Abstract
Lactococcus lactis DPC4275 is a bacteriocin-producing transconjugant of the industrial starter strain DPC4268. Strain DPC4275 was generated through conjugal transfer by mating DPC4268 with L. lactis MG1363 containing the 60-kb plasmid pMRC01, which encodes the genetic determinants for the lantibiotic lacticin 3147 and for a phage resistance mechanism of the abortive infection type. The many significant applications of this strain prompted a genetic analysis of its apparently unstable bacteriocin-producing phenotype. Increased levels of lacticin 3147 produced by DPC4275 were associated with the appearance of an 80-kb plasmid, designated pMRC02, which was derived from DNA originating from pMRC01 (60 kb) and a resident DPC4268 proteinase plasmid, pMT60 (60 kb). Indeed, pMRC02 was shown to be derived from the insertion of a 17-kb fragment of pMRC01, encompassing the lacticin 3147 operon, into pMT60. The presence of pMRC02 at a high copy number was found to correlate with increased levels of lacticin 3147 in DPC4275 compared to the wild-type containing pMRC01. Subsequent transfer of pMRC02 into the plasmid-free strain MG1363 by electroporation allowed a direct phenotypic comparison with pMRC01, also studied in the MG1363 background. Plasmid pMRC02 displayed phage resistance similar to that by pMRC01, although it was less potent, as demonstrated by a larger plaque size for phage c2 infection of MG1363(pMRC02). While this locus is flanked by IS946 elements, the sequencing of pMT60-pMRC01 junction sites established that this event was unlikely to be insertion sequence mediated and most probably occurred by homologous recombination followed by deletion of most of pMRC01. This was not a random occurrence, as nine other transconjugants investigated were found to have the same junction sites. Such derivatives of commercial strains producing increased levels of bacteriocin could be exploited as protection cultures for food applications.
The constant drive to improve food safety, coupled with growing consumer demands for natural solutions, has made bacteriocins the focus of intensive research in recent times (3, 23, 33). Several of these food-grade compounds have the ability to inhibit important food pathogens and food spoilage microorganisms, such as Listeria, Clostridium, Staphylococcus, and Enterococcus species (3). The most widely utilized bacteriocin to date is the broad-spectrum bacteriocin nisin, produced by certain strains of Lactococcus lactis. However, it has been reported that nisin-producing strains do not perform well as starter cultures, as they generally produce acid at slower rates than commercial starter cultures, have reduced heat resistance and proteolytic activity, and are more sensitive to bacteriophage attack (18, 19). To overcome this problem, mixed starters containing nisin-producing and nisin-resistant strains have been utilized (30).
The lantibiotic lacticin 3147 (28) is inhibitory to a variety of undesirable gram-positive bacteria, including Clostridium, Bacillus, Enterococcus, Listeria, Staphylococcus, and Streptococcus species. The genetic determinants responsible for the production of and immunity to this bacteriocin are encoded on a 60-kb plasmid, pMRC01. This plasmid was previously sequenced, and the open reading frames (ORFs) were identified and screened against a nonredundant protein database (7). In addition to the lacticin 3147 genetic determinants, pMRC01 also encodes phage resistance properties. Phenotypic analysis of this phage resistance mechanism indicated that it was of the abortive infection type (4) and appeared to target the phage lytic cycle at a point following phage DNA replication. The self-transmissible nature of this plasmid has facilitated the construction of over 30 food-grade lacticin 3147-producing starter strains via the natural approach of conjugal transfer of pMRC01 (4, 34, 35). One such lacticin 3147-producing starter is L. lactis DPC4275, a derivative of L. lactis DPC4268 that is widely used for cheddar cheese manufacture in Ireland, where it is recognized for its reliable fast acid-producing ability in dairy environments. In contrast to the aforementioned nisin-producing starters, DPC4275 retained the fast acid-producing capability and the phage-resistant properties of DPC4268. Consequently, DPC4275 (see Fig. 1) has been used successfully as the principal acid producer in the manufacture of cheddar cheese (35), low-fat cheddar (10), cottage cheese (21), and salami (5). Additional applications of this strain include its ability to further control cheese quality by reducing the levels of indigenous nonstarter lactic acid bacteria, which can often proliferate to high levels during the ripening of cheddar cheese (35, 36). The strong inhibitory activity displayed by DPC4275 against pathogens (35) represents a significant advantage in using this bacteriocin-producing starter strain. Indeed, the transconjugant strain was also successfully used as a protection culture to reduce the incidence of Listeria monocytogenes on the surfaces of mold-ripened cheese (33).
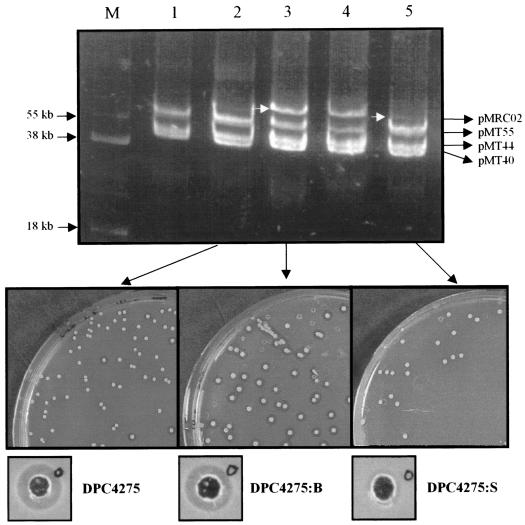
FIG. 1.
