Abstract
The effect of standard agricultural management on the genetic heterogeneity of nitrous oxide reductase (nosZ) fragments from denitrifying prokaryotes in native and cultivated soil was explored. Thirty-six soil cores were composited from each of the two soil management conditions. nosZ gene fragments were amplified from triplicate samples, and PCR products were cloned and screened by restriction fragment length polymorphism (RFLP). The total nosZ RFLP profiles increased in similarity with soil sample size until triplicate 3-g samples produced visually identical RFLP profiles for each treatment. Large differences in total nosZ profiles were observed between the native and cultivated soils. The fragments representing major groups of clones encountered at least twice and four randomly selected clones with unique RFLP patterns were sequenced to verify nosZ identity. The sequence diversity of nosZ clones from the cultivated field was higher, and only eight patterns were found in clone libraries from both soils among the 182 distinct nosZ RFLP patterns identified from the two soils. A group of clones that comprised 32% of all clones dominated the gene library of native soil, whereas many minor groups were observed in the gene library of cultivated soil. The 95% confidence intervals of the Chao1 nonparametric richness estimator for nosZ RFLP data did not overlap, indicating that the levels of species richness are significantly different in the two soils, the cultivated soil having higher diversity. Phylogenetic analysis of deduced amino acid sequences grouped the majority of nosZ clones into an interleaved Michigan soil cluster whose cultured members are α-Proteobacteria. Only four nosZ sequences from cultivated soil and one from the native soil were related to sequences found in γ-Proteobacteria. Sequences from the native field formed a distinct, closely related cluster (Dmean = 0.16) containing 91.6% of the native clones. Clones from the cultivated field were more distantly related to each other (Dmean = 0.26), and 65% were found outside of the cluster from the native soil, further indicating a difference in the two communities. Overall, there appears to be a relationship between use and richness, diversity, and the phylogenetic position of nosZ sequences, indicating that agricultural use of soil caused a shift to a more diverse denitrifying community.
After the last ice age, the nitrous oxide concentration increased in the atmosphere and remained constant (approximately 275 ppb) for about 10,000 years until the 19th century. Since then, the N2O concentration has increased significantly to approximately 315 ppb, and this has been predominately attributed to anthropogenic contributions. Due to its long estimated half-life (approximately 120 years), and a global warming potential about 310 times that of carbon dioxide, even a small N2O accumulation may cause destructive effects for centuries (4, 46).
Denitrification and nitrification are thought to be major sources of atmospheric nitrous oxide (20, 25, 37, 50). The capacity for denitrification is found among a wide variety of taxonomic groups within the Bacteria and Archaea (53). The reduction of nitrous oxide to molecular nitrogen, catalyzed by nitrous oxide reductase, is the last step in the complete denitrification pathway and represents a respiratory process in its own right, because many denitrifiers can grow at the expense of N2O as the sole electron acceptor (28). The plasmid-encoded nature of N2O reduction, at least in some strains, distinguishes this respiratory process from other steps in denitrification (43, 54).
Numerous environmental factors can vary the proportions of N2O and N2 produced, including soil moisture (3); pH (45); aeration (20, 37); carbon, nitrate, and nitrite availability (34, 45); pore structure (27); and freezing-thawing (25, 36) and drying-wetting (3, 37) events. N2O emissions from soils greatly increase with increasing N inputs by fertilization of agricultural soils (10). In most natural habitats, there is usually not enough nitrate to select the large populations of denitrifying organisms. Rather, these denitrifiers are thought to be effective aerobic competitors for carbon and may seldom use their denitrification capacity (45). Lack of carbon as an electron donor almost never prevents denitrification, although it is often not present in amounts that saturate the denitrification capacity. Under starvation conditions, N2O was found to be the main product of denitrification in a pure culture of Alcaligenes faecalis, indicating that an unbalanced supply of electron donor and acceptor may have profound effects on the N2O/N2 ratio (41, 45). Organic matter in soil also serves as a source of nitrate and drives oxygen removal by respiration, creating anaerobic microsites (45). Additionally, differences in oxygen threshold, carbon requirement, and kinetic parameters of various denitrifiers may influence competition with aerobic heterotrophs and thus also influence emission rates of nitrous oxide (7, 18).
Since only a minor portion of microbial diversity present in the environment is culturable by current techniques, analysis of the properties of a number of denitrifying soil isolates (8) probably provides a conservative estimate of the physiological diversity present in the environment (35). The majority of molecular studies investigating microbial diversity in natural communities have focused on sequence diversity, using primers that amplify 16S rRNA genes from bacterial and archaeal domains. The high phylogenetic diversity of denitrifiers makes the use of 16S ribosomal DNA (rDNA)-based approaches inappropriate for ecological study of this group. Hence, genes involved in the denitrification process, such as those coding for nitrite reductase (nirS and nirK) (2, 5, 31) or nitrous oxide reductase (nosZ), are used (35, 40).
In this study, we used PCR methods with subsequent cloning and sequencing to characterize the heterogeneity of nosZ gene fragments in soil samples taken from geomorphically similar plots but under different management: one cultivated with intensive agricultural practices and the other taken from natural plant succession that had never been cultivated. The N2O emissions from the cultivated soils are consistently more than double that from native soil (3, 8, 34).
MATERIALS AND METHODS
Soil sampling.
