Abstract
Genetic variants in the choline acetyltransferase (ChAT) gene have been suggested as risk factors for neurodegenerative Alzheimers Disease (AD). Here we tested the importance of genetic variants in the ChAT gene in normal cognitive function of elderly in a study sample of Danish twins and singletons (N=2070). The ChAT rs3810950 A allele, which has been associated with increased risk for AD, was found to be associated with a decrease cognitive status evaluated by a 5-component cognitive composite score (p=0.03 reg. coef. −0.30 CI 95% −0.57; −0.02), and the rs3810950 and rs8178990 ancestral GC haplotype was also associated with better cross sectional cognitive composite score (p=0.04 reg. coef. 0.59 CI 95% 0.03; 1.16). Growth curve model analyses applied to up to 10 years of follow-up data demonstrated that the rs3810950 A allele was associated with a lower cognitive composite score and MMSE at the lowest age (73 years of age), and was lower in the whole interval 73–82 although the absolute difference became smaller with age. Stratification by presence of the APOE ε4 allele revealed that rs3810950 AG/non-APOE ε4 carriers and rs3810950 AA/APOE ε4 carriers were associated with a lower cognitive composite score in younger elderly 73–83 years of age, similar to previous reports of association with AD.
Keywords: Dementia, Alzheimer, Aging, Genes, oldest old, MMSE, Cognitive function
Introduction
Sustaining cognitive abilities is a fundamental element for successful aging and is a major component of quality of life. Dementia is the most common neurodegenerative disorder affecting elderly. Most cases (60–70%) are diagnosed as Alzheimer disease (AD) whereas the rest are due mostly to vascular dementia (Bathum et al., 2006; Yip et al., 2002). Many candidate gene variants have been under investigation as risk factors for dementia, but well established contributors have been difficult to verify, e.g. because of low impact on the phenotype, the use of small sample sizes insufficient to detect association or sporadic chance findings. However, through meta-analyses statistically significant associations between genetic variants in a dozen genes and AD have been found (Bertram et al., 2007). As a prime example, the APOE allele ε4 has firmly been established as an important risk factor in AD susceptibility (Bertram et al., 2007) and recently large genome wide association studies (GWAS) of late onset AD further established associations with variants in the CLU, CR1 and PICALM genes (Lambert et al., 2009; Harold et al., 2009), which have been replicated in several independent studies.
Among the promising candidate genes involved in AD and cognitive functioning in the elderly is choline acetyltransferase gene (ChAT). Firstly, because ChAT encodes a key enzyme in the cholinergic system, secondly, because previous association studies indicate that genetic variants in ChAT are associated with the neurodegenerative disease AD. The central cholinergic system plays a pivotal role in cognitive functioning, such as learning, memory, and attention and is a target for acetylcholine inhibiters used for treatments of AD patients (Tang et al., 2008; Holmquist et al., 2007). The ChAT protein synthesises the neurotransmitter acetylcholine (ACh), and reduced levels of both ChAT and Ach are found in patients with AD (Holmquist et al., 2007). Genetic variants in the ChAT gene have been associated with AD in several studies and the A/G missense mutation, rs3810950, is particular interesting due to its functionality and because this variant was found to be associated with AD in Koreans, Americans and Europeans (Ahn et al., 2006; Ozturk et al., 2006; Mubumbila et al., 2002; Kim et al., 2004), although it failed to be replicated in Chinese (Tang et al., 2008). An on-line meta-analysis including nine studies show a marginal influence of the rs3810950 A allele on the risk of AD (OR= 1.17, 95% CI = 0.96; 1.44) (Bertram L et al., 2009), although it was not significant. ChAT gene polymorphisms have also been modestly associated with variation in cognitive function within the normal range (Ozturk et al., 2006), but this association has, this far, failed to be replicated by others (Piccardi et al., 2007; Scacchi et al., 2009; Tang et al., 2008).
The most consistent observation from the above mentioned studies was the impact of age on association. Thus, AD carriers of the rs3810950 A allele (AA and AG genotype) had statistically lower age at onset in Chinese (Tang et al., 2008), while AD patients carrying the rs3810950 AA genotype had statistically lower age at onset in Koreans (Kim et al., 2004). Also association between MMSE and ChAT gene variants became more significant when early onset cases were included (Ozturk et al., 2006).
In the present study we aim to scrutinize the relevance of variants in the ChAT gene further, by investigating the possible association between two functionally relevant variants in the ChAT gene and normal cognitive functioning measured both by the well established MMSE test and a 5-component cognitive composite score in two cohorts of elderly Danish individuals.
