Abstract
A highly sensitive quantitative PCR detection method has been developed and applied to the distribution analysis of human intestinal bifidobacteria by combining real-time PCR with Bifidobacterium genus- and species-specific primers. Real-time PCR detection of serially diluted DNA extracted from cultured bifidobacteria was linear for cell counts ranging from 106 to 10 cells per PCR assay. It was also found that the method was applicable to the detection of Bifidobacterium in feces when it was present at concentrations of >106 cells per g of feces. Concerning the distribution of Bifidobacterium species in intestinal flora, the Bifidobacterium adolescentis group, the Bifidobacterium catenulatum group, and Bifidobacterium longum were found to be the three predominant species by examination of DNA extracted from the feces of 46 healthy adults. We also examined changes in the population and composition of Bifidobacterium species in human intestinal flora of six healthy adults over an 8-month period. The results showed that the composition of bifidobacterial flora was basically stable throughout the test period.
The microflora of the gut has been investigated in great detail by use of anaerobic culture techniques, and intensive investigations have given significant information on the flora (5, 23). However, it is now recognized that not all organisms in the human intestinal flora have been cultivated (9). Moreover, classification and identification based on phenotypic traits do not always provide clear-cut results and are sometimes unreliable.
In recent years, analysis methods using 16S rRNA have been widely used in place of conventional culture methods for the structural analysis of intestinal flora (1, 25, 35). In complex mixed populations, 16S rRNA-targeted oligonucleotide probes have been applied to fluorescence in situ hybridization (FISH) as a culture-independent method (6, 10, 14). Techniques such as the clone library method and temperature gradient gel electrophoresis allow for the analysis of predominant bacteria that are difficult to culture but nevertheless represent a significant population (25a, 28, 32, 37). Although these 16S rRNA-gene-targeted methods have been applied successfully, PCR analysis using specific primers should make the analysis method capable of achieving the most sensitive results as well as providing ease and speed of use. So far, specific oligonucleotide primers have been designed for many bacterial species known to be present in the intestinal tract (12, 17, 18, 30, 34). Although conventional PCR does not permit quantitative detection of the target bacteria, real-time PCR with species-specific primers can provide a precise quantification method through the measurement of the amount of PCR products in each cycle as fluorescence (intensity of SYBR Green I) (11, 16, 26, 33).
Members of the genus Bifidobacterium are some of the most common organisms in the human intestinal tract (5, 23, 24, 29). It has been suggested that the Bifidobacterium species are important for maintaining general health, and many attempts have been made to increase populations of Bifidobacterium in the intestinal tract by supplying certain bifidobacterial strains and ingredients as food additives to stimulate growth of bifidobacteria (7, 8, 13, 15). Hence, the distribution of bifidobacteria in human intestinal microflora is highly interesting. To date, we have prepared 16S ribosomal DNA (rDNA)-targeted genus- and species-specific primers for all known species of bifidobacteria that inhabit the human intestinal tract (18, 19).
For this study, we developed a quantitative PCR method to detect bifidobacterial species in human intestinal tracts by combining real-time PCR with Bifidobacterium genus- and species-specific primers. The current PCR technique for fecal DNA was also used to investigate the distribution of bifidobacteria in the intestinal microflora of 46 healthy human adults. The changes in bifidobacterial flora in fecal samples collected from six healthy adults during an 8-month period were also examined.
MATERIALS AND METHODS
Reference strains and culture conditions.
The type strains of bifidobacteria were obtained from the American Type Culture Collection (Rockville, Md.) and the Japan Collection of Microorganisms (Wako, Japan). The strains were cultured anaerobically in GAM broth (Nissui Seiyaku, Tokyo, Japan) supplemented with 1% lactose at 37°C for 12 h. The bacterial count was determined microscopically with DAPI (4′,6-diamidino-2-phenylindole) staining, according to a previously described method (17). Serial 10-fold dilutions of the culture were also plated on GAM agar (Nissui Seiyaku). The plates were subsequently incubated at 37°C for 5 days in an anaerobic chamber (Takayama-kagaku, Tokyo, Japan), and cultural counts (CFU) were determined in triplicate.
Collection and preparation of fecal samples.
Forty-six healthy volunteers from our institute staff (41 males and 5 females; ages, 25 to 59 years [average, 37 ± 9]) provided fresh fecal samples. Forty volunteers provided samples only once (subjects Y-1 to Y-40), while six volunteers provided samples once a month for an 8-month period (subjects A to F). While no subject had received antibiotics, probiotics, or prebiotics during the 2 weeks prior to the sampling, volunteer A had received antibiotics up until 2 weeks prior to the first sampling. The samples were collected in sterile bags, refrigerated, and immediately taken to the laboratory.
Enumeration of bifidobacteria in fecal samples by culture method.