Soil samples were obtained in the beginning of October 2001 from the Michigan State University W. K. Kellogg Biological Station Long-Term Ecological Research site located at Hickory Corners, Mich. (http://lter.kbs.msu.edu). Soil samples were obtained from a conventionally tilled agricultural field and from a successional field that was never tilled. The cultivated field had been farmed for more than a century under various crop rotations, managed in accordance with regional agronomic practices. Since 1988, the cultivated field has been under a regimen characterized by high levels of fertilization, herbicide addition, annual tillage, and wheat-corn-soybean crop rotation. The native field has never been farmed and was cleared of trees in 1958. This field is generally covered with a diverse plant community dominated by herbaceous perennials. The cultivated and native fields, respectively, have geomorphically similar soils (Typic Hapludalfs, fine-loamy, mixed, mesic), but differ significantly in factors that are likely to influence denitrifier community composition (3, 7, 34, 45): pH (6.56 versus 5.7), bulk density (1.65 versus 1.35 g/cm3), total carbon (0.77 versus 1.97 g/100 g of dry soil), total nitrogen (0.077 versus 0.166 g/100 g of dry soil), and inorganic nitrogen pools (nitrate-N, 4.57 versus 0.93 μg/g of dry soil; and ammonium-N, 2.73 versus 8.75 μg/g of dry soil).
Six individual plots of the same treatments were each sampled six times, for a total of 36 soil cores per treatment (depth, 0 to 25 cm, diameter, 2.5 cm). Soil cores were homogenized by sieving through a 4-mm-diameter screen, and randomly collected soil aggregates from the batch of sieved soil were pooled in sterile tubes to form 0.25-g samples. These were immediately frozen to −80°C. Portions of samples were also stored at 4°C in order to determine moisture content by drying at 100°C for 48 h.
Nucleic acid extraction and analysis.
DNA was initially isolated from composite samples of increasing size (0.25, 0.5, 0.75, 1, 1.5, 2, 3, and 6 g) using Ultra Clean Soil DNA Isolation kits (MoBio Laboratories, Solana Beach, Calif.) as specified by the manufacturer. The A260/A280 ratio of the extracted DNA was 1.57 ± 0.14 (n = 20).
Composite samples for DNA isolations were formed by randomly selecting a number of tubes containing 0.25-g aggregates: e.g., a 3-g composite sample was formed from 12 tubes of 0.25-g soil aggregates. From DNA isolations of composite samples, eight nosZ 50-μl PCRs were combined to form a replicate and subjected to restriction fragment length polymorphism (RFLP) profiling. Three replicates (e.g., three composite soil samples) per soil management conditions and sample size were digested with the endonucleases HhaI, HaeIII, MspI, and TaqI (New England Biolabs). PCR products of those samples in which reproducibility and complexity of nosZ RFLP profiles among replicates could not be further improved by size increase were used for further analysis.
PCR amplification.
Existing nosZ sequences were retrieved from the National Center for Biotechnology Information (NCBI) sequence and genome databank using primer Short BLAST and DNA and amino acid sequence BLAST searches. The respective significance of E values of hits was also considered. All accessible nosZ sequences were aligned to evaluate the specificity and suitability of existing primers (17, 23, 39, 43). The sequences exhibiting short overlaps (200 bp) containing primer-binding sites were also aligned in separate alignments (35).
Fragments of nosZ genes were amplified with selected primer pair 661F and 1773R (39). Amplification was performed by using a mixture containing 20 pmol of each primer, 1.5 U of Taq DNA polymerase in storage buffer B (Promega, Madison, Wis.), 2.5 mM MgCl2, 0.1% Tryton (Promega, Madison, Wis.), 400-ng/μl bovine serum albumin (Boehringer Mannheim), 200 μM deoxynucleoside triphosphates (dNTPs; Invitrogen, San Diego, Calif.), and 1 μl of DNA in a final volume of 50 μl. The touchdown PCR was performed in a Perkin-Elmer 9600 thermal cycler (Norwalk, Conn.) and included 5 min at 95°C followed by 35 cycles of 60 s at 95°C, 60 s at 58°C, and 90 s at 72°C. The annealing temperature was shifted from 58°C to 53°C during the first 10 cycles and remained at 55°C during the remaining 25 cycles. Triplicate PCRs per sample were run, and aliquots were analyzed by electrophoresis on 2% (wt/vol) agarose gels (Gibco, BRL) followed by 10 min of staining with ethidium bromide (0.5 mg liter−1). Bands were visualized by UV excitation.
Cloning of nosZ PCR products from soil samples.
PCR products were purified by separation with low-melting-point agarose (2% [wt/vol]). Bands of the expected size (i.e., 1,100 bp) were excised and purified with the QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.) as specified by the manufacturer. Eluted nosZ PCR products (4 μl) were cloned by using the TOPO TA cloning kit (Invitrogen, San Diego, Calif.).
RFLP screening of nosZ clones.
The two libraries contained 288 randomly selected clones each and were organized on agar trays the size of microtiter plates. A 96-dot plater was used to inoculate clones from agar trays onto a microtiter plate containing 200 μl of Luria-Bertani (LB) freezing buffer supplemented with kanamycin (50 μg/ml). Clones were grown for 10 h at 36°C in an orbital shaker at 200 rpm. Using the multichannel pipettor, 2 μl of the resulting cell suspension was resuspended in a prepared 96-well 18-μl PCR mix. The inserts were amplified as described above, with the exception of a 10-min denaturation time and 25 PCR cycles. Aliquots (5 μl) were analyzed on 2.5% (wt/vol) agarose gels, and only clones bearing inserts of the expected size were analyzed further. The PCR products from selected clones were screened by RFLP. Known nosZ sequences were digested in silico using BCM Search Launcher utilities (http://searchlauncher.bcm.tmc.edu). In order to maximize resolution of RFLP analysis, PCR products (10 μl) were digested in separate reactions using 2.5 U of four selected restriction endonucleases (HhaI, HaeIII, and MspI at 37°C and TaqI at 65°C) overnight. The digested products were separated by gel electrophoresis in 3.5% (wt/vol) Metaphore agarose (FMC Bioproducts, Rockland, Maine) in freshly prepared 1× Tris-borate EDTA buffer at 4°C with 8 V cm−1 for 3.5 h. Digests were run in triplicate. The resulting RFLP patterns were manually aligned.