Materials and methods
Subjects
The subjects included in this study were drawn from participants from two population-based nationwide surveys conducted at the University of Southern Denmark: The Longitudinal Study of Ageing Danish Twins (LSADT) (Christensen et al., 2002; Skytthe et al., 2002) and The Danish 1905-birth-cohort Study (Nybo et al., 2003). LSADT is a longitudinal study of Danish twins aged 70 years and older. The study was initiated in 1995 and repeated every second year through 2005. A total of 689 participants provided a blood sample at the assessment in 1997. The Danish 1905-birth-cohort study is a prospective investigation of an entire birth cohort. The survey was initiated in 1998, when the participants were 92–93 years and followed by three follow-up studies of the participating survivors in 2000, 2003 and 2005. Of the 3,600 individuals still alive at intake, 2,262 participated, and 1,651 provided either a blood spot sample or a cheek swap at their first assessment in 1998. Each survey of the LSADT and 1905-birth-cohort studies comprises multidimensional face-to-face interviews focusing on health and lifestyle issues, as well as objective assessment of cognitive and physical abilities. Written informed consent was obtained from all participants and both studies were approved bythe Danish Scientific-Ethical Committees.
Cognitive functions
Cognitive functioning was assessed using the Mini Mental State Examination (MMSE) and a 5-component cognitive composite score (McGue & Christensen, 2001). The widely used MMSE ranges from 0 to 30 and can be graded as severely impaired for scores between 0 and 17, mildly impaired for scores between 18 and 23 and normal for scores between 24 and 30.
The 5-component cognitive composite measures were original selected to represent tasks that are sensitive to normative age changes but that can be reliably and briefly assessed by lay interviewers. The specific tasks included a fluency task, which involved the number of animals an individual could name in a 1-min interval, forward and backward digit span, and immediate and delayed recall of a 12-item list. The cognitive composite score was computed by taking the sum of the five standardized (using means and SDs from the initial LSADT assessment in 1998) measures and has been used in numerous publications for a decade (e.g. McGue & Christensen, 2002).
Genetic analysis
DNA was isolated from cheek swabs or blood spots, with the use of QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). DNA from whole blood was isolated using a salting out method. Genotyping of the two missense polymorphisms rs3810950 (exon 2 Thr/Ala) and rs8178990 (exon 5 Phe/Leu) were performed by allelic discrimination using pre-designed Taqman® SNP genotyping assays (Applied Biosystems). Reactions were conducted in a 10 μl volume using the conditions recommended by the manufacturer. PCR was performed in the Step One Plus™ Real-Time PCR system and genotypes called using the Step One™ Software version 2.1 (Applied biosystems). Genotyping of APOE ε4 alleles was performed as previously described (Bathum et al., 2006).
Statistical analysis
Statistical analysis was performed using STATA 10.1 (StataCorp, Texas, USA). Hardy-Weinberg equilibrium was calculated (df=1). Linear regression analysis was performed using initial intake measures and adjusted for age and sex in the combined LSADT and 1905-birth-cohort with 1 df on genotypes recoded 0, 1 and 2, where 0 are homozygotes for the major allele, 1 are heterozygotes, and 2 are homozygotes of the minor allele. Both the cognitive composite score and the MMSE were analysed as continuous variables. To account for the non-independence of twins from the LSADT study, twin pairs where both members participated were analysed in clusters of two, using the cluster option of the statistical software. Post-hoc analysis, were performed on two age defined cohort, i.e. the 1905-birth-cohort and a group consisting of 573 of 689 individuals from the LSADT cohort who were selected because they were 73–83 years of age and thus approximately a decade or more younger than participants from the older 1905-birth-cohort. Both cohorts were analysed by linear regression adjusted for age and sex. Corrections for multiple testing were not applied to these analyses of allele effects since an a priori hypothesis of association was assumed for each of the selected SNP alleles and for each stratification step. Tests of interaction between ChAT genotypes and sex or APOE ε4 alleles were performed using standard linear regression analysis. Haplotypes (1df) were deduced empirically using data from the unrelated individuals from the LSADT and 1905-birth-cohort in Haploview 4.1 (Barrett et al., 2005) and p-values, for Fishers exact test, of linkage analysis (LD) were calculated using Arlequin 3.1 (Excoffier et al., 2005). In order to analyse the longitudinal effects on cognitive composite score and MMSE and possibly maximize power, growth curve models were applied using all the data from intake and follow-up assessments in the combined LSADT and 1905-birth-cohort. A quadratic growth curve model fit the data significantly better than a linear growth curve model. Post-hoc analysis was applied using an additive (df=1) linear spline growth model with a knot at 82 years, to estimate the genetic effect on rate of change.
