Abstract
Four cDNAs encoding phosphoribosyl diphosphate (PRPP) synthase were isolated from a spinach (Spinacia oleracea) cDNA library by complementation of an Escherichia coli Δprs mutation. The four gene products produced PRPP in vitro from ATP and ribose-5-phosphate. Two of the enzymes (isozymes 1 and 2) required inorganic phosphate for activity, whereas the others were phosphate independent. PRPP synthase isozymes 2 and 3 contained 76 and 87 amino acid extensions, respectively, at their N-terminal ends in comparison with other PRPP synthases. Isozyme 2 was synthesized in vitro and shown to be imported and processed by pea (Pisum sativum) chloroplasts. Amino acid sequence analysis indicated that isozyme 3 may be transported to mitochondria and that isozyme 4 may be located in the cytosol. The deduced amino acid sequences of isozymes 1 and 2 and isozymes 3 and 4 were 88% and 75% identical, respectively. In contrast, the amino acid identities of PRPP synthase isozyme 1 or 2 with 3 or 4 was modest (22%–25%), but the sequence motifs for binding of PRPP and divalent cation-nucleotide were identified in all four sequences. The results indicate that PRPP synthase isozymes 3 and 4 belong to a new class of PRPP synthases that may be specific to plants.
PRPP is an important compound of intermediary metabolism. It is required for the de novo biosynthesis of purine and pyrimidine nucleotides and the pyridine nucleotide coenzyme NAD, as well as for the salvage of preformed purine, pyrimidine, and pyridine bases. PRPP is also used in the synthesis of His and Trp by plants and microorganisms (Hove-Jensen, 1988). The synthesis of PRPP is catalyzed by PRPP synthase (EC 2.7.6.1), which catalyzes the transfer of the β,γ-diphosphoryl moiety of ATP to the C-1 hydroxyl of Rib-5-P (Khorana et al., 1958): Rib-5-P + ATP → PRPP + AMP. The gene encoding PRPP synthase has been designated PRS. PRPP synthase is regarded as essential for all organisms, except for certain specialized mutants of Escherichia coli (Δprs) that can grow in the absence of PRPP synthase activity when supplied with compounds of the PRPP-requiring pathways (i.e. purine and pyrimidine nucleosides, NAD, His, and Trp; Hove-Jensen, 1988, 1989). Thus, all organisms contain at least one gene encoding PRPP synthase. Eukaryotes often have more than one PRS gene. Mammals such as human and rat have three PRS genes (Taira et al., 1987, 1989; Roessler et al., 1990; Taira et al., 1990), and the yeast Saccharomyces cerevisiae contains five genes, the products of which show a high degree of homology with PRPP synthases of mammals and bacteria (Carter et al., 1994, 1997). PRPP synthase-encoding genes have been cloned and characterized from a variety of organisms across a wide phylogenetic spectrum, including the gram-negative bacteria E. coli (Hove-Jensen, 1985; Hove-Jensen et al., 1986) and Salmonella typhimurium (Bower et al., 1988) and the gram-positive bacteria Bacillus subtilis (Nilsson and Hove-Jensen, 1987; Nilsson et al., 1989) and Bacillus caldolyticus (Krath and Hove-Jensen, 1996).
PRPP synthases from several organisms have been characterized in detail, including those of E. coli and S. typhimurium (Switzer, 1971; Hove-Jensen et al., 1986), B. subtilis (Arnvig et al., 1990), and mammals (Nosal et al., 1993; Tatibana et al., 1995). In general, these enzymes use ATP only as a diphosphoryl donor; the actual substrate is MgATP. The E. coli, S. typhimurium, and mammalian enzymes also require free Mg2+. All of the enzymes require Pi for activity, and the bacterial enzymes require it for stability (Switzer, 1969; Hove-Jensen et al., 1986). The PRPP synthases from bacteria and mammals are subject to inhibition by purine nucleotides, with ADP being the most potent inhibitor (Switzer and Sogin, 1973; Hove-Jensen et al., 1986). The enzymes from B. subtilis and mammals are inhibited by ADP as well as GDP (Arnvig et al., 1990; Ishijima et al., 1991; Nosal et al., 1993). ADP inhibits the enzyme competitively with ATP by binding to the active site. In addition, ADP is an allosteric inhibitor of bacterial and mammalian PRPP synthases. This effect has been studied primarily with the S. typhimurium enzyme by both kinetic analysis and equilibrium-binding studies (Switzer and Sogin, 1973; Gibson et al., 1982).
Structure-function of amino acid residues of PRPP synthase has been studied in some detail by chemical modification (Harlow and Switzer, 1990; Hilden et al., 1995), by the analysis of variant forms of the enzyme from bacteria (Bower et al., 1989) or humans (Becker et al., 1995), or by comparison of amino acid sequences from evolutionarily distant species (Hove-Jensen et al., 1986). Several amino acid residues have been implicated as important for structure or catalysis. Specifically, amino acid residues important in Rib-5-P binding (the PRPP-binding motif) have been identified (Willemoës et al., 1996), as well as a sequence important in the binding of divalent cation-nucleotide (Bower et al., 1989; Harlow and Switzer, 1990). The crystallization of the enzyme from B. subtilis is expected to greatly expand our knowledge in this field in the future (Bentsen et al., 1996).
Few reports have dealt with PRPP synthase from plants. They include analysis of homogenous or partially purified enzyme preparations of rubber tree latex (Gallois et al., 1997) or spinach (Spinacia oleracea) leaves (Ashihara, 1977a), as well as analysis of the regulation of PRPP synthesis (Ashihara and Komamine, 1974; Ashihara, 1977b). These reports indicate that PRPP synthase of plants may have properties that are different from the “classical” PRPP synthases of bacteria and mammals and that there is more than one type of enzyme. In this study we have analyzed a cDNA library of spinach for the presence of DNA fragments that could complement a bacterial prs–deletion. Analysis of nucleotide sequences and synthesized enzymes indicate that there are two classes of PRPP synthases in spinach.
MATERIALS AND METHODS
Microbiological Procedures
Escherichia coli strain MC1061 (Casadaban et al., 1983) was used as a source of plasmid DNA and HO773 (Δprs-4) served as the host for complementation (Post et al., 1996). Strains harboring the Δprs-4 allele lack PRPP synthase activity, which results in a requirement for purine and pyrimidine nucleosides, His, Trp, and NAD. All of these compounds, except NAD, are present in rich medium. Consequently, NAD was supplied to rich medium for growth of strain HO773. E. coli was grown in NZY medium containing NZ-amin and yeast extract (Hove-Jensen and Maigaard, 1993) with the addition, when necessary, of NAD (40 mg L−1) or ampicillin (50 or 100 mg L−1). Cell cultures were incubated at 37°C in an Aqua Shaker (A. Kühner, Inc., Birsfelden, Switzerland). Cell growth was monitored in an Eppendorf PCP6121 photometer at A436. An A436 of 1 (1-cm path length) corresponds to approximately 3 × 1011 cells L−1. To prepare an extract of E. coli cells, 100 mL of NZY medium was inoculated with 5 mL of an overnight culture and incubated for 18 h with shaking. Cells were harvested by centrifugation in a rotor (model SS34, Sorvall) at 5,000 rpm for 8 min at 4°C, washed twice in 0.9% NaCl, resuspended in 50 mm potassium phosphate buffer, 50 mm Tris-HCl (pH 7.6) or 50 mm Tris-HCl (pH 7.6), and disrupted by sonication in an ultrasonic disintegrator (model 150, Soniprep Measuring and Scientific Equipment, Ltd., London) for 60 s at 0°C. Debris were removed by centrifugation at 10,000 rpm for 15 min at 4°C.