A dilution series (10−1 to 10−8) was made in an anaerobic chamber, and 0.1-ml aliquots of the 105 to 108 dilutions were plated on Bifidobacterium-specific TOS agar (31). The ingredients of this medium (per liter) were as follows: Trypticase (BBL, Cockeysville, Md.), 10 g; yeast extract (Difco, Detroit, Mich.), 1 g; KH2PO4, 3 g; K2HPO4, 4.8 g; (NH4)2SO4, 3 g; MgSO4, 0.2 g; l-cysteine, 0.5 g; transgalactosylated oligosaccharides (Yakult Honsha Co., Tokyo, Japan), 10 g; and powdered agar, 15 g (Difco). These plates were subsequently incubated anaerobically at 37°C for 5 days, and all the colonies appearing on the 1st and 2nd highest dilution plates were picked in succession into GAM broth (Nissui Seiyaku). Template DNA was prepared from these cultures, and each isolate was identified by use of Bifidobacterium genus- and species-specific PCR primers (19). The CFU value of each species was calculated from the number of colonies and the result of the identification.
FISH analysis for bifidobacterial species.
The FISH analyses with genus- and species-specific 16S rRNA-targeted oligonucleotide probes were performed according to the method described by Langendijk et al., with some modification (14). Briefly, portions of each fecal sample were fixed with 3% paraformaldehyde at 4°C overnight. Then, 10 μl of the fixed-cell suspension of the appropriate dilution was applied to a glass slide and the cell smears were dehydrated in 96% ethanol for 10 min. After hybridization with specific probes at 45°C overnight, the slides were washed, dried, and mounted. The fluorescence measurements were performed with the Leica Q550FW system (Leica, Wetzlar, Germany) and the image analysis software Image-Pro Plus, v. 4 (Media-Cybernetics, Silver Spring, Md.). The following probes were used to enumerate bifidobacterial species: Bp153, 5′-GAG GAC CTT TGC CCA CCA-3′ (genus Bifidobacterium); PAD, 5′-GCG AAA ACT GAC CCT CG-3′ (Bifidobacterium adolescentis) (36); pBiCATg-4, 5′-ACA CCC CAT GCG AGG AGT-3′ (the B. catenulatum group); pBiLON, 5′-AGC CGT ATC TCT ACG ACC GT-3′ (B. longum); and pBiBIF, 5′-CCA CAA TCA CAT GCG ATC ATG-3′ (B. bifidum). These probes have been validated for each bifidobacterial species (T. Takada and K. Matsumoto, unpublished data).
DNA extraction from fecal samples.
Fecal samples (20 mg) were washed three times by suspension in 1.0 ml of phosphate-buffered saline and centrifugation of each preparation at 14,000 × g in order to remove PCR inhibitors. The fecal pellets were resuspended in 450 μl of extraction buffer (100 mM Tris-HCl, 40 mM EDTA, pH 9.0) and 50 μl of 10% sodium dodecyl sulfate. Three hundred milligrams of glass beads (diameter, 0.1 mm) and 500 μl of buffer-saturated phenol were added to the suspension, and the mixture was vortexed vigorously for 30 s using a FastPrep FP 120 instrument (BIO 101, Vista, Calif.) at a power level of 5.0. After centrifugation at 14,000 × g for 5 min, 400 μl of the supernatant was collected. Subsequently, phenol-chloroform extractions were performed and 250 μl of the supernatant was subjected to isopropanol precipitation. Finally, the DNA was suspended in 1 ml of TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
Specific primers for the B. adolescentis group.
The 16S rDNA sequences for 12 strains of B. adolescentis (positions 32 to 650 in the Escherichia coli numbering system) were basically determined by the method described previously (22), using an ABI PRISM 3100 DNA sequencer (Applied Biosystems, Foster City, Calif.). Five strains (MC-36, -37, -38, -39, and -41) were isolated previously (19) and seven strains (A-001, B-002, C-001, D-027a, D-009, E-040, and E-067) were isolated in this study. As shown in Fig. 1, these strains were divided into two genotypes, with six strains of genotype A (ATCC 15703T, MC-39, A-001, B-002, C-001, and D-027a) and seven strains of genotype B (MC-36, MC-37, MC-38, MC-41, D-009, E-040, and E-067). It was found that the strains of genotype B have point mutations in the primer target region of our previously developed primer (18). Although the strains of genotype B gave positive PCR results with the previously developed primer set BiADO-1 and BiADO-2, the populations of these strains were underestimated by real-time PCR. Therefore, we developed a new forward primer, BiADOg-1b, for detecting B. adolescentis genotype B, and changed the name of the genotype A-specific primer from BiADO-1 to BiADOg-1a (Table 1). For detection of the B. adolescentis group, consisting of genotypes A and B, the primers BiADOg-1a, BiADOg-1b, and BiADO-2 were used as a primer set in the same PCR mixture. The specificity of the primer set was confirmed by use of DNA extracted from 58 different strains, consisting of 12 strains of the B. adolescentis group, 31 strains of Bifidobacterium species, and 15 strains of non-Bifidobacterium species listed in a previous report (18).