Results
Two ChAT gene variants were assayed in the present study and genotypes for the rs3810950 variant were successfully obtained for 661 participants from the LSADT cohort and for 1308 participants from the 1905-birth-cohort. Genotypes for the rs8178990 variant were successfully obtained for 672 participants from the LSADT cohort and for 1374 participants from the 1905-birth-cohort. Both rs3810950 and rs8178990 polymorphisms were in Hardy-Weinberg equilibrium in unrelated LSADT (p=0.12 and p=0.13, respectively) individuals and the 1905-birth-cohort (p=0.47 and p=0.79, respectively). Descriptives, genotype and allele frequencies in the LSADT cohorts and the 1905-birth-cohort are presented in Table 1.
Table 1.
Descriptives in the Longitudinal Study of Aging Twins (LSADT) and the 1905-birth-cohort
| LSADT cohort | 1905 cohort | |
|---|---|---|
| Age | 78.8 years (73–95) | 92–93 years |
| Number of individuals | 689 | 1381 |
| % Females | 66 | 69 |
| Obtained sec./tert. educational level (in %) | 289 (42) | 457 (33) |
| Married (in %) | 282 (41) | 148 (11) |
| Middle or higher social class (in %) | 209 (30) | 416 (30) |
| Mean composite cognitive score | 0.95 | 0.34 |
| SD composite cognitive score | 3.33 | 3.45 |
| Mean MMSE | 25.90 | 21.95 |
| SD Mean MMSE | 3.68 | 5.64 |
| Median MMSE | 27 | 23 |
| % Nonimpaired | 80.7 | 48.7 |
| % Mildly impaired | 15.3 | 32.0 |
| % Severely impaired | 4.0 | 19.4 |
| rs3810950 No. ind. AA/AG/GG | 420/203/38 | 798/455/55 |
| rs3810950 minor allele (A) frequency (%) | 21.10 | 21.60 |
| rs8178990 No. ind. CC/CT/TT | 569/103/0 | 1189/179/6 |
| rs8178990 minor allele (T) frequency (%) | 7.66 | 6.95 |
| APOE No. ind. --/ − ε4/ ε4 ε4 | 445/202/18 | 1267/334/20 |
| APOE ε4 non-carriers: | ||
| rs3810950 No. ind. AA/AG/GG | 265/150/25 | 623/350/43 |
| rs8178990 No. ind. CC/CT/TT | 372/66/0 | 923/133/5 |
| APOE ε4 carriers: | ||
| rs3810950 No. ind. AA/AG/GG | 152/52/13 | 175/105/12 |
| rs8178990 No. ind. CC/CT/TT | 179/35/0 | 266/46/1 |
Cross-sectional analysis of ChAT alleles and cognitive function
Using an additive model adjusted for age and sex, and the sample comprised of the combined group of LSADT and 1905-birth-cohort participants we found that the minor rs3810950 A allele was statistically significantly associated with a lower cognitive composite score (p=0.034 reg. coef. −0.30 CI 95% −0.57; −0.02), but not with MMSE (p=0.67) (Table 2). The association was stronger in men (p=0.039 reg. coef. −0.53) than in women (p=0.21 reg. coef. −0.20 CI 95% −0.53; 0.12), but there was no statistical evidence that sex has an effect on the association between the rs3810950 A allele and cognitive composite score. The second ChAT variant, rs8178990, was not associated with either the cognitive composite score (p=0.92) or the MMSE (p=0.88) (Table 2).
Table 2.