DNA Methodology
The plasmids used were ppsaD, which encodes the D-subunit of barley PSI (Kjarulff and Okkels, 1993), and pSOD3, which encodes the mitochondrial superoxide dismutase of maize (White and Scandalios, 1989). Plasmid isolation and transformation in E. coli were performed as previously described (Mandel and Higa, 1970; Birnboim and Doly, 1979). Conditions for the use of restriction endonucleases (Amersham, Promega, and New England Biolabs) and DNA ligase (Promega) were as described by the vendors. Nucleotide sequences were determined by the chain-termination method (Sanger et al., 1977). Sequencing was performed with Sequenase DNA polymerase (version 2.0), the 7-deaza-dGTP reagent kit (U.S. Biochemical), and [α-33P]dATP (DuPont-New England Nuclear). Sequence ladders were established by gel electrophoresis in buffer gradient gels containing 8 m urea and 6% polyacrylamide (Sambrook et al., 1989). Alternatively, sequencing was performed at the Botanical Institute of the University of Copenhagen (Denmark) in a sequencer (model 377, Applied Biosystems) using the cycle-sequencing method with dye terminators, as recommended by the supplier (PRISM BigDye terminator cycle sequencing ready reaction kit, Applied Biosystems). Oligodeoxyribonucleotides used as primers were provided by Hobolth DNA Syntese (Hillerød, Denmark). Computer analysis of nucleotide and amino acid sequences were carried out with the DNA Strider program (Marck, 1988). Amino acid sequences were compared by using programs based on the BLAST algorithm (Altschul et al., 1990) at the National Center for Biotechnology Information Services. Amino acid sequences were aligned using the ClustalW program at the National Center for Biotechnology Information Services or by using the Multalign program (Barton, 1990). Phylogenetic analysis was performed by using the Analysis of Multiply Aligned Sequences program (Livingstone and Barton, 1993).
Cloning of Spinach PRS cDNA
A spinach (Spinacia oleracea) cDNA library, prepared from mRNA of young leaves of actively growing plants and contained in the excision proficient vector λZAPII, was used to generate approximately 107 ampicillin-resistant colonies containing excised plasmids, according to the procedure described by the supplier (Stratagene). Complementation of Δprs-4 was achieved by transformation of E. coli strain HO773 and plating on NZY medium containing ampicillin but lacking NAD. HO773 is a PRPP-less mutant strain due to deletion of the prs gene, which encodes PRPP synthase. Consequently, strain HO773 requires guanosine, uridine, His, Trp, and NAD. All of these compounds, except NAD, are present in a rich medium like NZY, and strain HO773 grows in rich medium supplemented with NAD. On the other hand, acquisition in HO773 of a PRS gene specifying active PRPP synthase makes the strain NAD prototrophic. The advantage of using strain HO773 in rich medium is that only minimal PRPP synthesis is required to make the strain NAD prototrophic.
Assay of PRPP Synthase Activity
PRPP synthase activity was assayed at 30°C by a modification of a procedure described previously (Arnvig et al., 1990). Bacterial cell extract (10 μL) was mixed with 40 μL of a reaction mixture (both prewarmed at 30°C) to yield the following final concentrations: 5 mm Rib-5-P, 3 mm [γ-32P]ATP (10 GBq/mol) prepared as described by Jensen et al. (1979), 5 mm MgCl2, 20 mm NaF, and either 50 mm Tris-HCl (pH 7.6) or 50 mm potassium phosphate buffer, and 50 mm Tris-HCl (pH 7.6). Samples (10 μL) were removed at intervals and mixed with 5 μL of 0.33 m HCOOH. This was applied to a polyethyleneimine-cellulose TLC sheet (Baker-flex, J.T. Baker). After drying, the chromatogram was developed in 0.85 m KH2PO4, which had been previously adjusted to pH 3.4 with 0.85 m H3PO4. PRPP synthase activity of chloroplasts and mitochondria was assayed by the same procedure, except that PEP (12.5 mm) and pyruvate kinase (2.5 mg L−1 of reaction mixture; Boehringer Mannheim) were included in the assay. Radioactivity was quantitated in an Instant Imager (model 2024, Packard, Meriden, CT). Protein concentration was determined by the bicinchoninic acid procedure (Smith et al., 1985) with chemicals provided by Pierce and with BSA as the standard.
Assay of Import of Polypeptides to Chloroplasts
DNA of the plasmids pBK842, pBK843, ppsaD, and pSOD3 were used as templates for in vitro mRNA transcription by T3 or T7 RNA polymerase. In vitro translation was carried out in the presence of [35S]Met (0.4 mm, 3.7 GBq mol−1, DuPont-New England Nuclear) in a rabbit reticulocyte lysate, as described by the supplier (Promega). In vitro translation products were used immediately or stored at −80°C. Pea (Pisum sativum) seedlings were grown in vermiculite at 21°C with 14-h/10-h light/dark cycles at the Department of Plant Physiology (Institute of Molecular Biology, University of Copenhagen, Denmark). Intact chloroplasts were isolated from 2-week-old pea shoots by homogenization and Percoll (Pharmacia) gradient centrifugation, as described by Cline et al. (1985). Isolated chloroplasts were resuspended in 50 mm Hepes-KOH (pH 8.0) and 0.33 m sorbitol at a density corresponding to a chlorophyll concentration of 1 g L−1.
An assay of import of polypeptides to chloroplasts was performed essentially as described by Cline et al. (1985). Import reactions were initiated by mixing 25 μL of labeled translation product with 0.275 mL of 50 mm Hepes-KOH (pH 8.0), 0.33 m sorbitol, 5 mm Met, 8 mm MgCl2, 8 mm ATP, and chloroplasts equivalent to 50 μg of chlorophyll. Reactions were incubated at 25°C for 30 min under fluorescent light. Chloroplasts were reisolated by centrifugation, resuspended in 120 μL of 50 mm Hepes-KOH (pH 8.0) and 0.33 m sorbitol, and divided into two parts. One part was incubated with protease thermolysin (Sigma) at a final concentration of 30 mg L−1. Following 40 min of incubation on ice, the protease activity was terminated by the addition of EDTA to 5 mm. The protease-treated and untreated samples were concentrated by centrifugation and resuspended in loading buffer, and polypeptides were separated by electrophoresis in a 14% SDS-polyacrylamide gel (Laemmli, 1970). As a molecular mass standard, the Rainbow Mr markers (Amersham) were used. Labeled bands were visualized by exposure of the gel to x-ray film (Hyperpaper, Amersham).
