime counterparts in seawater, but at much lower temperatures and higher salinities. Typically, CTC-stained cells in seawater account for only 1 to 10% of the total bacterioplankton assemblage (26, 44).
The high proportions of rRNA probe-detectable cells (range, 18 to 86%; mean and SEM, 60% ± 20%; n = 16) (Fig. 3B and 4) indicate that the majority of cells were active by that measure, maintaining protein synthesis machinery even at the coldest temperatures and salinities examined. These percentages are as high as those found for temperate coastal marine environments (8, 26) and are almost double those often reported for mesotrophic or oligotrophic systems, including cold polar regions (51) and the deep sea (12).
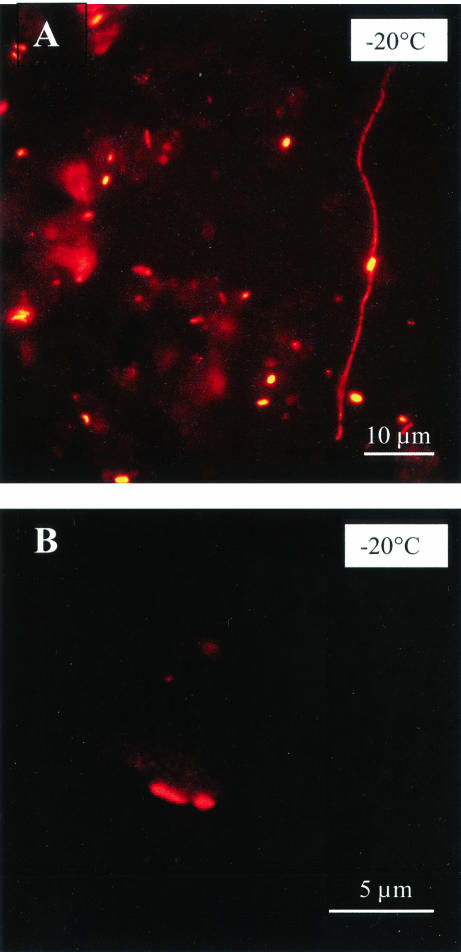
FIG. 4.
Images obtained by epifluorescence microscopy of particle-associated bacteria in isohaline-isothermal-melted samples of wintertime ice from the Chukchi Sea at −20°C. Hybridization with fluorescent probes is shown for Bacteria (A) and Archaea (B).
Contrary to the expectation of severe metabolic inhibitions at such extreme conditions, the proportions of active cells detected with either the CTC or the FISH method did not decrease (regardless of the differences between the methods, as observed previously by others [26]) when the cells were exposed to increasingly lower temperatures and higher salinities (Fig. 3B). This observation suggests that Arctic marine bacteria have specific adaptations for coping with the cold and salt encountered in sea ice or that exposure to increasingly harsh conditions as the winter progresses selects for active psychrophilic halophiles. From Antarctic studies, sea ice is already known to be one of the few environments favoring a predominance of psychrophiles over cold-tolerant microorganisms (19), but the link between psychrophilic and halophilic properties remains to be fully explored (36).
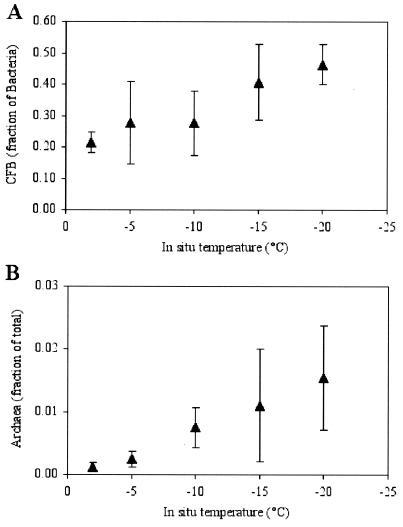
We have identified in this study one possible strategy for continued bacterial activity in wintertime sea ice: association with particles or surfaces (Fig. 3C and 4). Even though the proportions of attached cells remained relatively constant across the temperature gradient in the ice (Fig. 3A), the proportions of active (ARC and rRNA probe-detectable) cells that were attached increased toward the coldest ice horizons, where essentially all active cells were particle associated (Fig. 3C). Virtually no free-living cell was found to be CTC positive or to hybridize with the probes used for the −15 and −20°C ice samples. An increase in the proportions of CFB with decreasing temperature also was observed, such that these indicators for surface-associated populations (27) nearly dominated the bacterial community (46% of the total hybridizable cells) in the coldest ice horizon (Fig. 5A). These percentages (range, 16 to 82%; mean and SEM, 35% ± 17%; n = 16) are as high as or higher than those found for temperate seawater (8), reaffirming the numerical importance of CFB in high-latitude marine environments (24, 45, 51) and underscoring their hardiness.