FIG. 1.
Sequence alignment of 12 strains of B. adolescentis with the database sequence, showing sequence differences. Satokari et al. reported that B. adolescentis E-981074T (ATCC 15703T) harbors five copies of the rrn operon, and the sequences AF275881 (nru-1) and AF275882 (nru-5) differed at several base pairs (28). The annealing sites for the B. adolescentis-specific forward primer are boxed. Nucleotides differing from the type strain of B. adolescentis are shaded. These strains were divided into two genotypes, with six strains of genotype A (ATCC 15703T, MC-39, A-001, B-002, C-001, and D-027a) and seven strains of genotype B (MC-36, MC-37, MC-38, MC-41, D-009, E-040, and E-067).
TABLE 1.
PCR primers for detection of human intestinal bifidobacteria
| Target | Primer | Sequence | Product size (bp) |
|---|---|---|---|
| Bifidobacterium | g-Bifid-F | CTCCTGGAAACGGGTGG | 549-563 |
| g-Bifid-R | GGTGTTCTTCCCGATATCTACA | ||
| B. adolescentis groupa | BiADOg-1a | CTCCAGTTGGATGCATGTC | 279 |
| BiADOg-1b | TCCAGTTGACCGCATGGT | ||
| BiADO-2 | CGAAGGCTTGCTCCCAGT | ||
| B. angulatum | BiANG-1 | CAGTCCATCGCATGGTGGT | 275 |
| BiANG-2 | GAAGGCTTGCTCCCCAAC | ||
| B. bifidum | BiBIF-1 | CCACATGATCGCATGTGATTG | 278 |
| BiBIF-2 | CCGAAGGCTTGCTCCCAAA | ||
| B. breve | BiBRE-1 | CCGGATGCTCCATCACAC | 288 |
| BiBRE-2 | ACAAAGTGCCTTGCTCCCT | ||
| B. catenulatum group | BiCATg-1 | CGGATGCTCCGACTCCT | 285 |
| BiCATg-2 | CGAAGGCTTGCTCCCGAT | ||
| B. longumb | BiLON-1 | TTCCAGTTGATCGCATGGTC | 831 |
| BiLON-2 | GGGAAGCCGTATCTCTACGA | ||
| B. infantisb | BiINF-1 | TTCCAGTTGATCGCATGGTC | 828 |
| BiINF-2 | GGAAACCCCATCTCTGGGAT | ||
| B. dentium | BiDEN-1 | ATCCCGGGGGTTCGCCT | 387 |
| BiDEN-2 | GAAGGGCTTGCTCCCGA |
The B. adolescentis group consisted of B. adolescentis genotypes A and B. BiADOg-1a, BiADOg-1b, and BiADO-2 are used in the same PCR mixture. BiADOg-1a is the primer used to detect B. adolescentis genotype A and is synonymous with BiADO-1 (18). BiADOg-1b is the primer used to detect B. adolescentis genotype B.
B. longum, B. longum biotype longum; B. infantis, B. longum biotype infantis (27).
Real-time PCR.
Real-time PCR amplification and detection were performed in an ABI PRISM 7900HT Sequence Detection system (Applied Biosystems). The reaction mixture (10 μl) was composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates, a 1:100,000 dilution of SYBR Green I (Molecular Probes, Eugene, Oreg.), 11 ng of TaqStart antibody (ClonTech, Palo Alto, Calif.) per μl, 0.05 U of Taq DNA polymerase (Takara, Tokyo, Japan) per μl, 0.25 μM (each) specific primers, and 1 μl of ×1, ×10, and ×100 diluted template DNA. The amplification program consisted of one cycle of 94°C for 5 min, then 40 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 50 s, and finally one cycle of 94°C for 15 s. The fluorescent product was detected at the last step of each cycle. Following amplification, melting temperature analysis of PCR products was performed to determine the specificity of the PCR. The melting curves were obtained by slow heating at 0.2°C/s increments from 60 to 99°C, with continuous fluorescence collection. For determination of the number of bifidobacterial species present in each sample, fluorescent signals detected from two or three serial dilutions in the linear range of the assay were averaged and compared to a standard curve generated with standard DNA in the same experiment. For quantification of the genus Bifidobacterium, B. longum ATCC 15707T was used as the standard strain. The following bifidobacteria were also used as real-time PCR controls for species-specific quantification: B. adolescentis ATCC 15703T, B. angulatum ATCC 27535T, B. bifidum ATCC 29521T, B. breve ATCC 15700T, B. pseudocatenulatum JCM 1200T, B. longum ATCC 15707T, B. infantis ATCC 15697T, and B. dentium ATCC 27534T.