RESULTS
Isolation and Characterization of Spinach cDNA Specifying PRPP Synthase Activity
Complementation of the E. coli Δprs-4 allele by plasmids of the spinach cDNA library was performed as described in Methods. NAD-independent transformants appeared at a frequency of approximately 10−5 within the population of ampicillin-resistant transformants. The selection resulted in the isolation of 83 bacterial clones that grew in the absence of NAD. Plasmid DNA isolated from these clones was analyzed by restriction endonuclease digestion and by nucleotide sequencing, which revealed four different types of cDNA clones. They were designated PRS1, PRS2, PRS3, and PRS4, and their gene products were designated PRPP synthase isozymes 1, 2, 3, and 4, respectively. Each type of cDNA was represented in varying numbers of clones: 3 clones of PRS1, 12 clones of PRS2, 29 clones of PRS3, and 32 clones of PRS4. Seven other clones contained either PRS3 or PRS4 based on restriction endonuclease analysis. A representative of each class, pHO313 (PRS1), pBK842 (PRS2), pBK843 (PRS3), and pHO304 (PRS4), was chosen for further analysis. The insert of each of these four plasmids was sequenced on both strands with complete overlaps. Others were sequenced partially to establish which isozyme they encoded.
PRPP Synthase Activity Specified by PRS cDNA
Cell extracts of E. coli strain HO773, which had been transformed with the plasmids harboring each of the four PRS cDNAs, were analyzed for PRPP synthase activity (Table I). All of the extracts were able to catalyze PRPP synthesis at varying specific activities. PRPP synthesis occurred in a Rib-5-P-dependent manner (data not shown). Isozymes 1 and 2 were stimulated to a high degree by Pi, whereas isozymes 3 and 4 were independent of or slightly inhibited by Pi. Experiments performed under similar conditions with PRPP synthase of E. coli (strain HO773 harboring pHO11; Hove-Jensen, 1985) revealed an activity of 160 nmol min−1 mg−1 protein and 0.6 nmol min−1 mg −1 protein in the presence and absence of Pi, respectively. PRPP synthase activity of strain HO773 (Δprs) was undetectable (data not shown). We also analyzed the effect of ADP on PRPP synthase activity (Table I). Isozymes 1 and 2 appeared to be inhibited efficiently by ADP under these assay conditions, whereas isozymes 3 and 4 were essentially unaffected by ADP inhibition.
Table I.
PRPP synthase activity specified by PRS1, PRS2, PRS3, and PRS4 cDNAs
| Isozyme | Pia | Specific
Activityb
|
|
|---|---|---|---|
| −ADP | +ADP | ||
| nmol min−1 mg−1 protein | |||
| 1 | + | 191 | 74 |
| − | 14 | NDc | |
| 2 | + | 140 | 18 |
| − | 25 | ND | |
| 3 | + | 1060 | 880 |
| − | 1260 | 884 | |
| 4 | + | 80 | 72 |
| − | 89 | 99 | |
PRPP synthase activity was determined as described in Methods. Values are the averages of five determinations.
+, Activity assayed in the presence of 50 mm Pi; −, no Pi present.
−ADP, Activity determined by the standard assay in the presence of 1.0 mm ATP; +ADP, activity assayed in the presence of 1.0 mm ADP and 1.0 mm ATP.
ND, Not determined.
Nucleotide Sequence Analysis
The inserts harboring the four PRS cDNAs varied in length from 1285 to 1650 bp. Each sequence contained one open reading frame sufficient in length to code for a PRPP synthase polypeptide. A summary of the four mRNA sequences and some of their features are shown in Table II. The presumed translation initiation codons were preceded by 103 (PRS1), 154 (PRS2), 111 (PRS3), or 477 (PRS4) nucleotides. The PRS2 transcript contained a translation stop codon within the 154 nucleotides in-frame with the AUG codon, whereas PRS4 contained two translation stop codons in-frame with the AUG codon. In contrast, neither PRS1 nor PRS3 contained in-frame translation stop codons upstream of the presumed AUG initiation codon. This leaves the possibility open that the open reading frames of PRS1 and PRS3 are not complete. The sequences around the initiation AUG codon of PRS2, PRS3, and PRS4 resemble a consensus initiation sequence (AACAAUGGC), as suggested by Lütke et al. (1987). None of the mRNA sequences contained a poly (A+) sequence at the 3′ end. However, polyadenylation signals (Hunt et al., 1987) were found less than 30 nucleotides from the 3′ end of the PRS2 and PRS3 mRNA. Possible polyadenylation signals were found further upstream in PRS1 and PRS4 mRNA (data not shown).
Table II.
Summary of features of mRNAs specified by PRS1, PRS2, PRS3, and PRS4 cDNAs
| PRS cDNA | Nucleotide Sequencea | No. of Codons |
|---|---|---|
| 1 | 1-C … … 98 nt … … UGGAAUGGA . . 1003 nt UAA .170 nt U-1285 | 336 |
| 2 | 1-C 105 nt UAA 41 nt … . AUCAAUGGC . . 1180 nt UGA. 216 nt AAAUAA..14 nt.. C-1579 | 395 |
| 3 | 1-A … … . 106 nt … … … . .CCAAAUGGC . . .1213 ntUAA. 196 nt AAAUAU..25 nt.. C-1560 | 406 |
| 4 | 1-C. .365 nt. .UAG..69 nt. .UAA. .32 nt. .ACAAAUGGA … 949 nt UGA 215 nt G-1650 | 318 |
Translation initiation and termination codons are shown in bold and italics, respectively. Possible polyadenylation signal motifs are underlined. The nucleotide sequences of the four cDNAs have been deposited in the EMBL nucleotide sequence data bank under the accession nos. AJ006940 (PRS1), AJ006941 (PRS2), AJ006942 (PRS3), and AJ006943 (PRS4).
nt, Nucleotides.
Amino Acid Sequence Analysis
The calculated molecular masses of the polypeptides specified by each of the four cDNAs are: 36,582 D (PRPP synthase isozyme 1), 42,688 D (isozyme 2), 45,408 D (isozyme 3), and 35,362 D (isozyme 4). A comparison of the deduced amino acid sequences of the PRS-coding regions, together with the amino acid sequences of PRPP synthases from B. subtilis, E. coli, and human, are shown in Figure 1. Amino acid residues appear to be conserved along the entire length of each polypeptide, except for some additional amino acids at the N-terminal end of spinach isozymes 2 and 3. Specifically, the divalent cation-nucleotide-binding site (Fig. 1, region 1) and the PRPP-binding motif (Fig. 1, region 2) could be identified (Hove-Jensen et al., 1986; Bower et al., 1989). Comparison of the deduced amino acid sequences without the N-terminal extensions described above revealed 88% identity of isozyme 1 with isozyme 2. They differed at only 28 amino acids, and most of these differences were conservative. The identity of isozyme 3 with isozyme 4 was 75%, but there was only 22% to 25% identity of isozyme 1 or 2 with isozyme 3 or 4. A survey of the nucleotide databases revealed only two additional full-length PRPP synthase amino acid sequences of plant origin, PRPP synthase 1 and 2 of Arabidopsis (accession nos. X83764 and X92974). The identity of spinach isozyme 1 with Arabidopsis PRPP synthase 1 was 86%, whereas the identity of spinach isozyme 2 with Arabidopsis PRPP synthase 2 was 90%. In addition, the identity of spinach isozymes 1 and 2 with E. coli and human PRPP synthases 1, 2, and 3 was 44% to 48%, and with the B. subtilis enzyme the identity was 53%. In contrast, the identity of spinach isozymes 3 and 4 with E. coli, B. subtilis, and the human PRPP synthases was 20% to 24%.