FIG. 5.
Fractions of Bacteria that were CFB (A) and fractions of total cells that were Archaea (B) across the temperature gradient in wintertime sea ice. Error bars indicate the SEM (n = 3).
Archaea also were found to be present (Fig. 4B and 5B), although not abundant, in our sea-ice samples (range, 0.0 to 3.4%; mean and SEM, 0.7% ± 1%; n = 15); these percentages were similar to those reported for Arctic surface waters during autumn (0.1 to 2.6%) (51) and Antarctic surface waters during summer (0.2 to 1.3%; mean and SEM, 2.3% ± 2.4%) (34, 35). Even though these data represent the first evidence for archaea in sea ice, the finding of such low percentages in wintertime sea ice is somewhat contrary to expectation, given the high percentages of archaea (5 to 14%) found in at least some polar (Antarctic) waters during winter (34). However, similar to the behavior of CFB cells, we observed an increase in the proportions of archaea with decreasing temperature (Fig. 5B) (like others, we cannot exclude the possibility of probe cross-reactivity). If archaea prefer surfaces for growth—as indicated by the marked increase in their proportional abundance in Arctic nepheloid layers (51)—this finding supports our hypothesis that particle-associated cells have an advantage over free-living ones in extremely cold environments and can remain active.
Particle-associated bacteria are proportionally more active than free-living bacteria in many other marine habitats, including sediments and turbidity maxima (11), as well as various pelagic environments ranging from warm oligotrophic waters to highly productive coastal areas and cold polar waters (44, 47). Particularly at very low temperatures in sea ice, when brine volumes are reduced, the concentration of particulate matter in the brine can result in a high ratio of surface area to brine volume. Since only the active fraction of bacteria (and not the total number of cells) that were attached increased as the temperature decreased, surface association must present or reflect a distinct advantage to active cells as conditions become more severe.
A global correlation analysis of all of our wintertime ice samples (independent of temperature) reconfirmed the general importance of particulate matter to microbial life, in that highly significant correlations of total, attached, and metabolically active cell numbers were detected only with particulate variables (POC, PON, PIM, and POM but not DOC; all variables were scaled to ice volume) (Table 1). This finding suggests that the well-known relationship between activity and particulate matter also holds for sea-ice bacteria, even in the presence of high DOC concentrations in the ice (Table 1) (50). When variables were scaled to brine volume, correlations of bacterial densities with particulate variables became even more robust (higher r values and P values of <0.001) (Table 2) and additional significant relationships emerged, e.g., with concentrations of DOC and free-living bacteria. In fact, all brine-scaled variables were found to be tightly correlated with each other, emphasizing the importance of scaling to the actual microbial habitat (the brine). This analytical approach may be particularly appropriate for wintertime sea ice, when the physical concentration effects in freezing brine are accentuated. For example, brine scaling revealed no correlation between total bacterial counts and DOC in summertime (Antarctic) sea ice (a result attributed to the confounding effects of an active ice algal and grazer community in the more extensive brine inclusions of warmer ice) (50) but did reveal a significant correlation between concentrations of total bacteria and extracellular polymeric substances (EPS) in wintertime (Arctic) sea ice (28). The latter correlation, observed only for wintertime (and not springtime) ice, contributed to the conclusion that EPS play a cryoprotective role in winter (28).