Diversity indices.
Since any RFLP pattern may represent sequences from multiple phylogenetic groups (21), four tetrameric endonucleases per clone were used to maximize resolution. To evaluate richness and evenness, diversity statistics were calculated by using RFLP data as representations of different phylotypes. Phylotype richness (S) was calculated as the total number of distinct RFLP patterns in soil. The Shannon diversity index (22) was calculated as follows: H = -Σ(pi)(log2 pi), where p is the proportion of distinct RFLP patterns relative to the sum of all distinct patterns in one library. Evenness (22) was calculated from the Shannon diversity index, where E = H/Hmax and Hmax = log2 (S). Library coverage (C) was also used as a measure of captured diversity (14) and was calculated using C = 1 − n/N, where n is the number of clone types from a clone library that are encountered only once and N is the total number of clones analyzed. The species richness was estimated by two means. First, curve extrapolation was used to estimate the asymptotic value by fitting the data from plots containing species abundance data randomized 1,000 times (rarefaction) to a simple negative-exponential model (12, 19) and Eadie-Hofstee transformation (32), and second, the nonparametric Chao1 estimator (9) was calculated from randomized data as described by Hughes et al. (19). Log transformation was used to calculate Chao1 corresponding 95% confidence intervals (95% CIs) because the distribution of estimates is not normal (9). The analyses were performed with RECODE (R. Pestotnik, Josef Stefan Institute; http://www-f9.ijs/∼rok) in conjunction with Physics Analysis Workstation (PAW; http://wwwinfo.cern.ch/asd/paw) and EstimateS (version 6.0b1; R. Colwell, University of Connecticut; http://viceroy.eeb.uconn.edu/estimates). For the purpose of inputting the data into the programs, we treated each clone as a separate sample (19), and 1,000 randomizations were run. Since most of the problems in measuring bacterial diversity arise from the difficulties of defining phylotypes or operational taxonomic units (OTU), an alternative in the form of mean taxonomic distance (Dmean) between all pairs of isolates as a diversity index, combining proportional abundances with phylogenetic diversity, was also calculated: Dmean = 2 Σ d(i,j)/S(S + 1), where d(i,j) is one-nucleic-acid homology and S is the total number of clones (33, 48).
Sequencing of nosZ PCR products and phylogenetic analysis.
The fragments that represented major groups of clones with identical RFLP profiles encountered at least twice and four randomly selected clones with unique RFLP patterns encountered only once were sequenced. Clones were grown for 6 h, and nosZ fragments were amplified as described above, with exception of the M13 F-M13 R primer pair that binds to sites next to the insert in the plasmids. The amplified PCR products were purified with the QIAgen PCR purification kit (Qiagen, Chatsworth, Calif.), and DNA sequences were determined by direct sequencing with an ABI 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) using dye primer sequencing with primer pairs PCI and PCII (51) and 661F and 1773R (39). Contigs were assembled using Contig Assembly Program (http://www.cs.sunysb.edu), and preliminary studies were performed with BLAST (1).
The alignment of the predicted amino acid sequences with previously published sequences overlapping in the same region was performed with the Clustal W program (44) in the BioEdit program package (16). The basic alignment was created using default gap introduction and elongation weights. The resulting alignment was examined and realigned manually. Weights were increased incrementally to minimize the number and size of gaps while maximizing the extent of conserved blocks (15). Regions of ambiguous homology, translated primer binding sites, and insertions and deletions (indels) not present in all sequences analyzed were excluded, yielding an amino acid data set with 299 positions. Trees were reconstructed from distance matrices by using FITCH and neighbor joining (TREECON) (47) and the PHYLIP software package, version 3.6a2.1 (13), parsimony PROTPARS (PHYLIP), and maximum-likelihood MOLPHY (Institute Pasteur, http://bioweb.Pasteur.fr/seqanal/interfaces/prot_nucml.html). Two separate distance matrices were calculated by using PROTDIST (PHYLIP) with the Jones-Taylor-Thornton model and Dayhoff PAM 001 matrix (13) as the amino acid replacement models with randomized input order (“jumble” seven times). Neighbor-joining trees were reconstructed with four different models: Kimura two parameter, Poisson correction, Tajima, and Nei, as implemented in TREECON software (47). The model of Nei and Saitou was used with randomized species input order (“jumble,” random seed 7, seven times) options (13). FITCH trees were reconstructed using the global rearrangements and randomized species input order (“jumble,” random seed 7, seven times) options. Statistical evaluation of tree topologies was performed by bootstrap analysis with 1,000 resamplings for neighbor joining and parsimony as implemented in the PHYLIP and TREECON packages. After comparison of trees generated by different methods, a consensus tree was constructed by introducing multifurcations where the topology was not resolved.
Nucleotide sequence accession number.
The nosZ gene sequences have been deposited in the EMBL nucleotide sequence database under accession no. AJ550328 through AJ550375.
RESULTS
Representative nucleic acid extraction and amplification of nosZ gene fragments.
High-molecular-mass DNA (15 to 20 kb) was isolated from soil samples from both study sites. Amplification of nosZ fragments from the cultivated soils posed considerable difficulties (optical density at 260/280 nm [OD260/280] = 1.46 ± 0.10), and additional optimization of PCR was needed, resulting in the touchdown PCR protocol described above. Almost 50% larger total amounts of DNA were isolated from native field samples. Simple doubling of the amounts of DNA in PCRs did not produce significant improvement in amplification.