RESULTS
Real-time PCR detection.
DNA extracted from a known amount of B. longum ATCC 15707T was added in serial dilutions from 106 to 0 cells to a series of PCRs with Bifidobacterium genus-specific primers, and fluorescence was monitored throughout the reaction. As a result, the number of starting cells and the cycle number at which the product fluorescence becomes greater than a defined threshold were found to be linear over the range of DNA concentrations from 106 to 10 cells per PCR (r2 = 0.99). This indicates that the linear range for the procedures used in this study is over 106 cells per g of feces. Virtually identical results were obtained for eight Bifidobacterium species with their species-specific primers (data not shown).
Quantitative PCR detection of Bifidobacterium in culture medium.
The number of bifidobacteria in 1 ml of GAM broth was quantified by the real-time PCR method for nine species, using genus-specific primers for Bifidobacterium. The results were compared with those obtained by the direct counting method with DAPI staining and by the CFU counting method with culturing (Table 2). Similar bacterial counts were obtained for B. adolescentis, B. angulatum, B. breve, B. catenulatum, B. longum, B. infantis, and B. dentium by the quantitative PCR method and the DAPI staining method, with a difference of <0.2 log10. The number of B. bifidum and B. pseudocatenulatum determined by the PCR method was less than that determined by the DAPI staining method but similar to that determined by the culture method. The populations of B. infantis and B. dentium were enumerated to be lower by the colony counting method than by the other two methods.
TABLE 2.
Comparison of bifidobacterial populations in GAM broth, as determined by genus-specific quantitative PCR (qPCR), the direct counting method with DAPI staining (DAPI), and the culture method (CFU)
| Strain | Log10 bifidobacteria/ml, as determined by indicated method
|
||
|---|---|---|---|
| qPCR | DAPI | CFU | |
| B. adolescentis ATCC 15703T | 9.7 ± 0.1 | 9.9 ± 0.1 | 9.7 ± 0.1 |
| B. angulatum ATCC 27535T | 9.8 ± 0.1 | 9.6 ± 0.1 | 9.2 ± 0.1 |
| B. bifidum ATCC 29521T | 7.8 ± 0.2 | 8.6 ± 0.1 | 8.0 ± 0.1 |
| B. breve ATCC 15700T | 10.0 ± 0.2 | 9.8 ± 0.2 | 9.6 ± 0.1 |
| B. catenulatum ATCC 27539T | 9.8 ± 0.1 | 9.9 ± 0.2 | 9.5 ± 0.1 |
| B. pseudocatenulatum JCM 1200T | 9.5 ± 0.2 | 9.9 ± 0.1 | 9.6 ± 0.2 |
| B. longum ATCC 15707T | 9.6 ± 0.1 | 9.5 ± 0.1 | 9.5 ± 0.1 |
| B. infantis ATCC 15697T | 9.7 ± 0.1 | 9.7 ± 0.1 | 9.1 ± 0.1 |
| B. dentium ATCC 27534T | 9.8 ± 0.2 | 9.8 ± 0.1 | 9.1 ± 0.1 |
Quantitative PCR detection of Bifidobacterium species in fecal samples.
Real-time PCR analyses were performed to quantify individual Bifidobacterium species in six fecal samples obtained from healthy subjects, and the results were compared with those obtained by the FISH and culture methods (Table 3). Most of the bifidobacterial species detected by the FISH and culture methods were also detected at the same levels by quantitative PCR, except for CFU counts for samples C and D, which gave lower estimates. It is noteworthy that quantitative PCR allowed for detection of nonpredominant bifidobacterial species that could not be identified by the FISH or culture method.
TABLE 3.