Regression analysis of the effect of ChAT variants on cognitive composite scores and MMSE in the comprised LSADT and 1905-birth-cohort (N=2070). In single variation analysis an additive genetic model was applied. Haplotypes are defined by rs3810950 A/G and rs8178990 T/G
| Cognitive composite score reg. coef. (95% CI), p-value | MMSE reg. coef. (95% CI), p-value | |
|---|---|---|
| rs3810950 | −0.30 (−0.57; −0.02), 0.034 | 0.09 (−0.31;0.49), 0.43 |
| rs8178990 | −0.02 (−0.48;0.43), 0.92 | 0.05 (−0.67;0.57), 0.88 |
| APOE ε4 | −0.38 (−0.71; −0.06), 0.02 | −0.65 (−1.12; −0,17), 0.007 |
| APOE ε 4 carriers: | ||
| rs3810950 | −0.50 (−0.97; −0.04), 0.033 | 0.28 (−0.46;1.02), 0.46 |
| rs8178990 | 0.65 (−0.22;1.53), 0.14 | 0.97 (0.00;1,95), 0.05 |
| APOE ε 4 non-carriers: | ||
| rs3810950 | −0.23 (−0.56;0.10), 0.17 | −0.01 (−0.47;0.46), 0.98 |
| rs8178990 | −0.26 (−0.79;0.26), 0.32 | −0.41(−1.18;0.35), 0.29 |
| Haplotypes: | ||
| GC | 0.59 (0.03;1.16), 0.039 | 0.31 (−0.47;1.09), 0.43 |
| AC | −0.30 (−0.62;0.3), 0.076 | 0.41 (−0.28;0.68), 0.41 |
| GT | −0.09 (−0.55;0.37), 0.70 | −0.10 (−0.75; 0.54), 0.76 |
Stratification by APOE ε4 has been a common approach for previous studies on ChAT gene variants (Kim et al., 2004; Ahn et al., 2006; Ozturk et al., 2006), thus this approach was applied to the analyses. For the rs3810950 A allele both APOE ε4 carriers (p=0.03 reg. coef. −0.50 CI 95% −0.97; −0.04) and APOE ε4 non-carriers (p=0.17 reg. coef. −0.23 CI 95% −0.56; −0.10) had similar tendencies of lower cognitive function. However, although the effect was stronger among APOE ε4 carriers there was no significant evidence of interaction. Analysis of rs8178990 revealed that among APOE ε4 carriers the rs8178990 T allele was borderline associated with a better MMSE (p=0.05 reg. coef. 0.97 CI 95% 0.00; −1.95) and cognitive composite score (p=0.14 reg. coef. 0.65 CI 95% −0.22; −1.53) (Table 2). The effect of the APOE ε4 presence on the association between rs8178990 and MMSE was statistically significant (p=0.03) and there was a tendency of effect for the APOE ε4 presence/absence on the association between rs8178990 and cognitive composite score (p=0.08).
ChAT haplotypes and cognitive function
The two variants in the ChAT gene, rs3810950 and rs8178990, were in linkage disequilibrium in both cohorts (p<0.01, D′=1.00, r2= 0.02), and were situated in trans position with the most likely haplotypes of rs3810950 and rs8178990 being GC (estimated frequency 0.715), AC (estimated frequency 0.215) and GT (estimated frequency 0.07). Linear regression analysis of haplotypes showed that the ancestral GC haplotype was significantly associated with better cognitive composite score compared to the other haplotypes AC and GT (p=0.04 reg. coef. 0.59 CI 95% 0.03; −1.16) (Table 2), whereas the AC haplotype, defined by the rs3810950 A allele, showed tendencies of association with lower cognitive composite scores (p=0.08 reg. coef. −0.30 CI 95% −0.62; 0.03) (Table 2).
Age matched replicated association between ChAT genotypes, and cognitive function
The association between the rs3810950 A allele and lower cognitive composite score was significant when the study population was confined to individuals in approximately the same age range as used in previous studies. That is, the effect was significant using the LSADT cohort members of 73–83 years of age, (p=0.04 reg. coef. −0.55 CI 95% −1.09; −0.02), but it did not reach significance in the 1905-birth-cohort of 92–93 years of age when investigated separately (p=0.47). Since the association was evident for the younger elderly we continued with analyses of this cohort using a genotype based model and APOE ε4 stratification. Here rs3810950 AG/non-APOE ε4 carriers performed significantly worse on the cognitive composite score (p=0.05 reg. coef. −0.83 CI 95% −1.67; −0.01) and the MMSE (p=0.001 reg. coef. −3.57 CI 95% −5.62; −1.51) than those in the rs3810950 GG/non-APOE ε4 carriers reference group (Table 3). Also rs3810950 AA/APOE ε4 carriers had significantly lower cognitive composite score (p=0.03 reg. coef. −2.19 CI 95% −4.14; −0.24) and tendencies of lower MMSE (p=0.18 reg. coef. −0.56 CI 95% −1.37; −0.26) than those in the rs3810950 GG/APOE ε4 carriers reference group (Table 3).
Table 3.
Regression analysis of the effect of the rs3810950 variants on cognitive composite scores and MMSE in the age selected LSADT (73–83) cohort and the 1905-birth-cohort.