Plasmid profile analysis and bacteriocin production assays of L. lactis DPC4275 and its derivatives. Plasmid profiles are shown at the top as follows: M, marker (L. lactis UC317) (sizes indicated); lane 1, DPC4268 (parent); lane 2, DPC4275; lane 3, DPC4275:B (large-zone-producing strain); lane 4, DPC4275:B (large-zone-producing strain); lane 5, DPC4275:S (small-zone-producing strain). White arrows indicate the varying copy number of pMRC02 in DPC4275:B and DPC4275:S, respectively. Bacteriocin production assays are shown in the middle; colonies of the cultures indicated were overlaid with L. lactis DPC4268 as the sensitive indicator strain. Well diffusion assays at the bottom show the relative amounts of lacticin 3147 produced by DPC4275, DPC4275:B, and DPC4275:S, respectively, against the indicator L. lactis HP.
Given the extensive applications of DPC4275, the observation that the strain displayed variability with respect to the amount of bacteriocin produced warranted further investigation. This study describes the fluctuating levels of bacteriocin produced by DPC4275 and links the fluctuation to the appearance of an 80-kb cointegrate plasmid, pMRC02, formed by the incorporation of the lacticin 3147 genes into a resident plasmid. The phenotypic traits of this plasmid were examined in addition to the molecular event that led to its formation.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used for this study are listed in Table 1. Lactococcal strains were propagated at 30°C in M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% glucose (GM17) or lactose (LM17). Escherichia coli was grown at 37°C, with shaking, in Luria-Bertani medium (37). Stocks of all cultures were maintained at −20°C in 40% glycerol. Lactose indicator agar (24) and fast-slow differential agar (FSDA) (16) were also in this study. Solid media and soft agar overlays contained 1.5 and 0.7% agar, respectively. Where appropriate, ampicillin was added to media at 100 μg/ml for E. coli. LB medium was supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at concentrations of 1 mM and 40 μg/ml, respectively, as required. Selective plates containing lacticin 3147 were prepared as described by Ryan et al. (35). Briefly, an overnight culture of the wild-type lacticin 3147 producer, L. lactis DPC3147 (grown in GM17), was centrifuged at 10,000 × g for 10 min. The resulting supernatant (adjusted to pH 7.0 with concentrated NaOH) was filter sterilized by use of Millipore (Middlesex, England) HVLP filters (0.45-μm pore size) and was tempered to 45°C. Subsequently, the supernatant was added to an equal volume of double-strength lactose indicator agar (24) or GM17 agar at 45°C.
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Strain or phage | Description | Source or reference |
|---|---|---|
| Strains | ||
| L. lactis subsp. lactis strains | ||
| DPC4268 | Cheese starter | DPC culture collectiona |
| DPC5559 | DPC4268:pMRC01 transconjugant 1 | DPC culture collection |
| DPC5560 | DPC4268:pMRC01 transconjugant 2 | DPC culture collection |
| DPC5561 | DPC4268:pMRC01 transconjugant 3 | DPC culture collection |
| DPC4275 | DPC4268:pMRC01 transconjugant 4, starter strain | 4 |
| 35 | ||
| DPC5563 | DPC4268:pMRC01 transconjugant 5 | DPC culture collection |
| DPC5564 | DPC4268:pMRC01 transconjugant 6 | DPC culture collection |
| DPC5565 | DPC4268:pMRC01 transconjugant 7 | DPC culture collection |
| DPC5566 | DPC4268:pMRC01 transconjugant 8 | DPC culture collection |
| DPC4275:B | Large-zone Bac-producing derivative of DPC4275b | This study |
| DPC4275:S | Small-zone Bac-producing derivative of DPC4275 | This study |
| DPC4275:BB | Large-zone Bac-producing derivative of DPC4275:B | This study |
| DPC4275:BS | Small-zone Bac-producing derivative of DPC4275:B | This study |
| DPC4275:SB | Large-zone Bac-producing derivative of DPC4275:S | This study |
| DPC4275:SS | Small-zone Bac-producing derivative of DPC4275:S | This study |
| DPC4275:SNP | Non-Bac-producing derivative of DPC4275:S | This study |
| DPC3147 | pMRC01 parent strain | 27 |
| L. lactis subsp. cremoris strains | ||
| MG1363 | Plasmid-free derivative of NCDO712 | 11 |
| MG1363:pMRC01 | MG1363 transconjugant containing pMRC01 | 35 |
| MG1363:pMRC02 | MG1363 transformant containing pMRC02 | This study |
| HP | Lacticin 3147-sensitive indicator strain | DPC culture collection |
| E. coli strain | ||
| TOP10 | Cloning strain | Invitrogen |
| Bacteriophages | ||
| øc2 | Prolate-headed phage lytic for MG1363 | UCC phage collection |
| ø712 | Small isometric-headed phage lytic for MG1363 | UCC phage collection |
DPC, Dairy Products Research Centre, Teagasc, Moorepark, Fermoy, Co. Cork, Ireland.
Bac, bacteriocin.
Bacteriocin and bacteriophage assays.
Bacteriocin production was estimated by the deferred method as described by Tagg et al. (38). Relative levels of bacteriocin were assessed by the well diffusion assay (35). The bacteriophages used in this study are listed in Table 1. Lactococcal phages were propagated in their homologous hosts at 30°C in GM17 or LM17. Phage stocks were filter sterilized (0.45-μm-pore-size filters; Sartorius, Goettingen, Germany) and stored at 4°C. Plaque assays were performed according to the method described by Terzaghi and Sandine (39).