Comparison of quantitative PCR, the FISH method with specific probes, and the culture method for detection and quantification of bifidobacteria in fecal samples from six subjects
| Population | Log10 bifidobacteria/g of feces (wet weight)a for indicated volunteerb
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A
|
B
|
C
|
D
|
E
|
F
|
|||||||||||||
| qPCRc | FISHd | CFU | qPCR | FISH | CFU | qPCR | FISH | CFU | qPCR | FISH | CFU | qPCR | FISH | CFU | qPCR | FISH | CFU | |
| Bifidobacterium | 10.0 | 9.7 | 9.9 | 10.3 | 10.1 | 10.4 | 10.6 | 10.3 | 9.5 | 10.4 | 10.0 | 9.6 | 9.3 | 8.5 | 9.2 | 6.9 | − | 7.8 |
| B. adolescentis group | 10.0 | 9.5 | 9.8 | 10.1 | 9.9 | 10.1 | 10.6 | 10.1 | 9.5 | 10.3 | 9.9 | 9.5 | 8.8 | 8.0 | 9.0 | − | − | − |
| B. bifidum | − | − | − | − | − | − | 9.1 | 8.9 | − | − | − | − | − | − | − | − | − | − |
| B. catenulatum group | 9.0 | 9.1 | 9.3 | 8.7 | 8.5 | 9.2 | 9.6 | 9.1 | − | 9.3 | 9.1 | 8.3 | 8.8 | 8.0 | 8.7 | − | − | − |
| B. longum | 7.4 | − | − | 9.4 | 9.5 | 10.0 | 9.4 | 9.5 | − | 8.1 | 9.4 | 8.0 | 8.4 | 8.3 | 8.0 | 7.0 | − | 7.8 |
| B. breve | 7.9 | NT | 8.6 | − | NT | − | 8.4 | NT | − | − | NT | − | − | NT | − | − | NT | − |
| B. angulatum | − | NT | − | − | NT | − | − | NT | − | 6.9 | NT | − | 6.5 | NT | − | − | NT | − |
−, not detected; NT, not tested.
Fecal samples collected at the fifth month were used for comparison.
PCR, quantitative PCR. The detection limit of the quantitative PCR was 106 cells per g of feces.
The detection limit of the FISH was 108 cells per g of feces.
Distribution of Bifidobacterium species in the intestinal flora.
Table 4 shows the distribution of bifidobacterial species in the intestinal tracts of 46 healthy adult volunteers. Bifidobacterium species were detected in all samples, and the population of bifidobacteria per gram of feces (average ± standard deviation [SD]) was 9.4 ± 0.7 log10. The average number of species detected per individual in this study was 3.4 ± 1.1. The B. catenulatum group was commonly detected (detected in 89% of samples) and was present in a relatively larger population (8.9 ± 0.8 log10). The B. adolescentis group was found to be somewhat less common (83%) than B. longum and the B. catenulatum group, but was present in the greatest numbers (9.1 ± 0.9 log10). In contrast, B. longum was the species detected most commonly (96%), but exhibited populations (8.1 ± 0.7 log10) that were generally smaller than those of the B. adolescentis group and the B. catenulatum group (P < 0.01; unpaired t test). The B. adolescentis group was the most numerous species in 25 subjects, while the B. catenulatum group was predominant in 19 of the 46 subjects. Although B. longum was detected in almost all samples, it was the predominant species in only two subjects.
TABLE 4.
Distribution of Bifidobacterium species in the intestinal flora of human adults
| Subjectb | Log10 bifidobacteria/g of feces, measured by reaction with genus- or species-specific primera:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| g-Bifid | BiADOg | BiANG | BiBIF | BiBRE | BiCATg | BiLON | BiINF | BiDEN | |