TABLE 1.
Median, range, and Spearman rank order correlation coefficients for Arctic wintertime sea-ice chemical and bacterial variables (n = 16) scaled to ice volume
| Variable (U)a | Median (range) |
r value for the following cellsb:
|
|||
|---|---|---|---|---|---|
| Total | ARC | FL | ATT | ||
| DOC (mg of C liter−1) | 2.88 (1.37-69.2) | 0.59 | 0.50 | 0.54 | 0.55 |
| POC (mg of C liter−1) | 0.265 (0.0226-28.4) | 0.83** | 0.70* | 0.57 | 0.72* |
| PON (mg of N liter−1) | 0.045 (0.0018-3.6) | 0.85** | 0.74** | 0.61 | 0.76** |
| PIM (mg liter−1) | 4.04 (0.400-1,090) | 0.91** | 0.87** | 0.75** | 0.85** |
| POM (mg liter−1) | 2.64 (0.00-124) | 0.71** | 0.72** | 0.52 | 0.69** |
| Total cells (104 ml−1) | 7.97 (1.60-301) | 0.89** | 0.73** | 0.91** | |
| ARC (103 ml−1) | 1.97 (0.161-55.4) | 0.58 | 0.82** | ||
| FL cells (104 ml−1) | 4.23 (1.41-31.9) | 0.61 | |||
| ATT cells (104 ml−1) | 2.53 (0.193-291) | ||||
FL, free living; ATT, attached.
*, P< 0.01; **, P < 0.001.
TABLE 2.
Median, range, and Spearman rank order correlation coefficients for Arctic wintertime sea-ice chemical and bacterial variables (n = 16) scaled to brine volume
| Variable (U)a | Median (range) |
r value for the following cellsb:
|
|||
|---|---|---|---|---|---|
| Total | ARC | FL | ATT | ||
| DOC (mg of C liter−1) | 89 (10.3-3,540) | 0.75 | 0.69* | 0.80 | 0.74 |
| POC (mg of C liter−1) | 4.45 (0.491-1,450) | 0.88 | 0.77 | 0.75 | 0.87 |
| PON (mg of N liter−1) | 0.64 (0.037-138) | 0.84 | 0.76 | 0.71* | 0.82 |
| PIM (mg liter−1) | 209 (16.6-563,000) | 0.96 | 0.94 | 0.84 | 0.93 |
| POM (mg liter−1) | 101 (0.00-6,150) | 0.80 | 0.79 | 0.73 | 0.79 |
| Total cells (106 ml−1) | 3.36 (0.0867-154) | 0.92 | 0.85 | 0.97 | |
| ARC (104 ml−1) | 4.79 (0.0967-284) | 0.75 | 0.89 | ||
| FL cells (106 ml−1) | 1.82 (0.0952-16.3) | 0.81 | |||
| ATT cells (106 ml−1) | 1.06 (0.0394-149) | ||||
FL, free living; ATT, attached.
*, P< 0.01; all other P values were <0.001.
The paired features of low temperature and high salinity that characterize the inhabitable brine of wintertime sea ice also lead to increased viscosity. Free-living bacteria experience diffusive limitations on nutrient uptake, enzymatic reactions, and exchanges of metabolites in highly viscous fluids, limitations that surface-associated bacteria could overcome partially through direct access to adsorbed organic substances. Advection of DOC-rich brine across the surface would bring additional benefits relative to passive transport of free-living bacteria with the brine (29). However, the surface associations observed in this study also may reflect a secondary effect of a recently identified strategy for adaptation to cold briny habitats: cellular production of colloids or cryo- and/or osmoprotective compounds in the form of EPS (28). EPS would favor cell attachment (32) and a prevalence of attached active bacteria over free-living cells at the colder temperatures of sea ice, as observed in this study. The observed increase in the proportions of CFB (known for their abundant slime production) (27) with decreasing temperature and increasing salinity (Fig. 5A) further supports this hypothesis, as well as the suggestion (24, 45) that these organisms possess specific capabilities that enable them to thrive in the cold. An abundant CFB group utilizing high-molecular-weight organic compounds is consistent with work showing that this size class of organic matter is a large, biologically labile pool (1) not only in the ocean but also in sea ice.