Figure 1.
Multiple alignment of the deduced amino acid sequences of PRPP synthase from spinach, B. subtilis, E. coli, and human. The sequences are numbered from the presumed translation initiation Met. Sequences are aligned to give maximal identity, with hyphens indicating gaps introduced to improve the alignment. Spinach cDNAs were translated from the first Met codon. The sequence of isozyme 1 was extended upstream of this Met codon and this sequence is shown in lowercase. Relevant sequence motifs are indicated: Region 1, The divalent cation-nucleotide-binding site (Bower et al., 1989); Region 2, the PRPP/Rib-5-P-binding motif (Hove-Jensen et al., 1986; Willemoës et al., 1996). Possible consensus sequences for polypeptide maturation (So2 and -3 sequences) are underlined. Filled circles, Position with identical amino acid among all the sequences; open circles, position with conserved amino acid residues; So1, spinach isozyme 1; So2, spinach isozyme 2; So3, spinach isozyme 3; So4, spinach isozyme 4; Bs, B. subtilis (Nilsson et al., 1989); Ec, E. coli (Hove-Jensen et al., 1986); and Human1, human PRPP synthase 1 (Roessler et al., 1990).
The alignment (Fig. 1) revealed that, compared with isozyme 4, the deduced amino acid sequences of isozymes 2 and 3 contained N-terminal extensions of 76 and 87 amino acids, respectively. These N-terminal extensions contained several characteristics of a transit peptide required for transport of a polypeptide encoded by a nuclear gene into chloroplasts or mitochondria (Gavel and von Heijne, 1990; von Heijne and Nishikawa, 1991): The overall amino acid composition of this region is basic, the content of hydroxylated amino acids is high, and the sequence is initiated by MetAla. A cleavage-site consensus sequence for chloroplast transit peptides has been suggested as (Val/Ile)Xaa(Ala/Cys) ↓ Ala, where Xaa indicates any amino acid and the vertical arrow indicates the cleavage point. Often an Arg residue is found at a position between −10 and −6 (Gavel and von Heijne, 1990). Careful examination of the amino acid sequence of PRPP synthase isozyme 2 revealed a possible cleavage site at amino acid residues 40 to 43 (i.e. 40-ValLysCys ↓ Asn-43). Assuming cleavage at this position, we found Arg at position −10. In addition, the 42-amino acid N-terminal polypeptide generated by cleavage at this site contained three basic amino acid residues, no acidic residues, and 18 hydroxylated residues. Finally, the N-terminal amino acid residues were MetAla (Fig. 1). Altogether, these data indicated a location of PRPP synthase isozyme 2 in the chloroplast.
In contrast, PRPP synthase isozyme 3 contained a different motif for polypeptide maturation, which has been identified at the C-terminal end of some transit peptides of polypeptides transported to mitochondria: Arg at position −10, a hydrophobic residue at position −8, and Ser at position −5 relative to the C-terminal end of the cleaved transit peptide (Hendrick et al., 1989; von Heijne et al., 1989). This three-amino acid motif was found at the C terminus of the predicted transit peptide of isozyme 3 as 77-SerArgArgPheGlnMetSerSerAsnGlnGlu ↓ Asn-88, with relevant residues underlined and Glu as the C-terminal residue of the cleaved transit peptide. In addition, mitochondrial transit peptides are enriched in Arg and have a lower content of Ser and Thr than chloroplast transit peptides. Among the 87 amino acids removed by cleavage at the point suggested above, 17 were basic (9 of these were Arg), 4 were acidic, and 16 were hydroxylated. The N-terminal amino acids were MetAla. It is therefore possible that isozyme 3 is located within the mitochondrion.
The amino acid sequence of PRPP synthase isozyme 4 appeared to be complete (Fig. 1). Thus, isozyme 4 had a size comparable to that of “classical” PRPP synthases such as those of E. coli and human (i.e. without a transit peptide). This indicated that PRPP synthase isozyme 4 most likely is located in the cytosol.
It is possible that the coding sequence of PRS1 shown in Figure 1 is incomplete at the N-terminal encoding end. The amino acid sequence deduced from the region upstream of the suggested initiation AUG codon is included in Figure 1. This sequence appeared to be similar to a transit peptide because of a relatively high number of basic amino acids (6 of 34 residues) and Ser residues (7 of 34 residues), albeit fewer than those present in isozymes 2 and 3. The Arabidopsis PRS1 and PRS2 sequences do not reveal the presence of transit peptides at their N-terminal encoding ends.
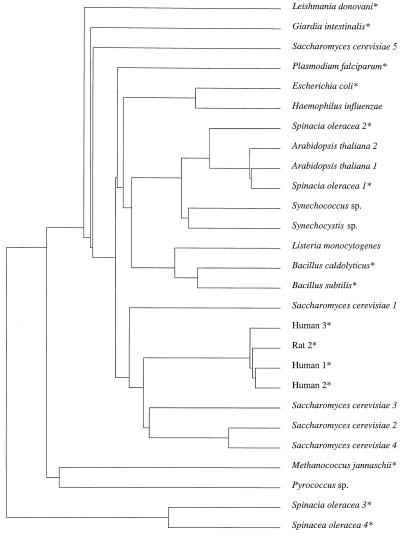
The phylogenetic relationship of the four spinach PRPP synthases with 23 additional PRPP synthase sequences from 16 other organisms was analyzed (Fig. 2). The phosphate-dependent spinach isozymes 1 and 2 are clustered as a subfamily with PRPP synthases 1 and 2 from Arabidopsis and the enzymes from the cyanobacteria Synechococcus sp. and Synechocystis sp., all of which are photosynthetic organisms. This subfamily of plant and cyanobacterial PRPP synthases are branched, with a second subfamily containing gram-positive bacterial PRPP synthases, indicating a close phylogenetic relationship. Together, these two subfamilies are related to the PRPP synthase subfamily of gram-negative bacteria and the parasite Plasmodium falciparum. A fourth subfamily contains the enzymes from mammals and four of the isoforms of the yeast Saccharomyces cerevisiae. Unrelated to these subfamilies are a number of enzymes from Archaea (Methanococcus jannaschii and Pyrococcus sp.), parasites (Leishmania donovani and Giardia intestinalis), and the fifth isoform of S. cerevisiae. In addition, the spinach phosphate-independent isozymes 3 and 4 appear to be relatively unrelated to the subfamilies of the other PRPP synthases described above. Therefore, it is possible that the spinach isozymes 3 and 4 and the enzymes from Archaea, L. donovani, and G. intestinalis may have diverged early during evolution.
Figure 2.