| A | 10.0 | 10.0 | — | — | 7.9 | 9.0 | 7.4 | — | — |
| B | 10.3 | 10.1 | — | — | — | 8.7 | 9.4 | — | — |
| C | 10.6 | 10.6 | — | 9.1 | 8.4 | 9.6 | 9.4 | — | — |
| D | 10.4 | 10.3 | 6.9 | — | — | 9.3 | 8.1 | — | — |
| E | 9.3 | 8.8 | 6.5 | — | — | 8.8 | 8.4 | — | — |
| F | 6.9 | — | — | — | — | — | 7.0 | — | — |
| Y-1 | 9.6 | — | — | — | — | 9.6 | 8.7 | — | — |
| Y-2 | 10.1 | 9.9 | — | 8.6 | 6.7 | 9.5 | 8.7 | — | — |
| Y-3 | 9.2 | 8.7 | 6.7 | 7.7 | — | 8.7 | 7.3 | — | — |
| Y-4 | 8.2 | 7.6 | — | — | — | 7.8 | 7.0 | — | — |
| Y-5 | 8.4 | 8.2 | — | — | — | 7.8 | 7.2 | — | — |
| Y-6 | 9.5 | 9.3 | — | — | — | 9.0 | 8.7 | — | 7.4 |
| Y-7 | 9.1 | 7.6 | — | — | — | 9.0 | 8.4 | — | — |
| Y-8 | 9.6 | 8.8 | — | — | 6.7 | 8.9 | 8.3 | — | — |
| Y-9 | 9.4 | 9.2 | — | 7.8 | 7.5 | — | 8.1 | — | — |
| Y-10 | 9.6 | 9.4 | — | 7.5 | — | 9.0 | 8.2 | — | 7.5 |
| Y-11 | 10.2 | 9.9 | — | 9.4 | — | 10.0 | 8.4 | — | — |
| Y-12 | 9.4 | 8.4 | — | — | — | 9.2 | 7.9 | — | — |
| Y-13 | 10.0 | 9.8 | — | — | — | 9.6 | 8.3 | — | — |
| Y-14 | 10.1 | 10.1 | — | 9.0 | — | 8.8 | 8.9 | — | — |
| Y-15 | 8.9 | 8.0 | — | — | — | 8.6 | 6.4 | — | — |
| Y-16 | 8.4 | 7.7 | — | — | — | — | 8.1 | — | — |
| Y-17 | 9.0 | 8.7 | — | — | — | 7.4 | 7.6 | — | — |
| Y-18 | 9.7 | 9.7 | — | — | — | 8.2 | 8.3 | — | — |
| Y-19 | 9.6 | 9.2 | — | 8.9 | 6.4 | 8.9 | 8.7 | — | 6.5 |
| Y-20 | 9.3 | 7.6 | — | 8.1 | — | 9.3 | 7.8 | — | — |
| Y-21 | 9.3 | 9.0 | — | 7.2 | — | 8.7 | 8.4 | — | 7.5 |
| Y-22 | 10.1 | 9.7 | — | — | — | 9.5 | 9.0 | — | — |
| Y-23 | 7.5 | — | — | — | — | 7.4 | — | — | — |
| Y-24 | 9.6 | 9.4 | — | 6.8 | — | 9.0 | 8.5 | — | — |
| Y-25 | 10.1 | — | — | 8.9 | — | 9.9 | 9.3 | — | — |
| Y-26 | 9.0 | 9.1 | — | — | — | 7.5 | 7.7 | — | — |
| Y-27 | 9.1 | — | — | — | 7.2 | 9.2 | 8.1 | — | — |
| Y-28 | 8.9 | 8.8 | 6.6 | — | — | 6.3 | 7.8 | — | — |
| Y-29 | 9.7 | 9.5 | — | — | — | 9.5 | 8.6 | — | — |
| Y-30 | 9.4 | 7.6 | — | — | — | 9.5 | 7.7 | — | — |
| Y-31 | 10.0 | — | — | — | — | 10.1 | — | — | — |
| Y-32 | 9.2 | 9.0 | — | — | — | — | 6.4 | — | — |
| Y-33 | 9.4 | 9.3 | — | — | — | 8.1 | 7.3 | — | — |
| Y-34 | 9.5 | — | — | — | 7.6 | 9.4 | 8.8 | 6.4 | — |
| Y-35 | 10.1 | 9.9 | — | 8.7 | — | 8.9 | 8.4 | 7.3 | — |
| Y-36 | 9.8 | 9.5 | 6.3 | — | — | — | 8.2 | — | — |
| Y-37 | 10.2 | — | — | — | — | 10.2 | 8.5 | — | — |
| Y-38 | 8.6 | 7.4 | — | — | — | 8.5 | 7.9 | — | — |
| Y-39 | 9.6 | 9.6 | — | — | — | 8.4 | 7.9 | — | — |
| Y-40 | 9.6 | 9.0 | — | — | — | 9.0 | 8.5 | — | — |
| No. positive (%) | 46 (100) | 38 (83) | 5 (11) | 13 (28) | 8 (17) | 41 (89) | 44 (96) | 2 (4.3) | 4 (8.7) |
| Mean ± SD | 9.4 ± 0.7 | 9.1 ± 0.9 | 6.6 ± 0.2 | 8.3 ± 0.8 | 7.3 ± 0.7 | 8.9 ± 0.8 | 8.1 ± 0.7 | 6.9 ± 0.7 | 7.2 ± 0.5 |
Primers g-Bifid, BiADOg, BiANG, BiBIF, BiBRE, BiCATg, BiLON, BiINF, and BiDEN are specific for genus Bifidobacterium, B. adolescentis group, B. angulatum, B. bifidum, B. breve, the B. catenulatum group, B. longum, B. infantis, and B. dentium, respectively. —, not detected.
Fecal samples for subjects A to F were collected during the fifth month.
Fluctuation of bifidobacterial flora in individuals.
