Abstract
SUMMARY
The Lyme disease spirochete Borrelia burgdorferi lacks the transcriptional cascade control of flagellar protein synthesis common to other bacteria. Instead, it relies on a post-transcriptional mechanism to control its flagellar synthesis. The underlying mechanism of this control remains elusive. A recent study reported that the increased level of BB0184 (CsrABb; a homolog of carbon storage regulator A) substantially inhibited the accumulation of FlaB, the major flagellin protein of B. burgdorferi. In this report, we deciphered the regulatory role of CsrABb on FlaB synthesis and the mechanism involved by analyzing two mutants, csrABb− (a deletion mutant of csrABb) and csrABb+ (a mutant conditionally over-expressing csrABb). We found that FlaB accumulation was significantly inhibited in csrABb+ but was substantially increased in csrABb−. In contrast, the levels of other flagellar proteins remained unchanged. Cryo-electron tomography and immuno-fluorescence microscopic analyses revealed that the altered synthesis of CsrABb in these two mutants specifically affected flagellar filament length. The leader sequence of flaB transcript contains two conserved CsrA-binding sites, with one of these sites overlapping the Shine-Dalgarno sequence. We found that CsrABb bound to the flaB transcripts via these two binding sites, and this binding inhibited the synthesis of FlaB at the translational level. Taken together, our results indicate that CsrABb specifically regulates the periplasmic flagellar synthesis by inhibiting translation initiation of the flaB transcript.
Keywords: Spirochete, Borrelia burgdorferi, Flagella, FlaB, Carbon storage regulator A
INTRODUCTION
Borrelia burgdorferi, the causative agent of Lyme disease, is a spirochete with characteristic flat-wave morphology (Charon and Goldstein, 2002;Goldstein et al., 1994). This organism has an unusual ability to swim in and invade highly viscous gel-like environments including host connective tissues (Li et al., 2010b;Charon and Goldstein, 2002;Moriarty et al., 2008; Szczepanski et al., 1990;Dunham-Ems et al., 2009;Kimsey and Spielman, 1990). The ability of B. burgdorferi to swim depends on the rotation of periplasmic flagella (PFs) (Motaleb et al., 2000;Sal et al., 2008;Li et al., 2010b;Goldstein et al., 1994). B. burgdorferi has 7 to 11 PFs that are subterminally attached at each pole of the cell cylinder and reside in the periplasmic space. These PFs form a tight-fitting ribbon that wraps around the cell cylinder body axis in a right-handed sense (Charon et al., 2009;Liu et al., 2009;Hovind-Hougen, 1984;Goldstein et al., 1996). In addition, the PFs have both skeletal and motility functions, i.e., aflagellated mutants are non-motile and are rod-shaped rather than having the flat-wave morphology (Motaleb et al., 2000; Sadziene et al., 1991;Sal et al., 2008). Motility is considered to be an important virulence factor of B. burgdorferi, as mutants defective in motility fail to invade human tissues and establish mammalian infection (Li et al., 2010b;Sultan et al., 2010;Charon and Goldstein, 2002).
In general, the PFs of spirochetes have a structure that is similar to the flagella of other motile bacteria such as Escherichia coli: A given PF is composed of a basal body, hook, and filament (Liu et al., 2009;Liu et al., 2010;Izard et al., 2008;Izard et al., 2009;Murphy et al., 2008;Kudryashev et al., 2010). Compared to the other flagellated bacteria, the flagellar filament of spirochetes is unique and is among the most complex of bacterial flagella (Li et al., 2000b; Charon and Goldstein, 2002). In most spirochete species, the filaments contain at least one flagellar sheath protein referred to as FlaA and three core proteins designated as FlaB1, FlaB2 and FlaB3 (Norris et al., 1988;Li et al., 2008;Li et al., 2010a;Li et al., 2000a). Our recent studies from the spirochete Brachyspira hyodysenteriae have revealed that each flagellin unit contributes to the stiffness of the PFs, and that this stiffness directly correlates with cell speed (Li et al., 2008). In contrast to other spirochetes, the flagellar filament of B. burgdorferi contains only one core protein termed FlaB (Fraser et al., 1997;Ge et al., 1998;Motaleb et al., 2004). FlaB has been shown to be the major flagellin protein, contributing to 10–14% of the total cell proteins (Motaleb et al., 2004).