Strain construction and DNA isolation and manipulations.
Solid surface mating experiments were performed as described by McKay et al. (25) and Harrington and Hill (13). Lactococcal plasmid DNA was isolated by the method of Anderson and McKay (2). Plasmid profiles were analyzed by electrophoresis on vertical 0.7% agarose gels (Gibco BRL, Life Technologies, Paisley, Scotland) for 4 h at 100 V. Plasmid sizes were estimated by comparison to the plasmids of L. lactis UC317. E. coli plasmid DNA was isolated by use of the QIAprep miniprep kit (Qiagen GmbH, Hilden, Germany). Restriction endonucleases, RNase, T4 DNA ligase, and shrimp alkaline phosphatase were purchased from Roche (Essex, United Kingdom) and were used according to the manufacturer's instructions. Electrotransformation of L. lactis was performed by the method of Holo and Nes (14). Primers for PCR were purchased from MWG-Biotech AG, Ltd. (Ebersberg, Germany) and are listed in Table 2. PCR reagents from Promega (Madison, Wis.) or Qiagen were used according to the manufacturers' instructions. In addition, the Expand Long Template PCR system from Roche was employed for long template reactions, according to the manufacturer's instructions. Fragments containing junction regions were amplified by inverse PCR as follows. Outward-oriented primers were designed for the pMRC01 sequence flanking each of the junction sites of pMRC02. Total pMRC02 DNA was digested with HpaI or EcoRV and recircularized at concentrations suitable for the formation of monomeric circles (6, 26). This recircularized DNA was then used as template for PCRs with the Expand Long Template PCR system, which allows for high-fidelity amplification of long templates. The PCR products generated were purified from gels by use of QIAquick or QIAEX II gel extraction kits (Qiagen), cloned into the PCR cloning vector pTOPO 2.1 by use of the TOPO TA cloning kit (Invitrogen BV, Groningen, The Netherlands), and sequenced.
TABLE 2.
Sequences of oligonucleotide primers used in PCRs and expected product sizes
| Primera | Sequence | Size (bp) of amplicon | Source or reference |
|---|---|---|---|
| prtP (F) | 5′-CGGTTATTGACAAGTGGC-3′ | 1,200 | 17 |
| prtP (R) | 5′-CGAGGCCATTGACCGTACC-3′ | 41 | |
| PLL+1 (F) | 5′-ATGGCATGCAAACATCTTACCATAGACGAA-3′ | 22,910 | 7 |
| PLL-23 (R) | 5′-TTCAGGATCCACGCAAAGACTAACCAAATA-3′ | ||
| PLL+40 (F) | 5′-ATCGTGTCGACTGGAGTGAATTTATTGTTA-3′ | 23,108 | 7 |
| PLL-4 (R) | 5′-CATTATCAGCTGAAAATTCTTGTCGACTGA-3′ | ||
| p24850 (F) | 5′-AAGGGAATTGAAGGGGTTCTGTAC-3′ | 12,666 | 7 |
| p37500 (R) | 5′-TCTTAAGTATAGGGCAA-3′ | ||
| p28000 (F) | 5′-ACAATTGGGAAAATACCTTGAAGAT-3′ | 3,281 | 7 |
| p31261 (R) | 5′-CTTGACCCATCTCTACCTGGTTCCT-3′ | ||
| PLL+40 (F) | 5′-ATCGTGTCGACTGGAGTGAATTTATTGTTA-3′ | 5,444 | 7 |
| p45500 (R) | 5′-TCCGACTCCACATACAAT-3′ | ||
| p46000 (F) | 5′-ACTTTGCCTGCAGTCTGAGCG-3′ | 5,489 | 7 |
| p51500 (R) | 5′-GATAAATTGGGATATTTAAGG-3′ | ||
| p51400 (F) | 5′-TCTGATAGGATCGCCTTAAG-3′ | 3,146 | 7 |
| p54500 (R) | 5′-TGTTGGTTGAAAAATTGTTG-3′ | ||
| p54400 (F) | 5′-TAAATTATAGCGATTGGTTG-3′ | 3,054 | 7 |
| p57500 (R) | 5′-TCTATCAGTGGTTAGCAAAG-3′ | ||
| p51400 (F) | 5′-TCTGATAGGATCGCCTTAAG-3′ | 6,106 | 7 |
| p57500 (R) | 5′-TCTATCAGTGGTTAGCAAAG-3′ | ||
| p54400 (F) | 5′-TAAATTATAGCGATTGGTTG-3′ | 8,912 | 7 |
| PLL-4 (R) | 5′-CATTATCAGCTGAAAATTCTTGTCGACTGA-3′ | ||
| PLL+1 (F) | 5′-ATGGCATGCAAACATCTTACCATAGACGAA-3′ | 2,853 | 7 |
| PLL-4 (R) | 5′-CATTATCAGCTGAAAATTCTTGTCGACTGA-3′ | ||
| p18150 (F) | 5′-AAGTGAGTGAAGAAGTCC-3′ | 4,850 | 7 |
| PLL-23 (R) | 5′-TTCAGGATCCACGCAAAGACTAACCAAATA-3′ | ||
| p21590 (F) | 5′-CAGTTGATAGTTATTTAG-3′ | 1,420 | 7 |
| PLL-23 (R) | 5′-TTCAGGATCCACGCAAAGACTAACCAAATA-3′ | ||
| p23800 (F) | 5′-GTTGTGAGCTGGTTTTTC-3′ | 7,450 | 7 |
| p31261 (R) | 5′-CTTGACCCATCTCTACCTGGTTCCT-3′ | ||
| p24270 (F) | 5′-CGGTTCAATGCTCAGTTT-3′ | 7,011 | 7 |
| p31261 (R) | 5′-CTTGACCCATCTCTACCTGGTTCCT-3′ | ||
| p37510 (F) | 5′-TTGCCCTATACTTAAGA-3′ | 4,782 | 7 |
| p42270 (R) | 5′-CCATGAGACCCTCTTACG-3′ | ||
| p37510 (F) | 5′-TTGCCCTATACTTAAGA-3′ | 2,598 | 7 |
| p40000 (R) | 5′-TAACAATAAATTCACTCC-3′ | ||
| p37510 (F) | 5′-TTGCCCTATACTTAAGA-3′ | 3,679 | 7 |
| p41170 (R) | 5′-CTGCCCTCGGAGTTCTAA-3′ | ||
| MT60A J1 (F) | 5′-TTTCACATTCCTCATTATAGAGTT-3′ | 1,673 | This study |
| p25021 J1 (R) | 5′-ATTGGCATGTTGACATTT-3′ | This study | |
| J2 280 J2 (F) | 5′-AGGACATACATATAGACA-3′ | 855 | This study |
| J2 1120 J2 (R) | 5′-AAGTCCTTTTTACTTTAT-3′ | This study | |