The observation of active bacteria at −20°C suggests that wintertime sea ice is more than a refugium for temporarily preserved life and brings the discussion of limits of life on Earth to a different level, with respect to its implications both for microbial physiology and for low-temperature carbon diagenesis. Significant amounts of terrestrial and marine organic carbon are transported with sediments in the upper layers of Arctic sea ice (16); the work reported here suggests that these sediments may play an important role in sustaining bacterial activity even during the coldest parts of the year. In the context of microbial physiology, limits no longer can be addressed as single-variable phenomena; at the phase change of liquid water to solid ice, the simultaneous effects of and responses to multiple stresses (e.g., temperature, salinity, and viscosity) must be considered (13). Future work will need to establish to what extent microbial activity is constrained by a decrease in water activity and an increase in ionic strength in the host solution compared to low-temperature effects on membrane and transport processes (49).
While this study demonstrates the important role of surface attachment for bacteria coping with extreme conditions, the exact nature of the benefits of attachment remains to be examined. Unclear at this stage is whether the benefits occurring at low temperatures are the same as those invoked for attached bacteria in warmer locations. The underlying mechanisms need to be explored in conjunction with studies of the types of surfaces relevant in the context of subzero activity, with particular focus on the role of organic exopolymers. Among other implications, the active bacteria reported here and their association with surfaces direct the search for life on frozen moons and planets (6) to particle-rich ice formations, be they in the form of lithogenic particles (Mars) or salt precipitates (Europa).
Acknowledgments
This research was supported by an NSF-OPP-LExEn award to J. W. Deming and H. Eicken and by the University of Washington Astrobiology Program. We are grateful for additional NSF support through the Barrow Arctic Science Consortium.
We thank D. Ramey for kind assistance. We also thank S. Carpenter for technical support, A. Stierle and C. Krembs for help in the field and laboratory, and L. Wells for discussion.
REFERENCES
- 1.Amon, R. M. W., H.-P. Fitznar, and R. Benner. 2001. Linkages among the bioreactivity, chemical composition, and diagenetic state of marine dissolved organic matter. Limnol. Oceanogr. 46:287-297. [Google Scholar]
- 2.Brown, M. V., and J. P. Bowman. 2001. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO). FEMS Microbiol. Ecol. 35:267-275. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter, E. J., S. Lin, and D. G. Capone. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho, C., and F. Azam. 1988. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature 332:441-443. [Google Scholar]
- 5.Christner, B. C. 2002. Incorporation of DNA and protein precursors into macromolecules by bacteria at −15°C. Appl. Environ. Microbiol. 68:6435-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chyba, C. F., and C. B. Phillips. 2001. Possible ecosystems and the search for life on Europa. Proc. Natl. Acad. Sci. USA 98:801-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and J. T. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, G. F. N., and W. F. Weeks. 1983. Equations for determining the gas and brine volumes in sea-ice samples. J. Glaciol. 29:306-316. [Google Scholar]
- 10.Cox, G. F. N., and W. F. Weeks. 1988. Numerical simulations of the profile properties of undeformed first-year sea ice during the growth season. J. Geophys. Res. 93:12449-12460. [Google Scholar]
- 11.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic Archaea and Bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deming, J. W. 2002. Psychrophiles and polar regions. Curr. Opin. Microbiol. 3:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Deming, J. W., and J. A. Baross. 2000. Survival, dormancy, and nonculturable cells in extreme deep-sea environments, p. 147-197. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 15.Eicken, H. 2003. From the microscopic to the macroscopic to the regional scale: growth, microstructure and properties of sea ice, p. 22-81. In D. N. Thomas and G. S. Dieckmann (ed.), Sea ice—an introduction to its physics, biology, chemistry and geology. Blackwell Science, London, England.