The regulation of bacterial flagellar synthesis and motility gene expression is complex. In Salmonella enterica serovar Typhimurium and E. coli, this synthesis and assembly has been shown to involve over 50 genes (Terashima et al., 2008;Aldridge and Hughes, 2002). In most externally flagellated bacteria, these genes are divided into three classes (class I, II and III), which are tightly regulated by a transcriptional cascade control mechanism (Macnab, 1992; Chevance and Hughes, 2008). Within this regulatory hierarchy, the class I genes initiate the expression of the class II genes, which encode the structural proteins involved in the assembly of the hook-basal body complexes, and two regulatory proteins, FliA (σ28) and FlgM (anti-σ28). FliA and FlgM control the expression of the class III genes including those that encode flagellin and chemotaxis proteins. FliA and FlgM remain bound as a complex in the cytoplasm during the basal body and hook synthesis. As FlgM exits the cells via the flagellum-export apparatus after the completion of the hook, FliA is free to initiate the transcription of the class III genes (Chevance and Hughes, 2008;Helmann, 1991;Macnab, 1992;Hughes et al., 1993).
Several lines of evidence indicate that B. burgdorferi does not employ the transcriptional cascade control mechanism to regulate its flagellar synthesis and assembly. This spirochete lacks homologs of FliA and FlgM, and there is no σ28 promoter consensus sequence evident in its genome (Fraser et al., 1997;Charon and Goldstein, 2002). In addition, all of the motility and chemotaxis genes analyzed to date are controlled by σ70, a house-keeping transcription factor (Charon and Goldstein, 2002;Ge and Charon, 1997;Ge et al., 1997;Yang and Li, 2009;Motaleb et al., 2011) . In contrast to other bacteria, recent studies of specific B. burgdorferi mutants indicate that this spirochete regulates flagellar synthesis by a post-transcriptional mechanism rather than via a transcriptional cascade (Motaleb et al., 2004;Ge et al., 1998;Sal et al., 2008). However, the mechanism involved in the post-transcriptional regulation remains unknown.
Studies of the Csr systems in various bacteria have established the importance of CsrA in both physiological and pathological functions, such as carbon metabolism, virulence, biofilm formation and motility [for reviews, see (Romeo, 1998;Babitzke et al., 2009;Lucchetti-Miganeh et al., 2008)]. The Csr system was first discovered in E. coli and was subsequently identified in many other bacterial species (Romeo, 1998;Romeo et al., 1993;Babitzke and Romeo, 2007). In E. coli, Csr system consists of the key determinant CsrA, an RNA binding protein, two non-coding RNA molecules (CsrB and CsrC), and a regulatory protein, CsrD (Suzuki et al., 2006). CsrA recognizes and binds to the consensus sequence RUACARGGAUGU, with a well-conserved binding motif (ARGGA) that often overlaps the Shine-Dalgarno (SD) sequence of a given transcript (Babitzke and Romeo, 2007;Baker et al., 2007;Brencic and Lory, 2009;Dubey et al., 2005;Timmermans and Van, 2010;Mercante et al., 2009). Binding of CsrA to this consensus site results in either the stabilization of mRNA or blocking of ribosome binding and, consequently, inhibition of translation initiation (Romeo, 1998;Wang et al., 2005;Wei et al., 2001).
A homolog of CsrA (CsrABb) was recently identified in B. burgdorferi (Sanjuan et al., 2009). Over-expression of CsrABb led to altered expression of several outer membrane antigens, reduction of FlaB, and a change in cell morphology, suggesting that CsrABb may have a potential regulatory role on the flagellation and motility of B. burgdorferi. In addition, we have recently shown that CsrABb is essential for animal infectivity (Sze and Li, 2010). The mechanism involved and the overall impact of CsrABb on B. burgdorferi flagellar synthesis and motility remains unknown. In this report, we described the regulatory mechanism of CsrABb on the synthesis of PFs (in particular the major flagellin protein, FlaB) and motility using genetic, biochemical, and cryo-electron tomography (cryo-ET) approaches. We found that CsrABb specifically regulates the flagellar synthesis of B. burgdorferi by inhibiting translation initiation of the flaB transcript.