| LSADT cohort 73–83 years of age (n=573) | 1905-birth-cohort 92–93 years of age (n=1381) | |||
|---|---|---|---|---|
| Cognitive composite score reg. coef. (95% CI), p-value | MMSE reg. coef. (95% CI), p-value | Cognitive composite score reg. coef. (95% CI), p-value | MMSE reg. coef. (95% CI), p-value | |
| Additive model: | ||||
| rs3810950 | −0.56 (−1.09; −0.02), 0.044 | −0.29 (−0.87;0.28), 0.31 | −0.12 (− −0.44;0.20), 0.46 | 0.26 (−0.27;0.80), 0.33 |
| Genotype based model*: | ||||
| rs3810950 AG | −0.59 (−1.25; −0.07), 0.080 | −0.17 (−0.84;0.49), 0.60 | 0.00 (−0.40;0.41), 1.00 | 0.54 (− −0.11;1.20), 0.11 |
| rs3810950 AA | −1.03 (−2.50;0.45), 0.17 | −0.83 (−2.49;0.83), 0.33 | −0.60 (−1.50;0.29), 0.19 | −0.25 (−1.75;1.25), 0.74 |
| APOE ε 4 carriers*: | ||||
| rs3810950 AG | −0.18 (−1.20;0.84), 0.73 | 0.34 (−0.74;1.43), 0.53 | −0.36 (−1.15;0.43), 0.37 | 1.16 (−0.18;2.50), 0.09 |
| rs3810950 AA | −2.19 (−4.14;0.24), 0.028 | −3.57 (−5.62; −1.51), 0.001 | −0.21 (−1.58;1.16), 0.76 | 2.06 (−0.16;4.29), 0.07 |
| APOE ε 4 non-carriers*: | ||||
| rs3810950 AG | −0.83 (−1.67; −0.00), 0.05 | −0.56 (−1.37;0.26), 0.18 | 0.11 (−0.37;0.58), 0.66 | 0.37 (−0.38;1.13), 0.33 |
| rs3810950 AA | −0.42 (−2.21;1.37), 0.64 | 0.50 (−0.91;1.90), 0.49 | −0.71 (−1.79;0.37), 0.20 | −0.91 (−2.71;0.88), 0.32 |
References are rs3810950 GG and rs8178990 CC, respectively
Longitudinal analysis of cognitive function
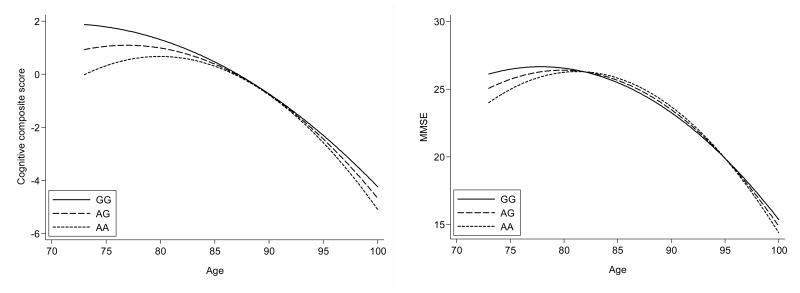
The participants in the comprised LSADT and 1905-birth-cohort were followed up to 10 years and were assessed up to 5 times with approximately two years intervals. When all assessments (Nobs=4812) were included in a quadratic growth curve model there was trend towards association between the rs3810950 A allele and lower cognitive composite score (p=0.056) and MMSE (p=0.046). As illustrated in figure 1 the model shows that the association was strongest up to approximately 82 years of age and after this age the association was less obvious. To simplify the estimates of the rate of change in cognitive function of the rs3810950 A allele an additive spline growth model was applied. These results showed that at 73 years of age the rs3810950 A allele was associated with lower cognitive composite score (p=0.006 coef −0.90 95%CI −1.55; −0.25) and up to the age of 82 the rs3810950 A allele was associated with lower decline of cognitive composite score (p=0.022 slope 0.09 CI 95% 0.013; 0.17). After 82 year of age there was no indication of association. Also at the age of 73 the rs3810950 A allele was associated with MMSE (p=0.021 coef −0.92 CI 95% −1.70; −0.14) and lower rate of decline up to the age of 82 (p=0.028; slope 0.12 CI 95% 0.013; 0.22). The quadratic growth curve model showed no association between the rs8178990 T allele and the decline of cognitive composite score (p=0.20) and MMSE (p=0.44).
Figure 1.
Quadratic growth curve model of average change of cognitive composite score and MMSE with age in rs3810950 genotype definded groups. Data are generated using the in the comprised LSADT and 1905-birth-cohort sample with multiple assessments (Nobs=4812).
Discussion
Sustaining cognitive abilities is a fundamental element in successful aging. Both environmental and genetic factors contribute to cognitive performance. In the present study, the two selected gene variants, rs3810950 and rs8178990, are located in the ChAT gene which is a candidate gene suspected to be associated with the neurodegenerative disorder Alzheimer’s. These genetic variants were thus investigated for association with normal cognitive function in two cohorts from population-based nationwide surveys. Here, we show that the rs3810950 A allele was associated with poorer performance on the cognitive composite score. This association was furthermore confirmed by the parallel association between a better cognitive composite score and the ancestral GC haplotype defined by the rs3810950 G allele and the rs8178990 C allele (Table 2). However, the haplotype analyses only estimated a minor additional effect to the rs3810950 A allele by the rs8178990 variant. In the cross sectional data both these minor alleles indicated association with higher MMSE. However, this finding was not significant and was most likely a result of the association with MMSE being less sensitive than that of the 5-component cognitive composite score. In the possibly more powerful longitudinal analyses, the rs3810950 A allele was associated with lower performance on both the cognitive composite score and MMSE. Genotype associations showed that rs3810950 A allele carriers performed worse on the cognitive composite score, equivalent to previously reported case control studies where the rs3810950 AG or AA genotypes were found to be more frequent in cases with AD (Mubumbila et al., 2002; Kim et al., 2004; Ahn et al., 2006; Ozturk et al., 2006; Mancama et al., 2007).