| J2 280 (F) | 5′-AGGACATACATATAGACA-3′ | 2,021 | 7 |
| p42270 (R) | 5′-CCATGAGACCCTCTTACG-3′ | ||
| p40081 | 5′-ATCGTTCCGACTGGAGTGAATTTATT-3′ | Inverse PCR | 7 |
| p38890 | 5′-TGTAGATGAGATGGCAACAATAACTG-3′ | ||
| p24270 | 5′-ACATTAAAACTGAGCATTGAACCG-3′ | Inverse PCR | 7 |
| p24850 | 5′-AAGGGAATTGAAGGGGTTCTGTAC-3′ |
F, forward; R, reverse.
Nucleotide sequence analysis.
Cloned PCR products were sequenced (MWG-Biotech custom DNA sequencing service) with a LI-COR 4200L automated DNA sequencer and dye primer chemistry according to a cycle sequencing protocol. Sequence data were compiled and analyzed with the DNAStar software package (Madison, Wis.), and database searches were performed with BLAST (1), using the latest release of the nonredundant databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
RESULTS
Plasmid pMRC01 has been conjugally transferred to various lactococcal starters in an effort to enhance the phage resistance of these strains. In the case of the starter strain L. lactis DPC4268, this resulted in the isolation of eight transconjugants, appearing to produce varying levels of lacticin 3147. These were designated DPC4275, -5559, -5560, -5561, -5563, -5564, -5565, and -5566 (4).
Bacteriocin production characteristics of transconjugants.
For assessment of the ability of the transconjugants to produce bacteriocin, isolated colonies of the eight transconjugant strains listed above were overlaid with a lawn of each of three bacteriocin-sensitive lactococci, L. lactis HP, L. lactis MG1363, and the parent strain, L. lactis DPC4268. Colonies of all transconjugants, except for DPC5563, produced zones of inhibition of variable size against some or all of the three indicator strains. DPC5563 did not produce sufficient bacteriocin to inhibit the indicator organisms selected for this assay. One of the eight transconjugants, DPC4275, was found to produce both large and small zones of inhibition when overlaid with the parental strain, DPC4268 (Fig. 1, DPC4275:B and DPC4275:S). Notably, when colonies producing large zones were selected and restreaked, the resultant colonies gave rise to zones that were again variable in size. Well diffusion assays were used to compare the relative levels of bacteriocin produced by these colonies. It was observed that colonies giving rise to large zones produced approximately twofold more bacteriocin than those producing small zones (Fig. 1). Upon comparison with other isolates, DPC4275 colonies producing large zones (Fig. 1) consistently produced higher levels of bacteriocin than large zone-producing colonies from other transconjugants (data not shown). Therefore, DPC4275 was chosen for further analysis and has subsequently proved to be an important starter strain for a variety of food safety and quality applications.
Interestingly, the presence of an enlarged (80 kb) plasmid was observed by plasmid profile analysis of bacteriocin-producing colonies of DPC4275. From colonies producing small zones, this replicon, designated pMRC02, was generally detected in a low copy number (based on observed intensities on agarose gels), while in strains producing large zones, pMRC02 was typically observed at a high intensity. In addition, we observed that when pMRC02 was present, particularly at a high copy number, one of the four native plasmids of DPC4268 (60 kb) was undetectable. This implicates the involvement of this replicon, designated pMT60, in the events leading to the formation of pMRC02.
Transfer and characterization of bacteriophage resistance encoded by pMRC02.
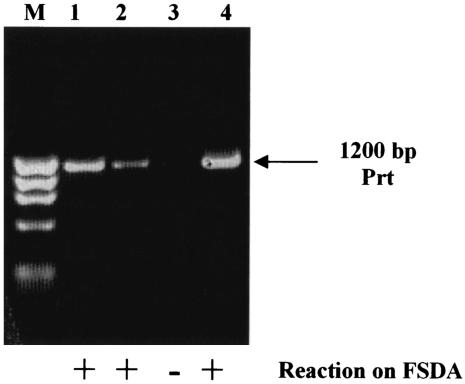
A more extensive study of pMRC02 required its transfer into a plasmid-free strain such as L. lactis MG1363. Attempts to conjugally transfer pMRC02 from DPC4275 into the plasmid-free strain were unsuccessful. This finding suggested that pMRC02 lacks the determinants for self-mobilization that are present on pMRC01. However, this plasmid was subsequently electroporated successfully into MG1363 by transforming the total plasmid DNA of DPC4275 and selecting on the basis of bacteriocin immunity. Previous studies in this laboratory confirmed that pMT60 encoded proteinase (Prt) activity (40). Plating of the resultant transformant, MG1363(pMRC02), on FSDA supplemented with glucose resulted in yellow colonies, indicating that pMRC02 was also Prt+. This result was confirmed by PCR with primers specific for the L. lactis proteinase genes (Table 2; Fig. 2).