RESULTS
Isolation of the csrABb− and csrABb+ mutants
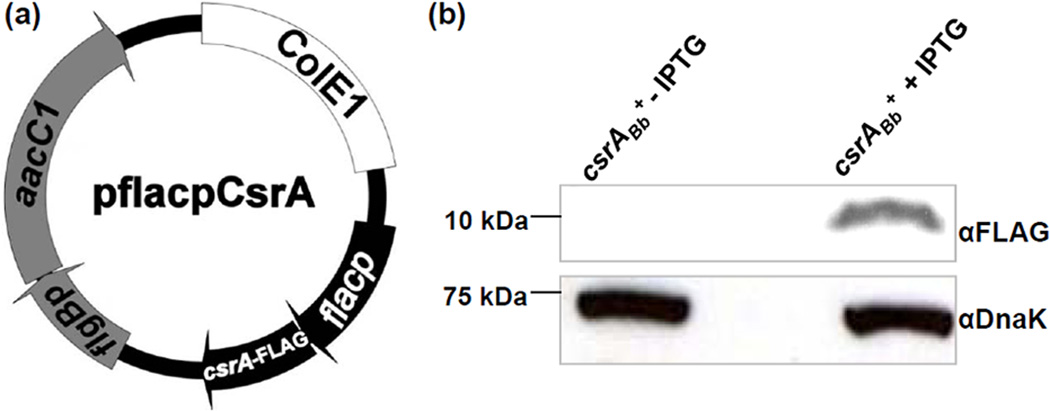
The csrABb− mutant was isolated as described before; it contains a deletion of the entire open reading frame bb0184 (Sze and Li, 2010). To isolate a strain that over-expresses CsrABb, pflacpCsrA (Figure 1a), an IPTG-inducible CsrABb plasmid, was electro-transformed into B. burgdorferi A3-LS, a derivative of B31A3 (a virulent and low passage strain) that carries a lacI gene (Gilbert et al., 2007). The transformants were selected on agar plates containing both gentamicin (40 µg/ml) and streptomycin (50 µg/ml). The presence of pflacpCsrA in antibiotic-resistant colonies was confirmed by PCR. One clone, csrABb+, was selected for further characterization. For the induction of CsrABb, 105 csrABb+ cells were inoculated into 10 ml of growth medium with antibiotics in the presence of 1 mM IPTG and incubated to the early stationary phase (approximately108 cells/ml). CsrABb synthesis was determined by western blots using an antibody directed against the FLAG epitope (Mofunanya et al., 2009), which was tagged to the C-terminus of CsrABb. We found that the addition of IPTG induced the synthesis of CsrABb, as the tagged FLAG was detected only after the induction in the csrABb+ strain (Figure 1b). The mass of CsrABb-FLAG was approximately 11 kDa as predicted. Thus, CsrABb was inducible in the csrABb+ strain and could be over-expressed in B. burgdorferi.
Figure 1.
Constructing the IPTG-inducible plasmid for the over-expression of CsrABb in B. burgdorferi and the detection of CsrABb in the csrABb+ strain by western blot. (a) Construction of the pflacpCsrA vector for the over-expression of csrABb. To conditionally over-express csrABb, the entire bb0184 gene was amplified by PCR. The obtained amplicon was fused to flacp (Gilbert et al., 2007) and then cloned into the shuttle vector pBSV2G (Tilly et al., 2009), yielding the pflacpCsrA plasmid. (b) Similar amounts of csrABb+ and IPTG-induced csrABb+ whole cell lysates were analyzed on SDS-PAGE. The presence of the FLAG-epitope-tagged CsrABb was detected using a specific antibody against the FLAG epitope. Immunoblots were developed using horseradish peroxidase secondary antibody with an ECL luminol assay as previously described (Sze and Li, 2010).