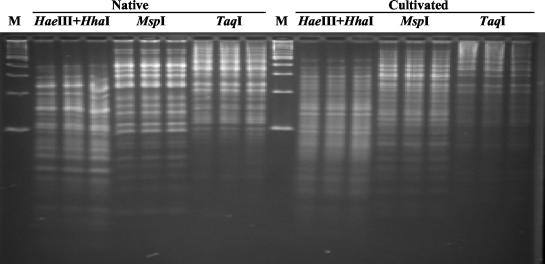
A collection of tubes containing 0.25-g randomly collected soil aggregates was randomly sampled to form composite samples of various sizes. The increase in soil sample size was proportional to the increase in similarities of total nosZ RFLP profiles per soil use. The largest variations were found in smaller samples (0.25 and 0.5 g), and discrimination between treatments was not feasible at that point. RFLP profiles of 1-, 1.5-, and 2-g samples from each soil use indicated increasing convergence of RFLP profiles from one soil use until triplicate 3-g composite samples produced visually identical RFLP profiles per soil use (Fig. 1). Further increase in composite soil sample size (6 g) did not produce any additional increase in complexity of RFLP profiles. Hence, a 3-g soil sample was used for subsequent studies. Large differences among native and cultivated soil nosZ RFLP profiles were observed in triplicate 3-g samples as a first indication of differences in the denitrifying microbial communities in the two soils (Fig. 1).
FIG. 1.
RFLP analysis of nosZ gene fragments from triplicate 3-g soil samples taken from a native field and a cultivated field. nosZ PCR products from both soils were digested with tetrameric restriction endonucleases HhaI and HaeIII in one reaction and MspI and TaqI in separate reactions. M, 100-bp molecular size marker.
RFLP analysis of clones.
Clone libraries from native and cultivated samples were formed with 288 clones from each. Only 146 and 210 clones from cultivated and native soil clone libraries, respectively, contained an insert of the expected size and were screened further. The RFLP analysis of clones revealed 104 and 86 unique RFLP groups in cultivated and native soil clone libraries, respectively. Among the 182 distinct nosZ RFLP patterns identified in the two soils, only 8 RFLP patterns were found in both soil libraries. The native soil clone library contained a dominant group of clones (32% of the library), whereas none of the groups from the cultivated soil library represented more than 5% of clones. The highest similarity of nucleic acid sequences between sequences with different RFLP patterns was 98.1%.
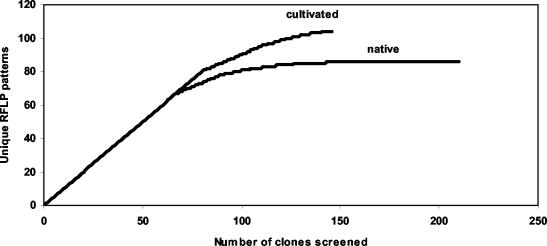
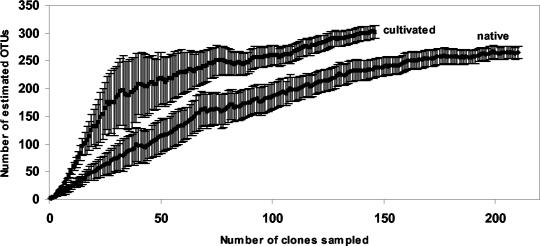
In order to determine how well the sampling captured the total diversity of RFLP patterns, two approaches were used. First, library coverage (14) was calculated to be 28.8% for the cultivated soil clone library, in contrast to 59% for the native soil clone library; and, second, the cumulative number of distinct RFLP patterns was plotted as a function of the number of clones screened (collector's curve) (Fig. 2). The latter approach is analogous to rarefaction analysis in estimating species richness from species abundance data (22). If continuous effort does not produce new distinct RFLP patterns, the rarefaction curve reaches an asymptotic value, the library coverage value, C, approaches 1 (14), and the libraries are assumed to be well sampled (38). The nonparametric Chao1 species richness estimator predicted existence of 301 ± 74 and 264 ± 78 unique nosZ OTU in cultivated and native soil, respectively. Using log transformation as described previously (9), the corresponding 95% CIs did not overlap (Fig. 3). The asymptotic values derived from curve extrapolation models were unreliable due to poor fit of the negative simple exponential and Eadie-Hofstee transformation models (data not shown), an observation consistent with previous results of Dunbar et al. (12). Evenness of population was 0.76 and 0.97 for native and cultivated soil, respectively, 1 being the case when all species are equally abundant.
FIG. 2.
Collector's curves indicating diversity of denitrifying bacteria as revealed by RFLP analysis of cloned nitrous oxide reductase gene fragments (nosZ) from native and cultivated soil samples. Fragments were digested with the tetrameric restriction endonucleases HhaI, HaeIII, MspI, and TaqI in separate reactions.
FIG. 3.
Chao1 estimates of native and cultivated field nosZ richness as a function of sample size. Error bars are 95% CIs and were calculated from the variance form derived by Chao (9) of the number of OTU (unique RFLP profiles) drawn in 1,000 randomizations at each sample size.
Phylogenetic analysis.