Statistical epistasis is often difficult to demonstrate in epidemiological studies, but to pursue this issue participants in the present study were grouped by the presence or absence of the APOE ε4 allele. Here, we observed in an age matched study that rs3810950 AG carriers performed statistically more poorly than the reference group (GG) when APOE ε4 was absent. In contrast the rs3810950 AA carriers perform statistically more poorly when APOE ε4 was present. The poorer cognitive performance of the rs3810950 AG/non APOE ε4 carriers was also found in case control studies with cases of AD (Ahn et al., 2006; Ozturk et al., 2006), and our finding that rs3810950 AA/APOE ε4 carriers tend to have poorer cognitive performance was likewise observed in a case control studies with cases of AD (Kim et al., 2004).
Our results suggest that the ChAT variant rs3810950 is not as strongly associated with cognitive function at age 92–93 as it is at age 73–83. An association between ChAT and the cognitive composite score was found in cross-sectional analysis of younger elderly (LSADT), but failed to be evident in a larger cohort of the oldest old (1905-birth-cohort). Longitudinal analysis further supported this pattern, as an association was observed only up to 82 years of age. Although it can not be ruled out that this finding of a statistically significant association in the younger elderly was due to a type I error, we suggest that the association between ChAT and cognitive function is age dependent. Replication in the age selected cohort of oldest old may thus not be the most appropriate cohort for a gene variant associated with lower cognitive performance or AD. However, recently we found that variants in the CLU and PICALM, that are suggested to be protective of AD, were also associated with better performance on the cognitive composite score scale in the oldest old (Mengel-From et al., 2011). It has been shown that the amount of ChAT positive neurons decrease with age (Bernard et al., 2009), and that the ChAT protein concentration is lower among patients diagnosed with AD compared to healthy elderly people (Holmquist et al., 2007). Therefore, it seems reasonable to assume that when the total level of ChAT protein declines with age, the contribution of ChAT variant Thr/Ala (rs3810950) to cognitive function in the oldest old weakens while the contribution is more apparent in the younger elderly. Our suggestion of an age dependent association should be taken into consideration in future association studies of the rs3810950 A allele and adds support to the suggestion that this allele may be considered as a risk factor of early onset of AD (Kim et al., 2004; Ozturk et al., 2006; Schwarz et al., 2003; Tang et al., 2008) rather than being a lifelong risk factor of AD. The age dependent association could explain why a study of Chinese participants who were on average 80.3 years old (+/− 8.9 years) (Tang et al., 2008), which is higher than the oldest participant in most of the other studies (Kim et al., 2004; Ozturk et al., 2006; Schwarz et al., 2003) failed to find an association between rs3810950 and AD. Interestingly, our longitudinal analyses showed that the rs3810950 A allele in addition to being associated with lower cognitive functioning was also associated with lower rate of decline up to 82 year of age. This observation is not easily interpreted but is likely a result of one or more phenomena e.g. selection by mortality, sample selection bias and a result of rs3810950 A allele carriers entering the study having a high level of compensatory factors. Or it could be that the rs3810950 A allele affects age of onset but not ultimately level of cognitive decline, so early onset decline more slowly. Further research is needed to fully understand the observation of lower rate of decline.
Both the rs3810950 and the rs8178990 genetic variants in ChAT are missense mutations in a multi-transcript gene (Harold et al., 2003). To the best of our knowledge, it is not known whether these variants alter the catalytic activity for synthesis of ACh, but other rare ChAT variants have been found in families with congenital myasthenic syndrom and have been shown to exhibit impaired or lack of catalytic activity (Ohno et al., 2001). Similar functional studies of the ChAT variants caused by the rs3810950 or rs8178990 variants may clarify the functionality of the rs3810950 and rs8178990 variant in Alzheimer’s and successful aging.
Acknowledgments
The study was supported by U.S. National Institute on Aging Researchgrant NIA-PO1-AG08761. The Danish Aging Research Center is supported by a grant from the VELUX Foundation and from The National Program for Research Infrastructure 2007 from the Danish Agency for Science Technology and Innovation. Brødrene Hartmanns Fond and Hørslev Fonden.
Footnotes
Financial Disclosure
The authors declare no conflicts of interest.