The over-expression of CsrABb alters cell shape and inhibits motility
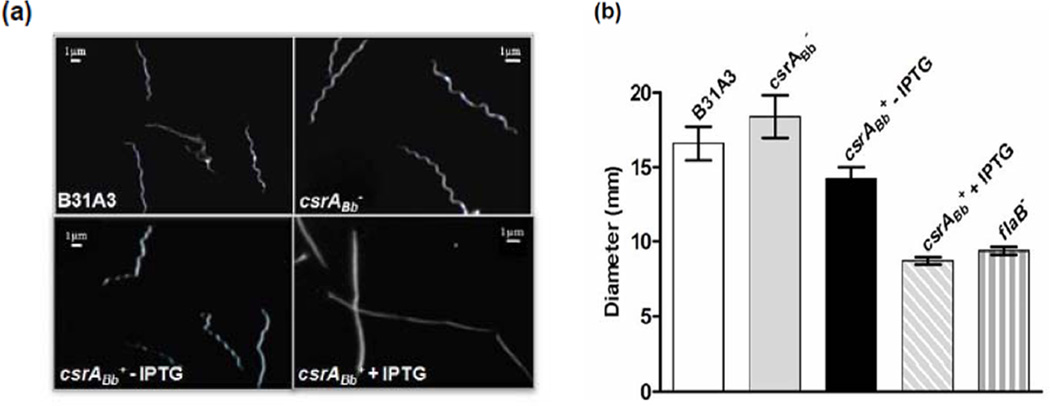
Before the addition of IPTG, the csrABb+ strain was motile and had the flat wave morphology as its parental A3-LS strain. However, after the induction with IPTG, the cells were rod-shaped and non-motile as revealed by dark-field microcopy (Figure 2a). This phenotype is similar to the previously described aflagellated mutants of B. burgdorferi and to those reported by Sanjuan et al. on a strain constitutively expressing CsrABb (Motaleb et al., 2000;Sal et al., 2008;Li et al., 2010b; Sanjuan et al., 2009). The impact of CsrABb on the motility was further evaluated using the swarm plate assay. As shown in Figure 2b, the diameter of swarm rings formed by the IPTG-induced csrABb+ strain was almost equivalent to the non-motile flaB− mutant, indicating that motility was significantly inhibited. Dark-field microscopy and bacterial motion tracking analyses further confirmed that the induced cells were completely non-motile (data not shown). Taken together, these results indicate that CsrABb markedly influences both cell shape and motility.
Figure 2.
The over-expression of CsrABb altered the cell morphology and motility of the csrABb+ strain. (a) Dark-field microscopic analysis of B31A3, csrABb, and csrABb+ with or without the induction of IPTG. (b) Swarm plate assay of B31A3, csrABb−, and csrABb+ with or without the induction of IPTG. The assays were carried out on 0.35% agarose containing 1:10 diluted BSK-II medium as previously described (Li et al., 2002;Motaleb et al., 2000). The data are presented as mean diameters (in millimeters) of swarms plus the standard errors from five plates.
CsrABb specifically inhibits FlaB accumulation
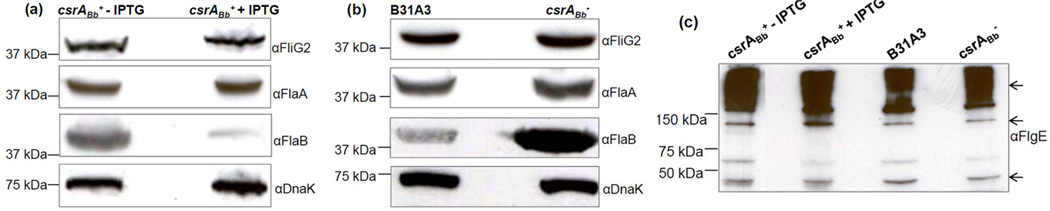
We determined the overall impact of CsrABb on the flagellation of B. burgdorferi. Western blot analysis was used to examine the amounts of various flagellar proteins in the csrABb+ and csrABb− mutants. After the induction (5 days) with IPTG, the amount of FlaB in the csrABb+ strain was decreased by approximately 75%, whereas the levels of the other flagellar proteins remained unchanged, including the flagellar sheath protein (FlaA), the flagellar motor switch protein (FliG2), and the flagellar hook protein (FlgE) (Figure 3a, c). Thus, the over-expression of CsrABb specifically inhibited the accumulation of FlaB. Based on the result that CsrABb represses the FlaB synthesis, we hypothesized that a mutant lacking CsrABb would have higher levels of FlaB than the wild type. Accordingly, we determined the amounts of the identical flagellar proteins in the csrABb− deletion mutant and its parental B31A3 strain (Sze and Li, 2010). We found that the amount of FlaB in this mutant was 4-fold greater than in the wild type, whereas the levels of FlaA, FliG2, and FlgE proteins remained unchanged (Figure 3b, c). These results indicate that CsrABb specifically modulates the overall synthesis of FlaB.
Figure 3.