Phylogenetic relationship of 27 PRPP synthase polypeptides from 16 different organisms. The unrooted tree was constructed as described in Methods. Numbers indicate isoforms of PRPP synthase. Asterisks indicate enzymes that have been shown experimentally to catalyze PRPP formation in vitro. A. thaliana 1 (accession no. X83764), A. thaliana 2 (accession no. X92974), B. caldolyticus (Krath and Hove-Jensen, 1996; accession no. X83708), B. subtilis (Nilsson et al., 1989; accession no. X16518), E. coli (Hove-Jensen et al., 1986; accession no. M13174), G. intestinalis (Kyradji and Bagnara, 1998; accession no. AF042173), H. influenzae (Fleischman et al., 1995; accession no. U32834), Human 1 (Roessler et al., 1990; accession no. D00860), Human 2 (Iizasa et al., 1989; accession no. Y00971), Human 3 (Taira et al., 1990; accession no. M57423), L. donovani (Hendrickson et al., 1993; accession no. M76553), Listeria monocytogenes (Gouin et al., 1994; accession no. M92842), M. jannaschii (Bult et al., 1996; J.N. McGuire, personal communication; accession no. U67576), P. falciparum (accession no. U54642), Pyrococcus sp. (Naeem et al., 1997; accession no. D79364), Rat 2 (Taira et al., 1987; accession no. M17259), S. cerevisiae 1 (Carter et al., 1994; accession no. X70069), S. cerevisiae 2 (Carter et al., 1994; accession no. X74414), S. cerevisiae 3 (Carter et al., 1994; accession no. X74415), S. cerevisiae 4 (Carter et al., 1994; accession no. Z35829), S. cerevisiae 5 (Hernando et al., 1998; accession no. X91067), Synechococcus sp. (Nagaya et al., 1993; accession no. D14994), and Synechocystis sp. (accession no. D64004).
Import of PRPP Synthase Isozyme 2 into Chloroplasts
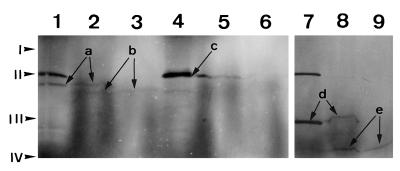
Polypeptides of PRPP synthase isozymes 2 and 3 produced in vitro were analyzed for import into pea chloroplasts by incubating the labeled translation products with purified chloroplasts. Protease thermolysin treatment of the import reactions was used to demonstrate the presence of polypeptides inside the chloroplasts. This protease degrades external proteins without affecting internal proteins (Cline et al., 1984). The D-polypeptide of barley PSI (Kjarulff and Okkels, 1993), which is encoded by a nuclear gene but located within chloroplasts, was included in the experiment. The results of the import analysis are shown in Figure 3. The molecular mass of the in vitro synthesized PRPP synthase isozyme 2 polypeptide was estimated as 43 kD (Fig. 3, lane 1), which is close to the molecular mass of 42.7 kD calculated from the deduced amino acid sequence. After incubation with chloroplasts under conditions that allow the import of precursor polypeptides, this 43-kD polypeptide was processed to one with a molecular mass of 39 kD (Fig. 3, lane 2). This lower-molecular-mass polypeptide was resistant to protease thermolysin digestion, indicating that it was inside of the organelle (Fig. 3, lane 3). Assuming that the cleaved N-terminal transit peptide had a molecular mass of approximately 4,000 D, the precursor polypeptide of isozyme 2 might be processed around amino acid 40. This is consistent with cleavage at the site suggested above (40-ValLysCys ↓ Asn-43). In contrast to isozyme 2, isozyme 3 was left unprocessed by chloroplasts and, consequently, all of the polypeptide was degraded by thermolysin (Fig. 3, lanes 4–6). As expected, the D-polypeptide of barley PSI was efficiently processed and imported by chloroplasts (Fig. 3, lanes 7–9).
Figure 3.
Import of polypeptides into chloroplasts. Incubation of radiolabeled, in vitro-synthesized polypeptides with isolated chloroplasts and SDS-PAGE were performed as described in Methods. An autoradiogram of the gel is shown. Lanes 1 to 3, Analysis of import of PRPP synthase isozyme 2; lanes 4 to 6, analysis of import of PRPP synthase isozyme 3; and lanes 7 to 9, analysis of import of the D-polypeptide of barley PSI. Lanes 1, 4, and 7, In vitro translation products; lanes 2, 5, and 8, reaction products of incubation of in vitro translation products with chloroplasts; lanes 3, 6, and 9, reaction products as in lanes 2, 5, and 8, respectively, but treated with the protease thermolysin. Arrows labeled “a” point to the bands representing the in vitro-synthesized 43-kD PRPP synthase isozyme 2 polypeptide. Arrows labeled “b” point to the bands representing the 39-kD processed PRPP synthase isozyme 2 polypeptide. The arrow labeled “c” points to the band representing the in vitro-synthesized 47-kD PRPP synthase isozyme 3 polypeptide. This band contained two polypeptides, with the lower one representing the PRPP synthase isozyme 3 polypeptide. Arrows labeled “d” point to the bands representing the in vitro-synthesized D-polypeptide of PSI (28 kD). Arrows labeled “e” point to the bands representing the processed D-polypeptide of PSI (21.5 kD). The band at 48.5 kD present in lanes 1, 4, and 7 is an artifact that originates from the presence of an endogenous template in the transcription-translation mixture, because the band was also present in transcription-translation reactions to which no template was added (data not shown). Positions of the molecular-mass markers are indicated at the left: I, BSA (66 kD); II, ovalbumin (46 kD); III, carbonic anhydrase (30 kD); and IV, trypsin inhibitor (21.5 kD).
The in vitro-labeled polypeptides of PRPP synthase isozymes 2 and 3 were also incubated with pea mitochondria isolated from 2-week-old pea shoots, as described by White and Scandalios (1987). An assay of import of polypeptides to mitochondria was performed as described by Hoff et al. (1995). Neither PRPP synthase isozyme 2 nor isozyme 3 polypeptides were imported and processed by the mitochondria by this procedure. The mitochondrial superoxide dismutase from maize was included in the experiment and was efficiently imported as expected (data not shown).
PRPP Synthase Activity in Pea Chloroplasts and Mitochondria
The chloroplast and mitochondria preparations isolated for transit peptide analysis were broken by sonic oscillation and assayed for PRPP synthase activity. Both organelles were able to catalyze the synthesis of PRPP in a Rib-5-P-dependent manner at low activities. The specific PRPP synthase activity in the chloroplast preparation was 3.6 nmol min−1 mg−1 protein, whereas the specific PRPP synthase activity of the mitochondria was 1.1 nmol min−1 mg−1 protein. It is possible that the activity in the mitochondrial preparation is underestimated because of the presence of a highly active ATPase, which may interfere with a quantitative determination of PRPP synthase activity.