Partial nosZ gene sequences (ca. 1,100 bp) from 48 clones were identified: 24 from each soil. Genes were given a designation beginning with treatment number (c-T1 for cultivated and n-T8 for native soil), followed by a clone number in a library (e.g., C54). Comparisons with the NCBI database by using a BLAST search revealed that all of the 48 sequences showed homology to known nosZ sequences. The nucleotide sequence similarities from pairwise comparisons of the cultivated and native soil clones were similar, ranging from 60.9 to 97.9% and from 65.9 to 98.7%, respectively. The average level of nucleotide similarity for cultivated soil clones was 74% ± 7%, in contrast to 12% higher sequence similarity of native soil clones (86% ± 7%). The average nucleotide similarity of combined cultivated and native soil sequences was 79% ± 9%. The average percentage of nucleotide similarity of all 105 nosZ gene sequences used in this study was 70% ± 10%. All clones exhibited <90% DNA sequence similarity to nosZ sequences from isolated denitrifying bacteria.
Percent similarities of all 105 nosZ sequences at the derived amino acid level were not significantly different from those at the DNA level ranging from 42 to 99% and averaged at 81% ± 12%. The difference in DNA sequence similarity observed between cultivated and native soil clones remained obvious also at the amino acid level, averaging at 77.4% ± 10% and 87.9% ± 10%, respectively. The calculated average DNA similarity was complementary to a corresponding mean taxonomic distance (Dmean) (Table 1).
TABLE 1.
Summary of data from RFLP and nucleotide analyses of nosZ gene fragments from denitrifier communities in native and cultivated soils
| Source | RFLP analysis results
|
Nucleotide sequence analysis results
|
|||||
|---|---|---|---|---|---|---|---|
| Sa | Estimated Seb | Hc | E (H/Hmax)d | C (%)e | Avg sequence similarity (%)f | Dmean (substitutions/100 sites)g | |
| Native | 86 (210) | 264 ± 74 | 4.87 | 0.76 | 59 | 85.9 ± 0.7 (100)h | 16.98 ± 13 |
| Cultivated | 104 (146) | 301 ± 78 | 6.47 | 0.97 | 28 | 74.6 ± 0.7 (100)h | 26.31 ± 18 |
Phylotype richness, S, was calculated as the total number of unique RFLP patterns in soil. Numbers in parentheses indicate the overall numbers of clones of the expected size screened by RFLP.
Estimated phylotype richness, Se, as estimated by nonparametric Chao1 estimator (9). The data are the mean ± standard error of the mean.
Shannon diversity index (22).
Evenness (22) of population.
Library coverage (14).
Average sequence similarity (± standard error of the mean) from pairwise comparisons of gene fragments.
The mean taxonomic distance (± standard error of the mean) from pairwise comparisons of gene fragments (32, 48).
Sequences with <90% identity to any published sequence recorded in the NCBI database.
The derived amino acid sequences of all 105 nosZ gene sequences were aligned, and approximately 15% of the 395 amino acid residues were conserved across all nosZ genes. The sequenced region includes 7 of the 11 conserved histidine residues reported previously (23, 40). Despite large differences at both the DNA and amino acid sequence levels, all seven of these histidine residues, H-129, H-130, H-132, H-178, H-346, H-391, and H-446, remained conserved. The conservation of these histidine residues suggests that the basic structural motifs necessary for proper functioning of the enzyme are maintained, indicating that sequences could produce functional enzymes.
The plasmid that harbored nosZ sequence from Ralstonia eutropha was used as the outgroup for phylogenetic distance analysis of the nosZ sequences, because it is the most distantly related, confirmed nosZ sequence exhibiting homology at the DNA and amino acid levels.
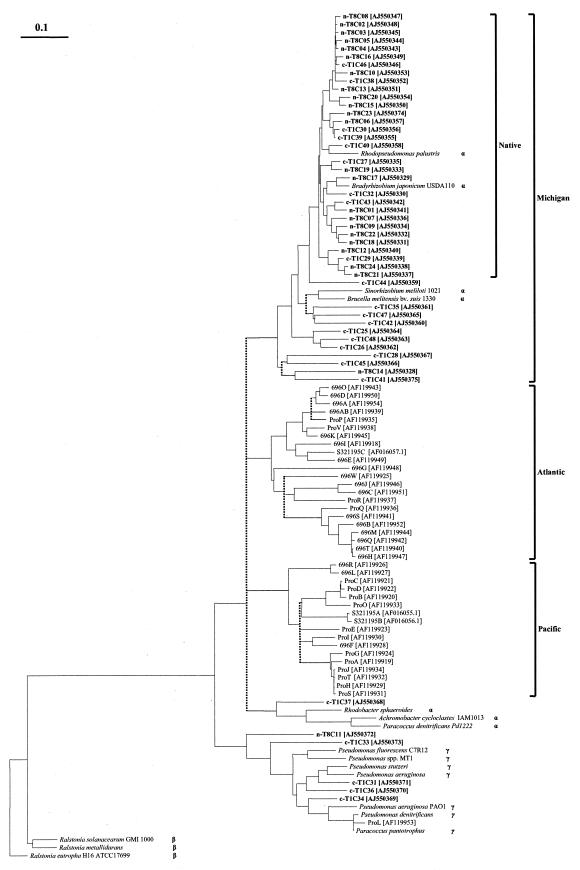
Sequences from native and cultivated fields formed a joined cluster (termed “Michigan soil”) within a cluster that contained proteobacterial sequences from Bradyrhizobium japonicum, Sinorhizobium meliloti, Brucella suis, and Rhodopseudomonas palustris. A more closely related subcluster (termed “native”) was formed, containing 91.7% of all native soil sequences and 37.5% of nosZ sequences from the cultivated soil, supported by all methods of tree reconstruction used. Almost half of the sequences from the cultivated soil (45.8%) and one sequence from the native soil were placed in a group containing nosZ sequences from cultivated bacteria S. meliloti and B. suis within the large Michigan cluster, whereas one sequence from cultivated soil was placed next to Rhodobacter sphaeroides. None of the sequences recovered in this study was placed into one of the two clusters containing sequences recovered from Atlantic or Pacific marine sediments (40). The remaining sequences from the cultivated and native soils (five and one, respectively) were related to γ-proteobacterial sequences represented by Pseudomonas spp.