Contributor Information
Jonas Mengel-From, Email: jmengel-from@health.sdu.dk, Danish Aging Research Center, Epidemiology Unit, Institute of Public Health, University of Southern Denmark, J.B. Winsløws Vej 9, DK-5000 Odense, Denmark phone: +45 6550 4082, Fax: +45 6550 3682. Department of Clinical Genetics, Odense University Hospital, Sdr. Boulevard 29, DK-5000 Odense, Denmark.
Kaare Christensen, Email: kchristensen@health.sdu.dk, The Danish Aging Research Center and The Danish Twin Registry, Epidemiology Unit, Institute of Public Health, University of Southern Denmark, J.B. Winsløws Vej 9, DK-5000 Odense, Denmark. phone: +45 6550 3049, Fax: +45 6550 3682. Department of Clinical Genetics, Odense University Hospital, Sdr. Boulevard 29, DK-5000, Odense, Denmark. Department of Clinical Biochemistry and Pharmacology, Odense University Hospital, Sdr. Boulevard 29, DK-5000, Odense, Denmark.
Mikael Thinggard, Email: mthingaard@health.sdu.dk, The Danish Aging Research Center, Epidemiology Unit, Institute of Public Health, University of Southern Denmark, J.B. Winsløws Vej 9B, DK-5000 Odense C, Denmark. Phone: +45 6550 3854, Fax: +45 6550 3682
Matt McGue, Email: mcgue001@umn.edu, Department of Psychology, University of Minnesota. University, N218 Elliott Hall, 75 East River Road, Minneapolis, Minnesota 55455 USA. phone: 612-625-2818, Fax: 612-626-2079. The Danish Aging Research Center, Epidemiology Unit, Institute of Public Health, University of Southern Denmark, J.B. Winsløws Vej 9, DK-5000 Odense, Denmark.
Lene Christiansen, Email: LChristiansen@health.sdu.dk, The Danish Aging Research Center, Epidemiology Unit, Institute of Public Health, University of Southern Denmark, J.B. Winsløws Vej 9, DK-5000 Odense, Denmark. phone: +45 6550 2843, Fax: +45 6550 3682.
Reference List
- Ahn JS, Ahn K, Kim JH, Kang BH, Kim E, Jo I, Kim DK. ApoE-epsilon 4-dependent association of the choline acetyltransferase gene polymorphisms (2384G>A and 1882G>A) with Alzheimer’s disease. Clin Chim Acta. 2006;368:179–182. doi: 10.1016/j.cca.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bathum L, Christiansen L, Jeune B, Vaupel J, McGue M, Christensen K. Apolipoprotein e genotypes: relationship to cognitive functioning, cognitive decline, and survival in nonagenarians. J Am Geriatr Soc. 2006;54:654–658. doi: 10.1111/j.1532-5415.2005.53554.x. [DOI] [PubMed] [Google Scholar]
- Bernard CE, Gibbons SJ, Gomez-Pinilla PJ, Lurken MS, Schmalz PF, Roeder JL, Linden D, Cima RR, Dozois EJ, Larson DW, Camilleri M, Zinsmeister AR, Pozo MJ, Hicks GA, Farrugia G. Effect of age on the enteric nervous system of the human colon. Neurogastroenterol Motil. 2009;21:746–e46. doi: 10.1111/j.1365-2982.2008.01245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen M, Mullin K, Blacker D, Tanzi R. The AlzGene Database. Alzheimer Research Forum; 2009. [Accessed [4 april 2011]]. Available at: http://www.alzgene.org. [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Christensen K, Gaist D, Vaupel JW, McGue M. Genetic contribution to rate of change in functional abilities among Danish twins aged 75 years or more. Am J Epidemiol. 2002;155:132–139. doi: 10.1093/aje/155.2.132. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den BH, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van BC, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Peirce T, Moskvina V, Myers A, Jones S, Hollingworth P, Moore P, Lovestone S, Powell J, Foy C, Archer N, Walter S, Edmonson A, McIlroy S, Craig D, Passmore PA, Goate A, Hardy J, O’Donovan M, Williams J, Liddell M, Owen MJ, Jones L. Sequence variation in the CHAT locus shows no association with late-onset Alzheimer’s disease. Hum Genet. 2003;113:258–267. doi: 10.1007/s00439-003-0960-2. [DOI] [PubMed] [Google Scholar]