DISCUSSION
Spinach contains at least four genes that specify PRPP synthase activity, as evaluated by screening a cDNA library for complementation of an E. coli Δprs allele. One enzyme (isozyme 3) was predicted to be located within the mitochondrion, and one enzyme (isozyme 4) was predicted to be located in the cytosol. The localization of isozyme 1 was uncertain, but the possibility exists that it is organelle localized. PRPP synthase isozyme 2 was localized within the chloroplast. Chloroplasts of Arabidopsis may contain several enzymes of the purine biosynthetic de novo pathway, including phosphoribosyl diphosphate amidotransferase, phosphoribosyl glycineamide synthase, phosphoribosyl glycineamide transformylase, and phosphoribosyl aminoimidazole synthase (Senecoff and Meager, 1993; Ito et al., 1994; Schnorr et al., 1994). Also, the PRPP consuming the purine salvage enzyme adenine phosphoribosyltransferase has been found in chloroplasts (Ashihara and Ukaji, 1985). In addition, the capacity of plastids of cowpea nodules to convert Rib-5-P to IMP has indicated the presence of PRPP synthase in this organelle (Atkins et al., 1997). Our finding of PRPP synthase activity in isolated pea chloroplasts confirmed the presence of this enzyme in chloroplasts. Chloroplasts of spinach leaves are able to synthesize Trp, which emphasizes the need for PRPP synthesis in this organelle (Zhao and Last, 1995).
We have confirmed the presence of PRPP synthase activity in pea mitochondria. Other reports have also provided evidence for the presence of PRPP synthase in mitochondria. Mitochondrial fractions of periwinkle and Jerusalem artichoke cell extracts are able to synthesize PRPP in a reaction that depends on ATP and Rib-5-P (Kanamori et al., 1980; Le Floc'h and Lafleuriel, 1983). It has been shown that mitochondria contain orotate phosphoribosyltransferase activity, which underlines the necessity of PRPP synthesis within this organelle (Kanamori et al., 1980). We were unable to demonstrate transport of PRPP synthase isozyme 3 into mitochondria, but the deduced amino acid sequence of this enzyme contains a potential transit peptide, indicating a location within mitochondria. The reason for lack of transport to isolated mitochondria remains unknown. Perhaps the PRS3 cDNA insert does not encode a full-length transit peptide, or maybe transport needs some additional factors that were not present in the assay.
Results of characterization of PRPP synthase purified from spinach leaves and from rubber tree latex have indicated that these enzymes are located in the cytoplasm (Ashihara, 1977a, 1977b; Gallois et al., 1997). They may be homologous to isozyme 4 described in the present work; however, the rubber tree latex enzyme is considerably larger in molecular mass (57 versus 35.4 kD calculated for isozyme 4). In addition, the activity of both of these enzymes, like that of isozyme 4, is independent of Pi. Le Floc'h and Lafleuriel (1983) also observed PRPP synthase activity in Jerusalem artichoke extracts, but this enzyme was dependent on the presence of Pi.
Mammals such as humans and rats contain PRPP synthase-associated proteins (Kita et al., 1994; Ishizuka et al., 1996). These proteins, which form oligomers with the catalytic PRPP synthase subunits, show a high degree of amino acid sequence similarity with human and rat PRPP synthases but apparently are without catalytic activity. It is possible that S. cerevisiae also contains PRPP synthase-associated proteins, because none of the five PRS genes of this organism complements the E. coli Δprs-4 allele (B. Hove-Jensen, unpublished results). The function of these proteins remains unknown. We have avoided this potential problem by selecting bacterial clones harboring cDNAs that complemented the Δprs-4 allele, and thus specified active PRPP synthase polypeptides.
Although all of our enzymatic analysis of spinach PRPP synthase was performed with crude preparations of enzymes that had been synthesized in E. coli, the data indicate that there are two different enzymatic forms of PRPP synthase. One form, comprising isozymes 1 and 2, resembles the “classical” PRPP synthases from E. coli and mammals. The maximal activities of these enzymes are dependent on the presence of Pi, and their activities are inhibited by ADP. The other form, comprising isozymes 3 and 4, appears to be independent of Pi and insensitive to ADP, at least under the assay conditions used here. This reciprocal relationship of effects between Pi and ADP on activity of the two enzyme forms resembles the behavior of certain variant forms of human PRPP synthase 1, which may be the molecular basis for some types of gout (Becker et al., 1995). These mutant forms are insensitive to ADP inhibition and simultaneously are activated by lower concentrations of Pi than the normal human PRPP synthase 1. In previous studies with plants the existence of PRPP synthase isozymes 1 and 2 might have been overlooked because of their requirement of Pi for activity and possibly also for stability.
The localization of the four PRPP synthases appears to be complex, with at least two organelle-localized enzymes and one cytosol-localized enzyme. It is likely that this complexity reflects the specialization of various organelles to biosynthetic pathways, with purine nucleotide synthesis occurring within both chloroplasts and mitochondria and with at least some amino acid synthesis pathways occurring within chloroplasts. In addition, the expression of the various PRS genes may by developmentally regulated if the demand for PRPP changes during cell growth.
ACKNOWLEDGMENTS
We are grateful to Tine Hoff for invaluable discussions and advice with import assays. We thank Bjarne Jochimsen for careful reading of the manuscript. Tonny D. Hansen and Anne L. Møller are acknowledged for pertinent technical assistance. We thank Charlotte Hansen for assistance with the automated nucleotide sequencing and Tine A. Eriksen for assistance with phylogenetic analysis.