FIG. 2.
Demonstration of proteinase-positive strains. Products were generated by PCR with primers specific for the L. lactis prtP gene. Lane 1, L. lactis DPC4268; lane 2, L. lactis DPC4275; lane 3, L. lactis MG1363(pMRC01); lane 4, L. lactis MG1363(pMRC02). M, Molecular weight marker IX (Roche Diagnostics). Reactions of strains on FSDA supplemented with glucose are indicated. +, proteinase-positive strains, observed by the presence of yellow colonies.
As mentioned previously, pMRC01 has been reported to encode abortive infection-type bacteriophage resistance in L. lactis MG1363 (4). The presence of pMRC02 in an L. lactis MG1363 background allowed direct comparison of any phage resistance properties mediated by this plasmid with those mediated by pMRC01, also in an MG1363 background. This was achieved by an assay using the small isometric-headed phage 712 and the prolate-headed phage c2 (Table 3). The resistance of MG1363(pMRC01) to phage c2 was demonstrated by the formation of pinpoint hazy plaques without a reduction in the efficiency of plaque formation, compared with the 3-mm-diameter clear plaques formed by the phage on the sensitive strain MG1363. Significantly, MG1363(pMRC02) presented a similar but less potent resistance, exhibiting 1-mm-diameter clear plaques without a reduction in the efficiency of plaque formation. However, both MG1363(pMRC01) and MG1363(pMRC02) appeared to be completely resistant to phage 712, which forms 2-mm-diameter clear plaques on MG1363. These findings suggest that the plasmid pMRC02 is derived from pMRC01.
TABLE 3.
Comparison of phage resistance of selected strains
| L. lactis strain | Characteristics for φc2 infection
|
Characteristics for φ712 infectiona
|
||||
|---|---|---|---|---|---|---|
| Titer (PFU/ml) | Plaque size (mm) | Plaque morphology | Titer (PFU/ml) | Plaque size (mm) | Plaque morphology | |
| MG1363 | 2.57 × 108 | 3 | Clear | 3.82 × 108 | 2 | Clear |
| MG1363(pMRC01) | 4.1 × 107 | Pinpoint | Hazy | |||
| MG1363(pMRC02) | 5.1 × 108 | 1 | Clear | |||
No plaques were visible for φ712 infection of MG1363(pMRC01) and MG1363(pMRC02).
Genetic analysis of the cointegrate plasmid pMRC02.
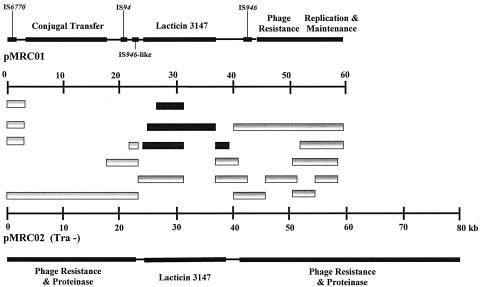
The molecular nature of the event which resulted in the formation of pMRC02 was further investigated by adopting a PCR approach whereby primers spanning the entire length of pMRC01 were used in reactions with pMRC01 and pMRC02 DNA as templates (primer sequences are available on request). When pMRC01 was used as template, PCR products of the expected sizes, based on nucleotide sequence data (7), were observed. However, when pMRC02 was used as template, products were only amplified when primers for a 20-kb region of the plasmid carrying the lacticin 3147 operons were used. Information generated from this approach determined that the conjugal transfer region, the putative phage resistance region, and the replication and maintenance regions of pMRC01, as described by Dougherty et al. (7), were absent from pMRC02 (Fig. 3). Interestingly, the functional IS946 elements flanking the lacticin 3147 module of pMRC01 did not appear to be involved in the cointegration event.
FIG. 3.
Comparison of the structures of pMRC01 (adapted from reference 7) and pMRC02. PCR analysis indicates the products generated from both plasmids (▪) and from pMRC01 alone (□).