- Holmquist L, Stuchbury G, Berbaum K, Muscat S, Young S, Hager K, Engel J, Munch G. Lipoic acid as a novel treatment for Alzheimer’s disease and related dementias. Pharmacol Ther. 2007;113:154–164. doi: 10.1016/j.pharmthera.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kim KW, Suh YJ, Park WY, Jhoo JH, Lee DY, Youn JC, Lee KH, Seo JS, Woo JI. Choline acetyltransferase G +4 A polymorphism confers a risk for Alzheimer’s disease in concert with Apolipoprotein E epsilon4. Neurosci Lett. 2004;366:182–186. doi: 10.1016/j.neulet.2004.05.041. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De DP, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van BC, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Mancama D, Mata I, Kerwin RW, Arranz MJ. Choline acetyltransferase variants and their influence in schizophrenia and olanzapine response. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:849–853. doi: 10.1002/ajmg.b.30468. [DOI] [PubMed] [Google Scholar]
- McGue M, Christensen K. The heritability of cognitive functioning in very old adults: evidence from Danish twins aged 75 years and older. Psychol Aging. 2001;16:272–280. doi: 10.1037//0882-7974.16.2.272. [DOI] [PubMed] [Google Scholar]
- McGue M, Christensen K. The heritability of level and rate-of-change in cognitive functioning in Danish twins aged 70 years and older. Exp Aging Res. 2002;28:435–451. doi: 10.1080/03610730290080416. [DOI] [PubMed] [Google Scholar]
- Mengel-From J, Christensen K, McGue M, Christiansen L. Genetic variations in the CLU and PICALM genes are associated with cognitive function in the oldest old. Neurobiol Aging. 2011;32:554. doi: 10.1016/j.neurobiolaging.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubumbila V, Sutter A, Ptok U, Heun R, Quirin-Stricker C. Identification of a single nucleotide polymorphism in the choline acetyltransferase gene associated with Alzheimer’s disease. Neurosci Lett. 2002;333:9–12. doi: 10.1016/s0304-3940(02)00955-2. [DOI] [PubMed] [Google Scholar]
- Nybo H, Petersen HC, Gaist D, Jeune B, Andersen K, McGue M, Vaupel JW, Christensen K. Predictors of mortality in 2,249 nonagenarians--the Danish 1905-Cohort Survey. J Am Geriatr Soc. 2003;51:1365–1373. doi: 10.1046/j.1532-5415.2003.51453.x. [DOI] [PubMed] [Google Scholar]
- Ohno K, Tsujino A, Brengman JM, Harper CM, Bajzer Z, Udd B, Beyring R, Robb S, Kirkham FJ, Engel AG. Choline acetyltransferase mutations cause myasthenic syndrome associated with episodic apnea in humans. Proc Natl Acad Sci U S A. 2001;98:2017–2022. doi: 10.1073/pnas.98.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk A, DeKosky ST, Kamboh MI. Genetic variation in the choline acetyltransferase (CHAT) gene may be associated with the risk of Alzheimer’s disease. Neurobiol Aging. 2006;27:1440–1444. doi: 10.1016/j.neurobiolaging.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccardi M, Congiu D, Squassina A, Manconi F, Putzu PF, Mereu RM, Chillotti C, Del ZM. Alzheimer’s disease: case-control association study of polymorphisms in ACHE, CHAT, and BCHE genes in a Sardinian sample. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:895–899. doi: 10.1002/ajmg.b.30548. [DOI] [PubMed] [Google Scholar]
- Scacchi R, Gambina G, Moretto G, Corbo RM. Variability of AChE, BChE, and ChAT genes in the late-onset form of Alzheimer’s disease and relationships with response to treatment with Donepezil and Rivastigmine. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:502–507. doi: 10.1002/ajmg.b.30846. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Eisele T, Diehl J, Muller U, Forstl H, Kurz A, Riemenschneider M. Lack of association between a single nucleotide polymorphism within the choline acetyltransferase gene and patients with Alzheimer’s disease. Neurosci Lett. 2003;343:167–170. doi: 10.1016/s0304-3940(03)00380-x. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Kyvik K, Holm NV, Vaupel JW, Christensen K. The Danish Twin Registry: 127 birth cohorts of twins. Twin Res. 2002;5:352–357. doi: 10.1375/136905202320906084. [DOI] [PubMed] [Google Scholar]
- Tang M, Rao D, Ma C, Guo Y, Han H, Ling K, Ling Y. Evaluation of choline acetyltransferase gene polymorphism (2384 G/A) in Alzheimer’s disease and mild cognitive impairment. Dement Geriatr Cogn Disord. 2008;26:9–14. doi: 10.1159/000140612. [DOI] [PubMed] [Google Scholar]
- Yip AG, Brayne C, Easton D, Rubinsztein DC. Apolipoprotein E4 is only a weak predictor of dementia and cognitive decline in the general population. J Med Genet. 2002;39:639–643. doi: 10.1136/jmg.39.9.639. [DOI] [PMC free article] [PubMed] [Google Scholar]