Abbreviation:
- PRPP
5-phospho-d-ribosyl α-1-diphosphate
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnvig K, Hove-Jensen B, Switzer RL. Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem. 1990;192:195–200. doi: 10.1111/j.1432-1033.1990.tb19214.x. [DOI] [PubMed] [Google Scholar]
- Ashihara H. Characterization of phosphoribosylpyrophosphate synthetase from spinach leaves. Z Pflanzenphysiol. 1977a;83:379–392. [Google Scholar]
- Ashihara H. Regulation of the activity of spinach phosphoribosylpyrophosphate synthetase by ‘energy charge’ and end products. Z Pflanzenphysiol. 1977b;85:383–392. [Google Scholar]
- Ashihara H, Komamine A. Regulatory properties of a plant phosphoribosylpyrophosphate synthetase. Plant Sci Lett. 1974;2:119–123. [Google Scholar]
- Ashihara H, Ukaji T. Presence of adenine phosphoribosyltransferase and adenosine kinase in chloroplasts of spinach leaves. Int J Biochem. 1985;17:1275–1277. doi: 10.1016/0020-711x(85)90020-5. [DOI] [PubMed] [Google Scholar]
- Atkins CA, Smith PM, Storer PJ. Reexamination of the intracellular localization of de novo purine synthesis in cowpea nodules. Plant Physiol. 1997;113:127–135. doi: 10.1104/pp.113.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GJ. Protein multiple sequence alignment and flexible pattern matching. Methods Enzymol. 1990;183:403–428. doi: 10.1016/0076-6879(90)83027-7. [DOI] [PubMed] [Google Scholar]
- Becker MA, Smith PR, Taylor W, Mustafi R, Switzer RL. The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J Clin Invest. 1995;96:2133–2141. doi: 10.1172/JCI118267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen A-K, Larsen TA, Kadziola A, Larsen S, Harlow KW. Overexpression of Bacillus subtilis phosphoribosylpyrophosphate synthetase and crystallization and preliminary x-ray characterization of the free enzyme and its substrate-effector complexes. Proteins. 1996;24:238–246. doi: 10.1002/(SICI)1097-0134(199602)24:2<238::AID-PROT10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Birnboim H, Doly J. A rapid alkaline extraction for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower SG, Harlow KW, Switzer RL, Hove-Jensen B. Characterization of the Escherichia coli prsA1-encoded mutant phosphoribosylpyrophosphate synthetase identifies a divalent cation-nucleotide binding site. J Biol Chem. 1989;264:10287–10291. [PubMed] [Google Scholar]
- Bower SG, Hove-Jensen B, Switzer RL. Structure of the gene encoding phosphoribosylpyrophosphate synthetase (prsA) in Salmonella typhimurium. J Bacteriol. 1988;170:3243–3248. doi: 10.1128/jb.170.7.3243-3248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD and others. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Carter AT, Beiche F, Hove-Jensen B, Narbad A, Barker PJ, Schweizer LM, Schweizer M. PRS1 is a key member of the gene family encoding phosphoribosylpyrophosphate synthetase in Saccharomyces cerevisiae. Mol Gen Genet. 1997;254:148–156. doi: 10.1007/s004380050402. [DOI] [PubMed] [Google Scholar]
- Carter AT, Narbad A, Pearson BM, Beck KF, Logghe M, Contreras R, Schweizer M. Phosphoribosylpyrophosphate synthetase (PRS): a new gene family in Saccharomyces cerevisiae. Yeast. 1994;10:1031–1044. doi: 10.1002/yea.320100805. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Martinez-Arias A, Shapira SK, Chou J. β-Galactosidase gene fusion for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Lubben TH, Keegstra K. Precursors to two nuclear-encoded chloroplast proteins bind to the outer envelope membrane before being imported into chloroplasts. J Biol Chem. 1985;260:3691–3696. [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM and others. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Gallois R, Prévôt J-C, Clément A, Jacob J-L. Purification and characterization of phosphoribosylpyrophosphate synthetase from rubber tree latex. Plant Physiol. 1997;115:847–852. doi: 10.1104/pp.115.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Gibson KJ, Schubert KR, Switzer RL. Binding of the substrates and the allosteric inhibitor adenosine 5′-diphosphate to phosphoribosylpyrophosphate synthetase from Salmonella typhimurium. J Biol Chem. 1982;257:2391–2396. [PubMed] [Google Scholar]
- Gouin E, Mengaud J, Cossart P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect Immun. 1994;62:3550–3553. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow KW, Switzer RL. Chemical modification of Salmonella typhimurium phosphoribosylpyrophosphate synthetase with 5′-(p-fluorosulfonylbenzoyl)adenosine. Identification of an active site histidine. J Biol Chem. 1990;256:5487–5493. [PubMed] [Google Scholar]
- Hendrick JP, Hodges PE, Rosenberg LE. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci USA. 1989;86:4056. doi: 10.1073/pnas.86.11.4056. 4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson NE, Allen T, Ullman B. Molecular characterization of phosphoribosyl-pyrophosphate synthetase from Leishmania donovani. Mol Biochem Parasitol. 1993;59:15–28. doi: 10.1016/0166-6851(93)90003-g. [DOI] [PubMed] [Google Scholar]
- Hernando Y, Parr A, Schweizer M. PRS5, the fifth member of the phosphoribosylpyrophosphate synthetase gene family in Saccharomyces cerevisiae, is essential for cell viability in the absence of either PRS1 or PRS3. J Bacteriol. 1998;180:6404–6407. doi: 10.1128/jb.180.23.6404-6407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilden I, Hove-Jensen B, Harlow KW. Inactivation of Escherichia coli phosphoribosylpyrophosphate synthetase by the 2′,3′-dialdehyde derivative of ATP. J Biol Chem. 1995;270:20730–20736. doi: 10.1074/jbc.270.35.20730. [DOI] [PubMed] [Google Scholar]
- Hoff T, Kirk KM, Meyer C, Caboche M. Isolation of two Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J Biol Chem. 1995;270:6100–6107. doi: 10.1074/jbc.270.11.6100. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B. Cloning and characterization of the prs gene encoding phosphoribosylpyrophosphate synthetase of Escherichia coli. Mol Gen Genet. 1985;201:269–276. doi: 10.1007/BF00425670. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B. J Bacteriol. 1988;170:1148–1152. doi: 10.1128/jb.170.3.1148-1152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hove-Jensen B. Phosphoribosylpyrophosphate (PRPP)-less mutants of Escherichia coli. Mol Microbiol. 1989;3:1487–1492. doi: 10.1111/j.1365-2958.1989.tb00134.x. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, Harlow KW, King CJ, Switzer RL. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- Hove-Jensen B, Maigaard M. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J Bacteriol. 1993;174:6852–6856. doi: 10.1128/jb.175.17.5628-5635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG, Chu NM, Odell JT, Nagy F, Chua N-H. Plant cells do not properly recognize animal gene polyadenylation signals. Plant Mol Biol. 1987;8:23–35. doi: 10.1007/BF00016431. [DOI] [PubMed] [Google Scholar]
- Iizasa T, Taira M, Shimada H, Ishijima S, Tatibana M. Molecular cloning and sequencing of human cDNA for phosphoribosylpyrophosphate synthetase subunit II. FEBS Lett. 1989;244:47–50. doi: 10.1016/0014-5793(89)81159-7. [DOI] [PubMed] [Google Scholar]
- Ishijima S, Kita K, Ahmad I, Ishizuka T, Taira M, Tatibana M. Expression of rat phosphoribosylpyrophosphate synthetase subunits I and II in Escherichia coli. J Biol Chem. 1991;266:15693–15697. [PubMed] [Google Scholar]