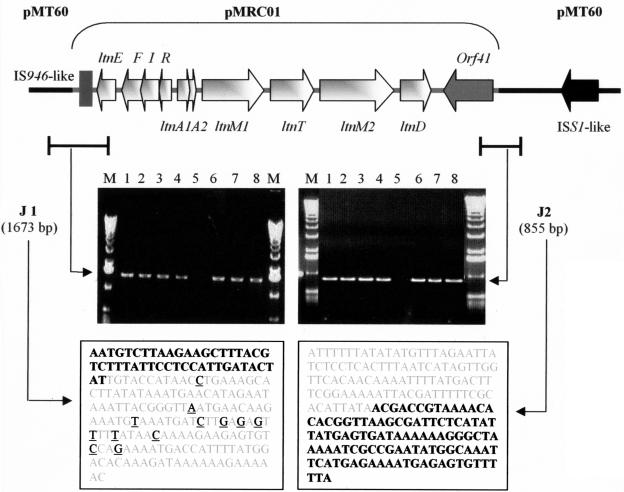
In order to elucidate the exact circumstances surrounding the insertion of the lacticin 3147 module into pMT60, it was necessary to sequence the junction sites of the integrated pMRC01 cassette. This was achieved by inverse PCR to isolate fragments containing the actual junctions of pMRC01 and pMT60 DNA. Amplicons were obtained for both junction site 1 (J1), at the 5′ end of the bacteriocin module of pMRC01, and junction site 2 (J2), at the 3′ end of the bacteriocin locus, which were then sequenced. The resulting sequences were compared to the pMRC01 sequence in addition to other existing sequences in the sequence databases (Fig. 4). J1 was contained in a 904-bp fragment, 343 bp of which was 99% identical to pMRC01 while the ensuing 116 bp displayed 89% identity to pMRC01. However, the remainder of this region showed no significant similarity to pMRC01 (with the exception of a 60-bp region within it, which was 98% identical to a region just upstream of ORF56), thus implying that this 445 bp was pMT60 DNA. J2 was isolated from a 2,279-bp fragment, the first 752 bp of which shared 100% identity with pMRC01 DNA. The subsequent 956 bp contained short regions of similarity to a number of lactococcal plasmids, including 86% similarity to a 90-bp region of pMRC01 and 85% similarity to a 153-bp segment of pSRQ900. Significantly, the remaining 571 bp exhibited very high similarity (98%) to the distal portion of the proposed 808-bp insertion sequence (IS) ISS1N from the L. lactis proteinase plasmid pSK111 (12). This IS element contains a 672-bp open reading frame, ORF-N1, flanked by 18-bp inverted repeats. Since J2 is contained on an EcoRV fragment, pMRC02 homology extends only from the EcoRV site within ORF-N1 to 135 bp beyond the 18-bp terminal repeat. This 18-bp inverted repeat on pMRC02 contains a single mismatch to the ISS1N repeat.
FIG. 4.
Schematic representation of the 17-kb fragment of pMRC01, encompassing the lacticin 3147 production and immunity gene clusters, inserted into pMT60, creating pMRC02. Arrows indicate ORFs and directions of transcription. Junction sites of pMRC02 are indicated (J1 and J2, respectively). Agarose gel electrophoresis shows PCR amplification of these junction sites by use of the primers listed in Table 2. Lanes 1, DPC5559; lanes 2, DPC5560; lanes 3, DPC5561; lanes 4; DPC4275; lanes 5, negative control; lanes 6, DPC5564; lanes 7, DPC5565; lanes 8, DPC5566. M, Molecular weight marker X (Roche Diagnostics). Text boxes contain the junction site sequences. pMT60 DNA is indicated in bold, while pMRC01 DNA is indicated in gray. Base pair differences between the junction sites and pMRC01 are underlined.
The sequence determination of J1 and J2 allowed primers to be designed for the previously unknown sequence of pMT60, with the goal of ascertaining if the same junction sites were present in all transconjugants generated. Besides the original eight transconjugants of DPC4268, three additional transconjugants were isolated from a separate conjugal mating. These strains were screened by PCR using the primers spanning J1 and J2 (Table 2; Fig. 4). All, with the exception of the nonproducing DPC5563, generated PCR products of 1,673 bp for J1 and 855 bp for J2 (Fig. 4). DPC4275-derived strains generated from the three-stage bacteriocin production assay were also screened by PCR with these primers in addition to primers specific for pMRC01. Again, all of these strains, with the exception of the nonproducing strain, generated products of the expected sizes, verifying both the existence of the same junction sites and the presence of the pMRC01 cassette in these strains. The nonproducing strain appeared to have lost both pMRC01 and pMRC02 and thus the ability to produce bacteriocin.
DISCUSSION
The transconjugant DPC4275 has previously been demonstrated to be quite a versatile starter strain (5, 10, 35) and protective culture (21, 34). For this report, the phenomenon of variable bacteriocin production by this transconjugant was studied in detail. Bacteriocin production was found to correspond to the appearance of an 80-kb plasmid, pMRC02, resulting from transfer of the lacticin 3147 locus from pMRC01 onto the resident proteinase plasmid, pMT60. This construct is bacteriocin producing and immune and proteinase positive and displays phage resistance similar to that of pMRC01.
Colonies producing large bacteriocin zones of inhibition appeared to contain a higher copy number of pMRC02 based on observation of an agarose gel. In addition, these colonies produced approximately twofold more bacteriocin than a small zone-producing colony, for which pMRC02 was visible at very low intensity or not visible at all in plasmid profiles. It is proposed that pMRC01, being inherently unstable in L. lactis DPC4268 due to plasmid incompatibility, possibly with pMT60, is maintained by forming the cointegrate plasmid, pMRC02. Mixed populations arise in culture, resulting in cells with various plasmid complements and copy numbers. As a result, individual colonies, when overlaid with another organism, may produce varying levels of lacticin 3147. However, the correlation between the presence of the cointegrate pMRC02 at a high copy number and a higher percentage of individual colonies producing large zones in a culture is consistent across the assays. Because of the more stable nature of this plasmid, it demonstrates a higher copy number and is therefore deemed responsible for the observed increase in bacteriocin production. In our opinion, it is the presence of this heterogeneous population of cells in a culture that gives rise to single colonies producing markedly different levels of lacticin 3147.