DISCUSSION
Large variations (0.1 to 10 g or even more) of sample size used for DNA extraction can be found in the literature (26, 30, 40) without establishing the appropriateness of the particular choice (24). In this study, we first determined the sample size that gave a consistent pattern with soil use and was not further influenced by increased sample size. That value was 3 g for these soils and primers. This finding is in good agreement with previous nitrifier ecology studies of the same soils (30). The sample size reported here is at least three times the size used in some other studies (2, 6, 31), further emphasizing the need to account for spatial variability of soil microbial communities. Use of composite soil samples from numerous tubes containing randomly collected soil aggregates and employing an even larger number of soil cores than was used in this study (n = 36 per treatment, 0 to 25 cm, 2.5-cm diameter) might help address methodological questions about how sampling strategies and representativeness of the sample influence the results of microbial biodiversity studies (24).
The RFLP analysis indicated lower nosZ gene diversity in the denitrifying community from the native field, which was also supported by the high average sequence similarity of native field clones in comparison to sequences from the cultivated field and low mean taxonomic distance (Table 1). Because CIs for the Chao1 estimator did not overlap, one can reject the null hypothesis at the significance level of 0.05 that there is no difference between the richness of cultivated and native field communities. Because Chao1 estimates for the cultivated field have not yet stabilized (Fig. 3), this suggests that further sampling will result in a greater difference in richness between the cultivated and native fields.
The two soils in this study are geomorphically similar, but are contrasting with respect to several parameters resulting from one field's agricultural use: e.g., pH, bulk density, total carbon, total nitrogen and inorganic nitrogen (fertilizer addition), tillage and plant cover, and herbicide addition, which likely have influenced the denitrifier community composition (7, 8, 49, 52). Oxygen threshold, carbon requirement, and kinetic parameters are known to vary between different denitrifiers (8, 45) and therefore also between different denitrifying communities (7, 8, 11, 34). Some culturable denitrifiers can compete well as aerobic heterotrophs for carbon resources and hence are more abundant irrespective of their denitrifying properties (45). Since pH is a strong negative selector for denitrifiers, the lower pH found in the native soil (5.7 versus 6.59) (8) may have contributed to the lower nosZ diversity in this soil. In addition, based on long-term measurements (from 1991 to 1999) (34), N2O flux from the cultivated soil is consistently more than double that from the native soil (3.22 ± 0.45 and 1.13 ± 0.11 g of N2O-N ha−1 day−1, respectively). The N2O emissions from the native soil were also significantly lower than those from the cultivated soil after periodic rainfall events (N2O/N2 mol ratio, 0.36 versus 0.9) (3), indicating the higher persistence of Nos activity under dry (aerobic) conditions in the native soil in comparison to cultivated soil. Additionally, denitrifying community enzymes from the cultivated field were found to be more sensitive to oxygen inhibition (7). It appears that despite less-favorable acidic pH of native soil, the relative activity of NosZ in that soil was higher as judged from the N2O/N2 ratio, but overall emissions of N2O were more than two times lower than that of the cultivated field. One could infer that higher NO3− concentration in cultivated soil has lead to higher flux of N2O from that soil. Consistently, the persistence of NosZ in cultivated soil was shown to be lower than that in native soil, despite the more acidic pH in native soil (3).
Perhaps lower availability of nitrate in the native soil relative to the cultivated field (0.93 versus 4.57 μg/g of dry soil) (3, 34) has selected for a community of denitrifiers able to maintain Nos activity under more aerobic conditions. Such a community could have a competitive advantage in exploiting the flush of carbon that occurs on soil wet-up, since it could use N2O as well as NO3− as an electron acceptor if oxygen were limiting (3, 10). The abovementioned data, RFLP, and phylogenetic analysis point to a possible lack of selection for a more specialized denitrifying community able to successfully consume N2O in the cultivated field soil.
Buckley et al. (6) found that these native and cultivated soils supported similar numbers of microorganisms, as determined by microscopic counts and CFU counts on R2A agar medium. Despite the similarities in population sizes, the total RNA yields from native soil were considerably higher than the total RNA yields from cultivated soil, indicating differences in ribosomal activities of the indigenous community (6). Similarly, we consistently isolated more than 50% larger amounts of total microbial DNA from native soil relative to cultivated soil. However, no detectable differences in the diversity or abundance of Chrenarchaeota (6) and ammonium-oxidizing bacteria (30) in the same cultured and native soil were observed despite considerable differences in the disturbance and amendment history associated with these two fields. Phylogenetic analysis (Fig. 4) revealed a shift in the nosZ denitrifying community, complementary to a recent report of Avrahami et al. (2), who observed a community shift to nirK denitrifying strains corresponding to fertilizer application, whereas the ammonia oxidizer community structure remained unchanged. This could perhaps be explained by different survival strategies (r or K) of the two microbial groups, although there is some recent evidence for existence of r/K strategists among nitrifiers as well (42). Denitrifying r strategists, e.g., Pseudomonas spp., respond to substrate availability with a high growth rate and thus compete for resources. Changes in their community structure in response to environmental conditions are therefore expected, whereas slow growing K strategists such as ammonium oxidizers, by preserving their ribosomes, modulate activity rather than their community structure in response to change in nutrient supply (e.g., fertilizer). It could also be argued that cultivation of soil regularly opens new environmental niches, especially carbon resources. Availability of organic compounds enables denitrifiers to grow and compete as aerobic heterotrophs, hence supporting the higher diversity found in cultivated soil (52).