- 16.Eicken, H. 2003. The role of Arctic sea ice in transporting and cycling terrigenous organic matter, p. 46-53. In R. Stein and R. W. Macdonald (ed.), The organic carbon cycle in the Arctic Ocean. Springer-Verlag KG, Berlin, Germany.
- 17.Eicken, H., C. Bock, R. Wittig, H. Miller, and H.-O. Poertner. 2000. Nuclear magnetic resonance imaging of sea ice pore fluids: methods and thermal evolution of pore microstructure. Cold Reg. Sci. Technol. 31:207-225. [Google Scholar]
- 18.Eicken, H., J. Weissenberger, I. Bussmann, J. Freitag, W. Schuster, F. Valero Delgado, K.-U. Evers, P. Jochmann, C. Krembs, R. Gradinger, F. Lindemann, F. Cottier, R. Hall, P. Wadham, M. Reisemann, H. Kousa, J. Ikavalko, G. H. Leonard, H. Shen, S. F. Ackley, and L. H. Smedsrud. 1998. Ice tank studies of physical and biological sea-ice processes, p. 363-370. In H. T. Shen (ed.), Ice in surface waters. Proceedings of the 14th International Symposium on Ice. A. A. Balkema, Rotterdam, The Netherlands.
- 19.Helmke, E., and H. Weyland. 1995. Bacteria in sea ice and underlying water of the eastern Weddell Sea in midwinter. Mar. Ecol. Prog. Ser. 117:269-287. [Google Scholar]
- 20.Huston, A. L., and J. W. Deming. 2002. Relationships between microbial extracellular enzymatic activity and suspended and sinking particulate organic matter: seasonal transformations in the North Water. Deep-Sea Res. Part II 49:5211-5225. [Google Scholar]
- 21.Junge, K., C. Krembs, J. W. Deming, A. Stierle, and H. Eicken. 2001. A microscopic approach to investigate bacteria under in-situ conditions in sea-ice samples. Ann. Glaciol. 33:304-310. [Google Scholar]
- 22.Junge, K., H. Eicken, and J. W. Deming. 2003. Motility of Colwellia psychrerythraea strain 34H at subzero temperatures. Appl. Environ. Microbiol. 69:4282-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junge, K., H. Eicken, and J. W. Deming. A microscopic approach to investigate bacteria under in-situ conditions in Arctic lake ice: initial comparisons to sea ice. In R. Norris and F. Stootman (ed.), Bioastronomy 2002: life amongst the stars. International Astronomical Union Symposium Series, in press. Astronomical Society of the Pacific, San Francisco, Calif.
- 24.Junge, K., J. F. Imhoff, J. T. Staley, and J. W. Deming. 2002. Phylogenetic diversity of numerically important bacteria in Arctic sea ice. Microb. Ecol. 43:315-328. [DOI] [PubMed] [Google Scholar]
- 25.Karl, D. M., D. F. Bird, K. Bjorkman, T. Houlihan, R. Shackelford, and L. Tupas. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286:2144-2147. [DOI] [PubMed] [Google Scholar]
- 26.Karner, M. B., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 28.Krembs, C., H. Eicken, K. Junge, and J. W. Deming. 2002. High concentrations of exopolymeric substances in wintertime sea ice: implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep-Sea Res. Part I 9:2163-2181. [Google Scholar]
- 29.Logan, B. E., and D. K. Kirchman. 1991. Uptake of dissolved organic matter by marine bacteria as a function of fluid motion. Mar. Biol. 111:175-181. [Google Scholar]
- 30.Maruyama, A., and M. Sunamura. 2000. Simultaneous direct counting of total and specific microbial cells in seawater, using a deep-sea microbe as target. Appl. Environ. Microbiol. 66:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maykut, G. A. 1986. The surface heat and mass balance. NATO ASI Ser. Ser. B 9-164:395-463. [Google Scholar]
- 32.Meiners, K, R. Gradinger, J. Fehling, G. Civitarese, and M. Spindler. 2003. Vertical distribution of exopolymer particles in sea ice of the Fram Strait (Arctic) during autumn. Mar. Ecol. Prog. Ser. 248:1-13. [Google Scholar]