Sequencing of the junction sites of pMT60 and pMRC01 DNA confirmed that a region of approximately 17 kb of pMRC01 had been inserted into pMT60. This 17-kb fragment contains 12 ORFs, starting just upstream of ORF30 and continuing to position 40827, just upstream of ORF41 (opposite orientation). ORF31 through ORF40 form part of the lacticin 3147 locus (7, 22). Comparison of the predicted protein sequence of ORF41 with the databases shows the highest similarity to conserved hypothetical proteins from a number of organisms, including Sinorhizobium meliloti and Agrobacterium tumefaciens, in addition to weak homologies to a number of helicase proteins. Interestingly, this protein also contains conserved domains of proteins from the helicase family. Helicases unwind double-stranded DNA, RNA, or DNA-RNA hybrids into two single strands (9) and are involved in a variety of processes, including DNA replication, repair, and recombination. Therefore, ORF41 could play a part in the integration event itself, which may explain its retention on the pMRC01 fragment inserted into pMT60. ORF30, the only other ORF which is not involved in lacticin 3147 production or immunity, appears to be an IS946 element which has several mutations, including a frame shift, and also lacks the right inverted repeat (7). Thus, this IS element is thought to be nonfunctional. The phage resistance phenotype displayed by pMRC02 differs from that of pMRC01, in that φc2 consistently produces larger plaques on L. lactis MG1363(pMRC02) than on MG1363(pMRC01). Therefore, we suggest that the phage resistance encoded by pMRC02 is likely to result from an ORF residing on the native pMT60 plasmid.
The presence of a putative ISS1-like element was noted for pMT60 approximately 1,700 bp from the 3′ end of the inserted fragment. Only the distal portion of this IS element was present on the cloned inverse PCR product; however, the available nucleotide sequence was 98% homologous to ISS1N, which is located just downstream of prtM on the L. lactis proteinase plasmid pSK111 (12). Interestingly, ISS1 elements have been shown to be almost identical to IS946 (31), the IS elements which flank the bacteriocin module on pMRC01. Therefore, the presence of two identical functional and one nonfunctional IS element in the vicinity of the bacteriocin coding region of pMRC01 would initially imply that the insertion of the 17-kb pMRC01 fragment into pMT60 is an IS-mediated event. If this were indeed the case, it would be expected that a copy of an IS element would exist at either one or both of the junction sites (20, 32). However, examination of the junction site sequences clearly shows that there are no IS elements present at the exact joins between pMT60 and pMRC01 DNA. Furthermore, while the exact pMRC01-pMT60 junction is evident for J2, the integration point on the J1 side is more ambiguous. J1 is 99% homologous to pMRC01, exhibiting just a single base mismatch over 343 bp, up to position 23951, which is still over 100 bp upstream from the nonfunctional IS946 element. However, the next 112 bp show only 89% homology to pMRC01, as a result of several single base pair differences in the sequence, while there is no homology with J1 after position 23838 on pMRC01 (Fig. 3).
The sequence data generated imply that the event resulting in the insertion of the lacticin 3147 genes into pMT60 occurred by homologous recombination (29). Fusion of pMT60 and pMRC01 in their entirety would result in a 120-kb plasmid, which is likely to be highly unstable. Consequently, it is probable that all but those ORFs conferring the bacteriocin production and immunity phenotype were lost, resulting in an 80-kb plasmid and explaining the nonduplication of sequence at the junction sites. Also, the sections of pMT60 surrounding J1 and J2 exhibit small regions of similarity to parts of pMRC01, further substantiating this hypothesis. It is important, however, that it is not known whether the entire pMT60 sequence is present in pMRC02. The nature of this event appears to be very precise, since integration occurs at the same point for all bacteriocin-producing transconjugants examined, of which three were isolated from a separate mating experiment. It is interesting that although pMRC01 has been successfully transferred to 32 starter cultures to date (7; unpublished data), the formation of a cointegrate plasmid was observed only for DPC4268.
It is well known that bacteriocins from lactic acid bacteria can be encoded by mobile genetic elements such as transposons and plasmids. The genetic determinants governing the synthesis of the lantibiotic nisin are encoded on a large composite transposon which also encodes proteins for sucrose utilization (15, 27). Moreover, lacticin 481, another lactococcal lantibiotic, is flanked by IS elements (8), which can result in it “hopping” to other locations. Likewise, in this report we have demonstrated the integration of the lacticin 3147 locus into the resident proteinase plasmid of a commercial starter, generating a desirable bacteriocin-producing derivative. Moreover, the mechanism by which this acquisition occurred was probably not due to classical IS activity but more likely occurred through a single-crossover homologous recombination followed by a deletion of all but the bacteriocin-producing region of pMRC01.