FIG.4.
Neighbor-joining tree of partial nosZ gene products (299 of 391 residues). The consensus tree was reconstructed based on neighbor-joining, parsimony, FITCH, and maximum-likelihood (MOLPHY) analyses. Unresolved nodes are displayed as multifurcations and are indicated by dashed lines. Clones obtained from this experiment are shown in boldface for native (n-T8) and cultivated (c-T1) field soil. Accession numbers are given in brackets for the cultivated species listed: Rhodopseudomonas palustris [NZ_AAAF01000014], Bradyrhizobium japonicum USDA110 [AJ002531], Sinorhizobium meliloti 1021 [NC_003037], Brucella melitensis bv. suis 1330 [AE014528], Rhodobacter sphaeroides [AF125260], Paracoccus denitrificans Pd1222 [AJ010260], Pseudomonas fluorescens C7R12 [AF056319], Pseudomonas sp. strain MT1 [AB054991], Pseudomonas stutzeri [M22628.1], Pseudomonas aeruginosa [X65277.1], P. aeruginosa PAO1 [NC_002516], Pseudomonas denitrificans [AF016059], Paracoccus pantotrophus [AF016058], Ralstonia solanacearum GMI1000 [AL646084], Ralstonia metallidurans [NZ_AAAI01000000], Ralstonia eutropha H16 ATCC17699 [X65278], and Achromobacter cycloclastes IAM1013 [Y15161].
Phylogenetic tree reconstruction of nosZ partial gene products correlates well with previously described existence of three major groups of nosZ sequences that correspond to α-, β-, and γ-Proteobacteria (29). As yet, no putative nitrous oxide reductase gene cluster has been identified in gram-positive bacteria or archaea, suggesting that another class or classes of nitrous oxide reductase sequences remain undiscovered (29). In addition, existing nosZ sequences (although some exhibited only a 230-bp overlap with the gene region used in this study) were used to construct an additional phylogenetic tree (data not shown). The branching pattern of that tree was similar and complementary to previously published results on nosZ gene diversity in forest soil (35), marine sediments (40), and pure cultures (29).
The consensus nosZ tree revealed several important patterns (Fig. 4). First, the vast majority of clones that formed the Michigan soil cluster did not branch with any known denitrifying bacteria, indicating that the two soils contain uncharacterized denitrifying organisms. Second, the majority of sequences from the native field formed a distinct cluster (native) containing only nine clones from the cultivated field indicating a close relationship among these sequences. Consequently, it is unlikely that further sequencing would unravel a highly similar community structure in both fields. This is also supported by observed higher DNA similarity among sequences from the native field and the RFLP analysis results. Clones from cultivated field were more distantly related to each other and 65% were found outside the native soil cluster. Third, the branching order of Pacific, Atlantic, and Michigan environmental clusters was influenced by the method of tree reconstruction, rendering the consensus tree less supportive to the proposed role of habitat-specific evolution in diversification of denitrifying communities as was previously suggested (5, 40). Fourth, tree organization indicated that the two denitrifying nosZ soil communities are related and with some overlap. Consistent with previous findings (2), the effects of agricultural treatments caused a shift in the denitrifying community rather than dramatic changes.
Overall, the native soil showed a less diverse denitrifying community than the cultivated soil. This finding is supported by the collector's curves, Chao1 estimator, and library coverage calculations, as well as diversity indices, sequence identities, and the phylogenetic tree. Differences in disturbance and amendment history of the two soils are the most probable causes for the observed shift in the microbial community, thus providing a possible basis for the previously reported functionally different denitrifier communities that inhabit the two soils (3, 7, 34). Further research should be directed toward obtaining quantitative information on abundance of the different nosZ genes in both soils by quantitative PCR or any other quantitative molecular detection method (e.g., in situ hybridization or quantitative dot blot hybridization).
Acknowledgments
We are indebted to Verónica Grüntzig and Héctor L. Ayala-del-Río for their generous help and advice throughout the experiments and their contribution to this project. We also thank Rok Pestotnik for sharing the RECODE program and for useful discussions.
This work was supported by Slovene Ministry of Science, School and Sport grant no. S18-490-002/19104/98 to B.S. and NSF grant no. DEB-0075564 and DOE grant no. DE-FG02-98ER62535 from the Biotechnology Investigations Ocean Margins Program.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Avrahami, S., R. Conrad, and G. Braker. 2002. Effects of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergsma, T. T., G. P. Robertson, and N. E. Ostrom. 2002. Influence of soil moisture and land use history on denitrification end products. J. Environ. Qual. 31:711-717. [DOI] [PubMed] [Google Scholar]
- 4.Bouwman, A. F. 1990. Soils and the greenhouse effect. Wiley, Chichester, United Kingdom.
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley, D. H., J. H. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Chrenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavigelli, M., and G. P. Robertson. 2000. The functional significance of denitrifier community composition in terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 8.Cavigelli, M., and G. P. Robertson. 2001. Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biol. Biochem. 33:297-310. [Google Scholar]
- 9.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 30:101-110. [PubMed] [Google Scholar]
- 10.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dendooven, L., and J. M. Anderson. 1994. Dynamics of reduction enzymes involved in the denitrification process in pasture soil. Soil Biol. Biochem. 26:1501-1506. [Google Scholar]
- 12.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 14.Good, I. J. 1958. The population frequency of species and the estimation of the population parameters. Biometrics 40:237-246. [Google Scholar]
- 15.Hall, B. G. 2001. Phylogenetic trees made easy. A how-to manual for molecular biologists. Sinauer Associates, Inc., Sunderland, Mass.