- 33.Mock, T., and R. Gradinger. 1999. Determination of Arctic ice algal production with a new in situ technique. Mar. Ecol. Prog. Ser. 177:15-26. [Google Scholar]
- 34.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Y. Wu, and E. F. Delong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, A. E., K. Y. Wu, C. L. Moyer, D. M. Karl, and E. F. Delong. 1999. Evidence for circumpolar distribution of planktonic Archaea in the Southern Ocean. Aquat. Microb. Ecol. 18:263-273. [Google Scholar]
- 36.Nichols, D. S., J. Olley, H. Garda, R. R. Brenner, and T. McMeekin. 2000. Effect of temperature and salinity stress on growth and lipid composition of Shewanella gelidimarina. Appl. Environ. Microbiol. 66:2422-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, P. B. 2000. A habitat for psychrophiles in deep Antarctic ice. Proc. Natl. Acad. Sci. USA 97:1247-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priscu, J. C., E. E. Adams, W. B. Lyons, M. A. Voytek, D. W. Mogk, R. L. Brown, C. P. McKay, C. D. Takacs, K. A. Welch, C. F. Wolf, J. D. Kirshtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2144. [DOI] [PubMed] [Google Scholar]
- 39.Rasband, W. S., and D. S. Bright. 1995. NIH Image: a public domain image processing program for the Macintosh. J. Microbeam Anal. 1995:137-149. [Google Scholar]
- 40.Reimnitz, E., P. W. Barnes, and W. S. Weber. 1993. Particulate matter in pack ice of the Beaufort Gyre. J. Glaciol. 39:186-198. [Google Scholar]
- 41.Rivkina, E. M., E. I. Friedmann, C. P. McKay, and D. A. Gilichinsky. 2000. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 66:3230-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothchild, L. J., and R. L. Mancinelli. 2001. Life in extreme environments. Nature 409:1092-1101. [DOI] [PubMed] [Google Scholar]
- 43.Rublee, P., S. Merkel, and M. Faust. 1983. The transport of bacteria in the sediments of a temperate marsh. Estuar. Coast. Shelf Sci. 16:501-509. [Google Scholar]
- 44.Sherr, B. F., P. del Giorgio, and E. B. Sherr. 1999. Estimating abundance and single-cell characteristics of actively respiring bacteria via the redox-dye, CTC. Aquat. Microb. Ecol. 18:117-131. [Google Scholar]
- 45.Simon, M., F. O. Gloeckner, and R. Amann. 1999. Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat. Microb. Ecol. 18:275-284. [Google Scholar]
- 46.Skidmore, M. L., J. M. Fought, and M. J. Sharp. 2000. Microbial life beneath a high Arctic glacier. Appl. Environ. Microbiol. 66:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, D. C., M. Simon, A. L. Alldredge, and F. Azam. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139-142. [Google Scholar]
- 48.Stierle, A. P., and H. Eicken. 2002. Sedimentary inclusions in Alaskan coastal sea ice: small-scale distribution, interannual variability and entrainment requirements. Arct. Antarct. Alp. Res. 34:103-114. [Google Scholar]
- 49.Thomas, D. N., and G. S. Dieckmann. 2002. Antarctic sea ice—a habitat for extremophiles. Science 295:641-644. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, D. N., G. Kattner, R. Engbrodt, V. Giannelli, H. Kennedy, C. Haas, and G. S. Dieckmann. 2001. Dissolved organic matter in Antarctic sea ice. Ann. Glaciol. 33:297-303. [Google Scholar]
- 51.Wells, L. E., and J. W. Deming. 2003. Abundance of Bacteria, the Cytophaga-Flavobacterium cluster and Archaea in cold oligotrophic waters and nepheloid layers of the Northwest Passage, Canadian Archipelago. Aquat. Microb. Ecol. 31:19-31. [Google Scholar]
- 52.Wettlaufer, J. S. 1999. Crystal growth, surface phase transitions and thermomolecular pressure. NATO ASI Ser. Ser. I 56:39-67. [Google Scholar]