Acknowledgments
This research was partly funded by grant aid under the Food Sub-Programme of the Operational Programme for Industrial Development (1997-2000), which was administered by the Irish Department of Agriculture, Food and Forestry and is supported by national and EU funds.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 4.Coakley, M., G. F. Fitzgerald, and R. P. Ross. 1997. Application and evaluation of the phage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl. Environ. Microbiol. 63:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffey, A., M. P. Ryan, R. P. Ross, E. Arendt, and G. Schwarz. 1998. Use of a broad host-range bacteriocin-producing Lactococcus lactis transconjugant as an alternative starter for salami manufacture. Int. J. Food Microbiol. 43:231-235. [DOI] [PubMed] [Google Scholar]
- 6.Collins, F. S., and S. M. Weissman. 1984. Directional cloning of DNA fragments at a large distance from an initial probe: a circularization method. Proc. Natl. Acad. Sci. USA 81:6812-6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 8.Dufour, A., A. Rince, P. Uguen, and J. P. Le Pennec. 2000. IS1675, a novel lactococcal insertion element, forms a transposon-like structure including the lacticin 481 lantibiotic operon. J. Bacteriol. 182:5600-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egelman, E. H. 1998. Bacterial helicases. J. Struct. Biol. 124:123-128. [DOI] [PubMed] [Google Scholar]
- 10.Fenelon, M. A., M. P. Ryan, M. C. Rea, T. P. Guinee, R. P. Ross, C. Hill, and D. Harrington. 1999. Elevated temperature ripening of reduced fat cheddar made with or without lacticin 3147 starter culture. J. Dairy Sci. 82:10-22. [Google Scholar]
- 11.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haandrikman, A. J., C. van Leeuwen, J. Kok, P. Vos, W. M. de Vos, and G. Venema. 1990. Insertion elements on lactococcal proteinase plasmids. Appl. Environ. Microbiol. 56:1890-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrington, A., and C. Hill. 1992. Plasmid involvement in the formation of a spontaneous bacteriophage insensitive mutant of Lactococcus lactis. FEMS Microbiol. Lett. 75:135-141. [DOI] [PubMed] [Google Scholar]
- 14.Holo, H., and I. F. Nes. 1989. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 15.Horn, N., S. Swindell, H. Dodd, and M. Gasson. 1991. Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol. Gen. Genet. 228:129-135. [DOI] [PubMed] [Google Scholar]
- 16.Huggins, A. R., and W. E. Sandine. 1984. Differentiation of fast and slow acid producing strains of lactic streptococci. J. Dairy Sci. 67:1674-1679. [Google Scholar]
- 17.Kok, J., K. J. Leenhouts, A. J. Haandrikman, A. M. Ledeboer, and G. Venema. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipinska, E. 1973. Use of nisin-producing lactic streptococci in cheese-making. Bull. Int. Dairy Fed. 73:1-24. [Google Scholar]
- 19.Lipinska, E. 1977. Nisin and its applications, p. 103-130. In M. Woodbine (ed.), Antimicrobials and antibiosis in agriculture. Butterworths, London, United Kingdom.
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McAuliffe, O., C. Hill, and R. P. Ross. 1999. Inhibition of Listeria monocytogenes in cottage cheese manufactured with a lacticin 3147-producing starter culture. J. Appl. Microbiol. 86:251-256. [DOI] [PubMed] [Google Scholar]
- 22.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Identification and overexpression of ltnI, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129-138. [DOI] [PubMed] [Google Scholar]
- 23.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 24.McKay, L. L., K. A. Baldwin, and E. A. Zottola. 1972. Loss of lactose metabolism in lactic streptococci. Appl. Microbiol. 23:1090-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKay, L. L., K. A. Baldwin, and P. M. Walsh. 1980. Conjugal transfer of genetic information in group N streptococci. Appl. Environ. Microbiol. 40:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauch, P. J., and W. M. de Vos. 1992. Transcriptional regulation of the Tn5276-located Lactococcus lactis sucrose operon and characterization of the sacA gene encoding sucrose-6-phosphate hydrolase. Gene 121:55-61. [DOI] [PubMed] [Google Scholar]
- 28.Rea, M. C., and T. M. Cogan. 1994. Buttermilk plants: the Irish version of kefir. Ir. Scientist 2:7. [Google Scholar]
- 29.Reimmann, C., and D. Haas. 1993. Mobilization of chromosomes and non-conjugative plasmids by cointegrative mechanisms, p. 137-173. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 30.Roberts, R. F., E. A. Zotolla, and L. L. McKay. 1992. Use of nisin-producing starter cultures suitable for cheddar cheese manufacture. J. Dairy Sci. 75:2353-2363. [Google Scholar]
- 31.Romero, D. A., and T. R. Klaenhammer. 1990. Characterization of insertion sequence IS946, an iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J. Bacteriol. 172:4151-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero, D. A., and T. R. Klaenhammer. 1993. Transposable elements in lactococci: a review. J. Dairy Sci. 76:1-19. [DOI] [PubMed] [Google Scholar]
- 33.Ross, R. P., M. Galvin, O. McAuliffe, S. M. Morgan, M. P. Ryan, D. P. Twomey, W. J. Meaney, and C. Hill. 1999. Developing applications for lactococcal bacteriocins. Antonie Leeuwenhoek 76:337-346. [PubMed] [Google Scholar]
- 34.Ross, R. P., C. Stanton, C. Hill, G. F. Fitzgerald, and A. Coffey. 2000. Novel cultures for cheese improvement. Trends Food Sci. Technol. 11:96-104. [Google Scholar]
- 35.Ryan, M. P., M. C. Rea, C. Hill, and R. P. Ross. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 62:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan, M. P., R. P. Ross, and C. Hill. 2001. Strategy for manipulation of cheese flora using combinations of lacticin 3147-producing and -resistant cultures. Appl. Environ. Microbiol. 67:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trotter, M., R. P. Ross, G. F. Fitzgerald, and A. Coffey. 2002. Lactococcus lactis DPC5598, a plasmid-free derivative of a commercial starter, provides a valuable alternative host for culture improvement studies. J. Appl. Microbiol. 93:134-143. [DOI] [PubMed] [Google Scholar]
- 41.Vos, P., G. Simons, R. J. Siezen, and W. M. de Vos. 1989. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J. Biol. Chem. 264:13579-13585. [PubMed] [Google Scholar]