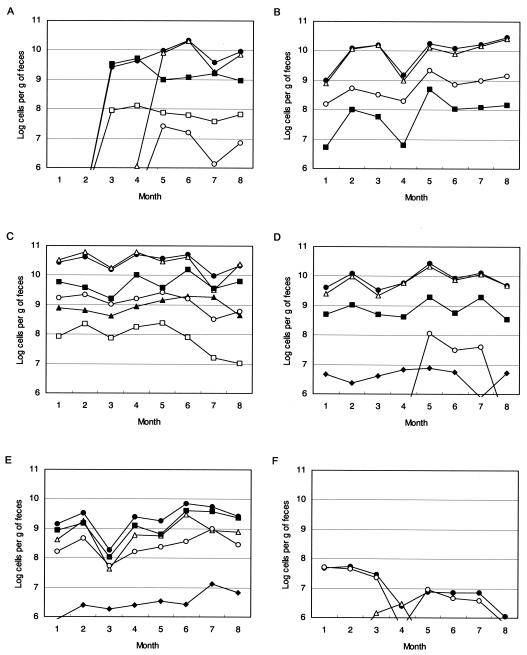
Fecal samples were collected monthly from six healthy adults for 8 months to examine changes in the population level of each Bifidobacterium species (Fig. 2). The highest population and species diversity was observed for serial sample C compared to those of the other five samples. The composition of the bifidobacterial flora was found to be stable throughout the test period. Different bifidobacterial floras were observed for samples B, D, and E; however, the composition of bifidobacteria remained unchanged. On the other hand, considerable changes in the population and composition of bifidobacterial species were observed for serial sample A. Bifidobacterium species were not detected for the first 2 months, but were found at population levels of 1010 thereafter. The B. catenulatum group was the most numerous species at the third and fourth months, but was replaced by the B. adolescentis group after the fifth month. The population of bifidobacteria was much lower in sample F than in the other five samples, and a simple bifidobacterial flora was observed.
FIG. 2.
Population of Bifidobacterium species in fecal samples collected from six human subjects over an 8-month period. (A) Volunteer A; (B) volunteer B; (C) volunteer C; (D) volunteer D; (E) volunteer E; (F) volunteer F. Symbols: •, Bifidobacterium; ▵, B. adolescentis group; ▪, B. catenulatum group; ○, B. longum; ▴, B. bifidum; □, B. breve; ♦, B. angulatum.
DISCUSSION
For this study, quantitative detection of bifidobacteria in the human intestinal flora was achieved by real-time PCR methods combined with Bifidobacterium genus- and species-specific primers. Since quantitative PCR targets extracted DNA, the number of 16S rRNA genes encoded by the genome, differences in DNA extraction efficiency, and point mutations in the target region of a primer may influence measurements. However, the cell counts determined by the quantitative PCR method were almost similar to those determined by the DAPI counting method (Table 2). This suggests that the sensitivity of measurement by these assays may not vary greatly depending on the species or strain. The number of bifidobacteria based on the culture method tended to be lower than estimates by quantitative PCR and the DAPI counting method. Such underestimates may be explained by the presence of bifidobacterial cells lacking colony-forming ability, the occurrence of cell aggregation, and the selection bias of the medium.
Real-time PCR analysis with species-specific primers of fecal samples from six healthy adults gave basically similar results to those obtained by FISH and culture methods (Table 3). Although we used the previously developed primers BiADO-1 and BiADO-2 to detect B. adolescentis (18), quantitative PCR in this case sometimes gave lower estimates than FISH and culture methods. Therefore, we determined the 16S rDNA sequences of 12 strains of B. adolescentis and found the presence of genotype B, having point mutations in the primer target region (Fig. 1). Although the issue of whether the two genotypes belong to the same species requires further study, we refer to them as the B. adolescentis group, for which we have developed a modified primer set (Table 1).
As shown in Table 3, the PCR method was able to detect nonpredominant species, in contrast to the FISH and culture methods, due to differences in detection limits. The real-time PCR method detected target bifidobacterial species when present at concentrations of over 106 cells per g of feces, while the conventional culture method, which identifies bacterial strains isolated from bifidobacteria-selective media, can analyze only the more numerous bacteria in the sample. Since the predominant Bifidobacterium species are usually present at levels of 109 to 1010 cells per g in healthy human feces, the detection limit of the culture method for subdominant bifidobacterial species is usually about 108 cells per g. The reliable detection limit for FISH analysis has been reported to be 108 cells per g (6). Therefore, the specific PCR method should be 10 to 100 times more sensitive than the culture and FISH methods.
Many studies have been reported on the distribution of Bifidobacterium species (2-4, 21, 24, 25a, 28). In the adult intestinal flora, B. adolescentis and B. longum have been reported as the major bifidobacterial species (4, 21, 24, 28). In addition to these two species, we have clarified that the B. catenulatum group is commonly detected in adults; however, our previous PCR analysis failed to show the population levels of the species (18). This quantitative PCR study clearly showed the populations and frequencies of each bifidobacterial species (Table 4), confirming that the B. catenulatum group, the B. adolescentis group, and B. longum are the three predominant bifidobacterial species present in the intestinal flora of human adults. B. breve and B. infantis have been reported to be the major typical species of the intestinal tract of infants (2, 3), but this study also detected these two species in a few adult subjects. Thus, the current real-time PCR method provides an accurate and highly sensitive detection method for bifidobacterial species. This technique has also made it possible to analyze fecal flora in a larger number of samples. In this context, the present study will ensure a much improved understanding of the distribution of bifidobacterial species in the intestinal flora of human adults.