- Ishizuka T, Kita K, Sonoda T, Ishijima S, Sawa K, Suzuki N, Tatibana M. Cloning and sequencing of human complementary DNA for the phosphoribosylpyrophosphate synthetase-associated protein 39. Biochim Biophys Acta. 1996;1306:27–30. doi: 10.1016/0167-4781(96)00030-9. [DOI] [PubMed] [Google Scholar]
- Ito T, Shiraishi H, Okada K, Shimura Y. Two amidophosphoribosyltransferase genes of Arabidopsis thaliana expressed in different organs. Plant Mol Biol. 1994;26:529–533. doi: 10.1007/BF00039565. [DOI] [PubMed] [Google Scholar]
- Jensen KF, Houlberg U, Nygaard P. Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-α-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal Biochem. 1979;98:254–263. doi: 10.1016/0003-2697(79)90138-6. [DOI] [PubMed] [Google Scholar]
- Kanamori I, Ashihara H, Komamine A. Subcellular distribution and activity of enzymes involved in uridine-5′-monophosphate synthesis in Vinca rosea cells. Z Pflanzenphysiol. 1980;96:7–16. [Google Scholar]
- Khorana HG, Fernandes JF, Kornberg A. Pyrophosphorylation of ribose 5- phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J Biol Chem. 1958;230:941–948. [PubMed] [Google Scholar]
- Kita K, Ishizuka T, Ishijima S, Sonoda T, Tatibana M. A novel 39-kDa phosphoribosylpyrophosphate synthetase-associated protein of rat liver. Cloning, high sequence similarity to the catalytic subunits, and a negative regulatory role. J Biol Chem. 1994;269:8334–8340. [PubMed] [Google Scholar]
- Kjarulff S, Okkels JS. Cloning and sequencing of a full-length cDNA clone encoding the PSI-D subunit of photosystem I from barley. Plant Physiol. 1993;101:335–336. doi: 10.1104/pp.101.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krath BN, Hove-Jensen B. Bacillus caldolyticus prs gene encoding phosphoribosyldiphosphate synthase. Gene. 1996;176:73–79. doi: 10.1016/0378-1119(96)00222-3. [DOI] [PubMed] [Google Scholar]
- Kyradji S, Bagnara AS (1998) Characterisation of the gene encoding phosphoribosylpyrophosphate synthetase (PRS) of Giardia intestinalis. Mol Biochem Parasitol (in press) [DOI] [PubMed]
- Laemmli UK. Cleavage of structual proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Floc'h F, Lafleuriel J. The role of mitochondria in the recycling of adenine into purine nucleotides in the Jerusalem artichoke (Helianthus tuberosus L.) Z Pflanzenphysiol. 1983;113:61–71. [Google Scholar]
- Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem R, Morikawa M, Imanaka T. Gene cloning and characterization of recombinant ribose phosphate pyrophosphokinase from a hyperthermophilic Archaeon. J Ferment Bioeng. 1997;83:412–418. [Google Scholar]
- Nagaya M, Aiba H, Mizuno T. A cyanobacterial gene encoding a protein with extensive homology to mammalian phosphoribosylpyrophosphate synthetase. Biosci Biotechnol Biochem. 1993;57:1958–1959. doi: 10.1271/bbb.57.1958. [DOI] [PubMed] [Google Scholar]
- Nilsson D, Hove-Jensen B. Phosphoribosylpyrophosphate synthetase of Bacillus subtilis. Cloning, characterization and chromosomal mapping of the prs gene. Gene. 1987;53:247–255. doi: 10.1016/0378-1119(87)90013-8. [DOI] [PubMed] [Google Scholar]
- Nilsson D, Hove-Jensen B, Arnvig K. Primary structure of the tms and prs genes of Bacillus subtilis. Mol Gen Genet. 1989;218:565–571. doi: 10.1007/BF00332425. [DOI] [PubMed] [Google Scholar]
- Nosal JM, Switzer RL, Becker MA. Overexpression, purification, and characterization of recombinant human 5phosphoribosyl-1-pyrophosphate synthetase isozymes I and II. J Biol Chem. 1993;268:10168–10175. [PubMed] [Google Scholar]
- Post D, Switzer RL, Hove-Jensen B. The defective phosphoribosyl diphosphate synthase in a temperature-sensitive prs-2 mutant of Escherichia coli is compensated by increased enzyme synthesis. Microbiology. 1996;142:359–365. doi: 10.1099/13500872-142-2-359. [DOI] [PubMed] [Google Scholar]
- Roessler BJ, Bell G, Heidler S, Seino S, Becker M, Palella TD. Cloning of two distinct copies of human phosphoribosylpyrophosphate synthetase cDNA. Nucleic Acids Res. 1990;18:193. doi: 10.1093/nar/18.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr KM, Nygaard P, Laloue M. Molecular characterization of Arabidopsis thaliana cDNAs encoding three purine biosynthetic enzymes. Plant J. 1994;6:113–121. doi: 10.1046/j.1365-313x.1994.6010113.x. [DOI] [PubMed] [Google Scholar]
- Senecoff J, Meager R. Isolating the Arabidopsis thaliana genes by suppression of Escherichia coli mutants. Plant Physiol. 1993;102:387–399. doi: 10.1104/pp.102.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Switzer RL. Regulation and mechanism of phosphoribosylpyrophosphate synthetase. I. Purification and properties of the enzyme from Salmonella typhimurium. J Biol Chem. 1969;244:2854–2863. [PubMed] [Google Scholar]
- Switzer RL. Regulation and mechanism of phosphoribosylpyrophosphate synthetase. III. Kinetic studies of the reaction mechanism. J Biol Chem. 1971;246:2447–2458. [PubMed] [Google Scholar]
- Switzer RL, Sogin DC. Regulation and mechanism of phosphoribosylpyrophosphate synthetase. V. Inhibition by end products and regulation by adenosine diphosphate. J Biol Chem. 1973;248:1063–1073. [PubMed] [Google Scholar]
- Taira M, Iiaza T, Shimada H, Kudoh J, Shimizu N, Tatibana M. A human testis-specific mRNA for phosphoribosylpyrophosphate synthetase that initiates from a non-AUG codon. J Biol Chem. 1990;265:16491–16497. [PubMed] [Google Scholar]
- Taira M, Iizasa T, Yamada K, Shimada H, Tatibana M. Tissue-differential expression of two distinct genes for phosphoribosyl pyrophosphate synthetase and existence of the testis-specific transcript. Biochim Biophys Acta. 1989;1007:203–208. doi: 10.1016/0167-4781(89)90040-7. [DOI] [PubMed] [Google Scholar]
- Taira M, Ishijima S, Kita K, Yamada K, Iizasa T, Tatibana M. Nucleotide and deduced amino acid sequences of two distinct cDNAs for rat phosphoribosyl-pyrophosphate synthetase. J Biol Chem. 1987;262:14867–14870. [PubMed] [Google Scholar]
- Tatibana M, Kita K, Taira M, Ishijima S, Sonoda T, Ishizuka T, Iizasa T, Ahmad I. Mammalian phosphoribosyl-pyrophosphate synthetase. Adv Enzyme Regul. 1995;35:229–249. doi: 10.1016/0065-2571(94)00017-w. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Nishikawa K. Chloroplast transit peptides. FEBS Lett. 1991;278:1–3. doi: 10.1016/0014-5793(91)80069-f. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmenn G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- White JA, Scandalios JG. In vitro synthesis, importation and processing of Mn-superoxide dismutase (SOD-3) into maize mitochondria. Biochim Biophys Acta. 1987;926:16–25. doi: 10.1016/0304-4165(87)90178-4. [DOI] [PubMed] [Google Scholar]
- White JA, Scandalios JG. Deletion analysis of the maize mitochondrial superoxide dismutase transit peptide. Proc Natl Acad Sci USA. 1989;86:3534–3538. doi: 10.1073/pnas.86.10.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemoës M, Nilsson D, Hove-Jensen B. Effects of mutagenesis of aspartic acid residues in the putative phosphoribosyl diphosphate binding site of Escherichia coli phosphoribosyl diphosphate synthetase on metal ion specificity and ribose 5-phosphate binding. Biochemistry. 1996;35:8181–8186. doi: 10.1021/bi9528560. [DOI] [PubMed] [Google Scholar]
- Zhao J, Last RL. Immunological characterization and chloroplast localization of the tryptophan biosynthetic enzymes of the flowering plant Arabidopsis thaliana. J Biol Chem. 1995;270:6081–6087. doi: 10.1074/jbc.270.11.6081. [DOI] [PubMed] [Google Scholar]