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Hoeren, F. U., B. C. Berks, S. J. Ferguson, and J. E. McCarthy. 1993. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans. New and conserved structural and regulatory motifs. Eur. J. Biochem. 218:49-57. [DOI] [PubMed] [Google Scholar]
- 18.Holtan-Hartvig, L., P. Dörsch, and L. R. Bakkern. 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3− and N2O reduction. Soil Biol. Biochem. 32:833-843. [Google Scholar]
- 19.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang, S., and K. Hanaki. 2000. Effects of oxygen concentration and moisture content of refuse on nitrification, denitrification and nitrous oxide production. Bioresour. Technol. 71:159-165. [Google Scholar]
- 21.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magurran, A. E. 1988. Ecological diversity and its measurement, p. 7-47. Princeton University Press, Princeton, N.J.
- 23.McGuirl, M. A., L. K. Nelson, J. A. Bollinger, Y.-K. Chan, and D. M. Dooley. 1998. The nos (nitrous oxide reductase) gene cluster from the soil bacterium Achromobacter cycloclastes: cloning, sequence analysis, and expression. J. Inorg. Biochem. 70:155-169. [DOI] [PubMed] [Google Scholar]
- 24.Morris, C. E., M. Bardin, O. Berge, P. Frey-Klett, N. Fromin, H. Girardin, M.-H. Guinebretière, P. Lebaron, J. M. Thiéry, and M. Troussellier. 2002. Microbial biodiversity: approaches to experimental design and hypothesis testing in primary scientific literature from 1975 to 1999. Microbiol. Mol. Biol. Rev. 66:592-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, C., M. Martin, R. J. Stevens, R. J. Laughlin, C. Kammann, J. C. G. Ottow, and H.-J. J.äger. 2002. Processes leading to N2O emissions in grassland soil during freezing and thawing. Soil Biol. Biochem. 34:1325-1331. [Google Scholar]
- 26.Nüsslein, K., and J. M. Tiedje. 1999. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65:3622-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parry, S., P. Renault, C. Chenu, and R. Lensi. 1999. Denitrification in pasture and cropped soil clods as affected by pore space structure. Soil Biol. Biochem. 31:493-501. [Google Scholar]
- 28.Pichinoty, F., M. Mandel, B. Greenway, and J.-L. Gracia. 1977. Étude de 14 bactéries dénitrifiantes appartenant au groupe Pseudomonas stutzeri isolées du sol par culture d'enrichissement en présence d'oxyde nitreux. Ann. Inst. Pasteur Microbiol. 115:419-430. [PubMed] [Google Scholar]
- 29.Philipott, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 30.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forrested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raaijmakers, J. G. W. 1987. Statistical analysis of the Michaelis-Menten equation. Biometrics 43:793-803. [PubMed] [Google Scholar]
- 33.Rao, C. R. 1980. Diversity and dissimilarity coefficients: a unified approach. Theor. Popul. Biol. 21:24-43. [Google Scholar]
- 34.Robertson, G. P., E. A. Paul, and R. R. Harwood. 2000. Greenhouse gases in intensive agriculture: contributions of individual gases to the radiative forcing of the atmosphere. Science 289:1922-1925. [DOI] [PubMed] [Google Scholar]
- 35.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Röver, M., O. Heinemeyer, and E. A. Kaiser. 1998. Microbial induced nitrous oxide emissions from arable soil during winter. Soil Biol. Biochem. 30:1859-1865. [Google Scholar]
- 37.Rudaz, A. O., E. Wälti, G. Kyburz, P. Lehmann, and J. Fuhrer. 1999. Temporal variation in N2O and to N2 fluxes from permanent pasture in Switzerland in relation to management, soil water content and soil temperature. Agric. Ecosyst. Environ. 73:83-91. [Google Scholar]
- 38.Sanders, H. L. 1968. Marine benthic diversity: a comparative study. Am. Nat. 102:243-282. [Google Scholar]
- 39.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 40.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schalk-Otte, S., R. J. Seviour, J. G. Kuenen, and M. S. M. Jetten. 2000. Nitrous oxide production by Alcaligenes faecalis during feast and famine regimens. Water Res. 34:2080-2088. [Google Scholar]
- 42.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwintner, C., M. Sabaty, B. Berna, S. Cahors, and P. Richaud. 1998. Plasmid content and localization of the genes encoding the denitrification enzymes in two strains of Rhodobacter sphaeroides. FEMS Microbiol. Lett. 165:313-321. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, New York, N.Y.
- 46.Trogler, W. C. 1999. Physical properties and mechanisms of formation of nitrous oxide. Coord. Chem. Rev. 187:303-327. [Google Scholar]
- 47.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for construction and drawing of evolutionary trees for Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 48.Watve, M. G., and R. M. Gangal. 1996. Problems in measuring bacterial diversity and a possible solution. Appl. Environ. Microbiol. 62:4299-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wieland, G., R. Neumann, and H. Backhaus. 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 67:5849-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf, I., and R. Russow. 2000. Different pathways of formation of N2O, to N2 and NO in black earth soil. Soil Biol. Biochem. 32:229-239. [Google Scholar]
- 51.Zhou, J., A. V. Palumbo, and J. M. Tiedje. 1997. Sensitive detection of a novel class of toluene-degrading denitrifiers, Azoarcus tolulyticus, with a small-subunit rRNA primers and probes. Appl. Environ. Microbiol. 63:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, J., B. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zumft, W. G. 1992. The denitrifying procaryotes, p. 443-582. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.). The procaryotes. Springer, Heidelberg, Germany.
- 54.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]