It is widely recognized that the total numbers of Bifidobacterium are relatively constant at the genus level (6, 20). However, only a small number of reports have addressed long-term observations at the species level (20, 28). For the 8-month analysis for the present study, the population and composition of each bifidobacterial species were essentially stable in five of six cases. Although a significant change in bifidobacterial flora was observed for sample A, it should be noted that this subject had received antibiotics 2 weeks prior to the first sampling. We hope that further studies using real-time PCR will clarify how treatment with antibiotics or intake of probiotics affects the human intestinal flora.
Conclusion.
For this study, we developed a quantitative PCR detection method to investigate the distribution of bifidobacterial species in the human intestinal tract by using DNA extracted from fecal samples. Genus- and species-specific primers for bifidobacteria were used in conjunction with real-time PCR, allowing accurate quantification of the target bacteria. The advantage of this specific PCR technique is that the method is approximately 10 to 100 times more sensitive than the culture and FISH methods. Furthermore, the PCR method does not require anaerobic conditions, allows the preservation of DNA in the freezer, and enables the international shipping of DNA. Therefore, quantitative PCR may enable us to make more extensive and detailed analyses of intestinal microflora. Such molecular biological techniques may provide important clues for the understanding of the relationship between intestinal flora and health and disease in humans.
Acknowledgments
We thank K. Nomoto for his valuable advice.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. [DOI] [PubMed] [Google Scholar]
- 3.Biavati, B., P. Castagnoli, F. Crociani, and L. D. Trovatelli. 1984. Species of the Bifidobacterium in the feces of infants. Microbiologica 7:341-345. [PubMed] [Google Scholar]
- 4.Biavati, B., P. Castagnoli, and L. D. Trovatelli. 1986. Species of the genus Bifidobacterium in the feces of human adults. Microbiologica 9:39-45. [PubMed] [Google Scholar]
- 5.Finegold, S. M., V. S. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 6.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescence in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 8.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen, H. J., G. R. Gibson, P. Elfferich, G. C. Raangs, A. C. Wildeboer-Veloo, A. Argaiz, M. B. Roberfroid, and G. W. Welling. 2000. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125-129. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kageyama, A., M. Sakamoto, and Y. Benno. 2000. Rapid identification and quantification of Collinsella aerofaciens using PCR. FEMS Microbiol. Lett. 183:43-47. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitajima, H., Y. Sumida, R. Tanaka, N. Yuki, H. Takayama, and M. Fujimura. 1997. Early administration of Bifidobacterium breve to preterm infants: randomised controlled trial. Arch. Dis. Child Fetal Neonatal 76:F101-F107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, Y.-K., K. Nomoto, S. Salminen, and S. L. Gorbach. 1999. Handbook of probiotics. Wiley Interscience, New York, N.Y.
- 16.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 20.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsuoka, T., K. Hayakawa, and N. Kimura. 1974. Die faekalflora bei menschen. II. Mitteilung: die zusammensetzung der bifidobakerien flora der verschiedenen altersgruppen. Zentbl. Bakteriol. Parasitenkd. Infektionskr. Abt. 1 Orig. Reihe A 226:469-478. [PubMed] [Google Scholar]
- 22.Miyake, T., K. Watanabe, T. Watanabe, and H. Oyaizu. 1998. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 42:661-667. [DOI] [PubMed] [Google Scholar]
- 23.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutai, M., and R. Tanaka. 1987. Ecology of Bifidobacterium in the human intestinal flora. Bifidobacteria Microflora 6:33-41. [Google Scholar]
- 25.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2001. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45:39-44. [DOI] [PubMed] [Google Scholar]
- 27.Sakata, S., M. Kitahara, M. Sakamoto, H. Hayashi, M. Fukuyama, and Y. Benno. 2002. Unification of Bifidobacterium infantis and Bifidobacterium suis as Bifidobacterium longum. Int. J. Syst. Evol. Microbiol. 52:1945-1951. [DOI] [PubMed] [Google Scholar]
- 28.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. W. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scardovi, V. 1984. Genus Bifidobacterium Orla-Jensen, 1924, 472, p. 1418-1434. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 30.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 31.Sonoike, K., M. Maeda, and M. Mutai. 1986. Selective agar medium for counting viable cells of bifidobacteria in fermented milk. J. Food Hygienic Soc. Jpn. 27:238-244. [Google Scholar]
- 32.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto, T., M. Morotomi, and R. Tanaka. 1992. Species-specific oligonucleotide probes for five Bifidobacterium species detected in human intestinal microflora. Appl. Environ. Microbiol. 58:4076-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]