The level of FlaB was altered in the IPTG-induced csrABb+ strain and the csrABb− mutant. (a) The over-expression of CsrABb repressed the level of FlaB. (b) The level of FlaB was increased in the csrABb− mutant. (c) The level of FlgE remained unchanged in both the csrABb− mutant and the IPTG-induced csrABb+ strain. Similar amounts of whole cell lysates from B31A3, csrABb−, and the csrABb+ strain with or without IPTG were analyzed on SDS-PAGE. The presence of FlaA, FlaB, FlgE, FliG2, and DnaK were detected using specific antibodies against these proteins. Immunoblots were developed as described in Figure 1. In 3(c), arrows point to the monomer of FlgE and high-molecular-weight FlgE complex previously reported (Sal et al., 2008).
CsrABb binds to the flaB transcript leader sequence
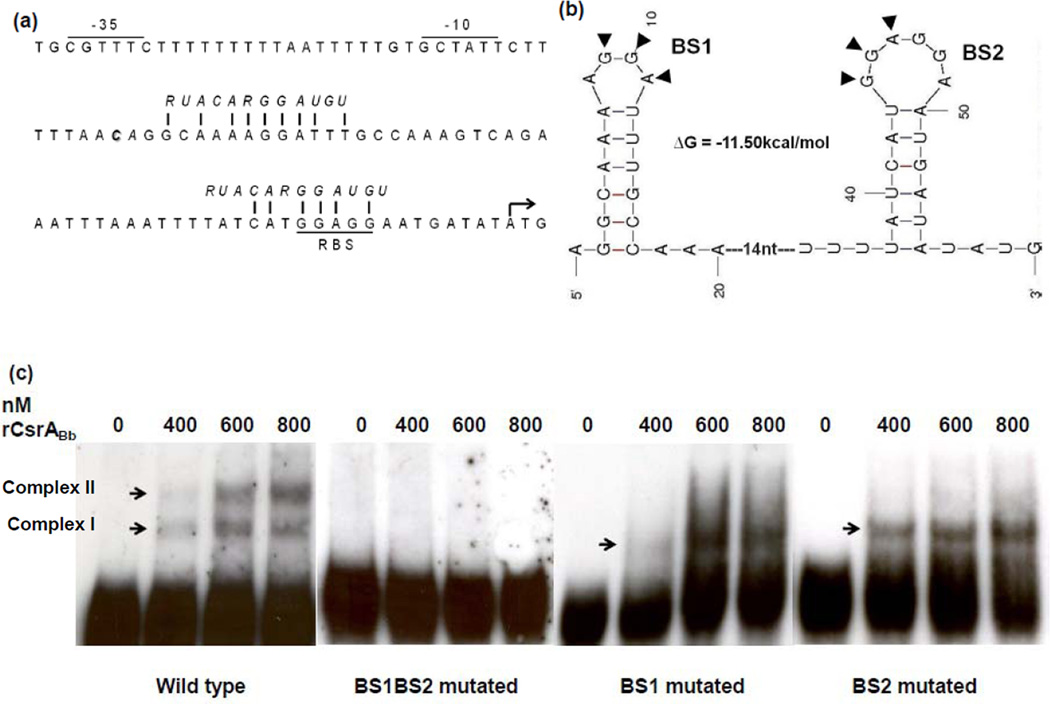
CsrA is an RNA-binding protein and affects its targeted genes by binding to the consensus sequence (RUACARGGAUGU), which typically appears in the leader sequence of a given transcript (Lenz et al., 2005;Dubey et al., 2005;Babitzke and Romeo, 2007). The flaB transcript contains a 56 nt untranslated leader (UTL) sequence. We found that the flaB UTL region contains two potential CsrA binding sites with a conserved sequence (ANGGA), where one of the binding sites is 2 nt after the transcription start site, and the other overlaps the SD sequence (Figure 4a). RNA secondary structure prediction revealed that these two potential CsrA binding sequences formed two RNA hairpin loops with the GGA motifs (Figure 4b). As the E. coli CsrA binds with high affinity to GGA motifs within hairpin loops (Dubey et al., 2005), the predicted two loops within the flaB UTL can be authentic binding sites for CsrABb.
Figure 4.
CsrABb binds to the 5’-untranslated leader (UTL) of the flaB transcript. (a) The upstream region of the flaB gene. The −10 and −35 regions of the flaB promoter and the ribosome-binding site (RBS) are underlined; * represents the transcriptional start site; the ATG start codon is marked with an arrow; the CsrA binding consensus sequence is italicized, and the vertical lines represent the conserved residues that match the E. coli CsrA-binding consensus sequence. (b) Computer-predicted secondary structure of the flaB mRNA leader sequence. The secondary structure and the free energy value were determined using the mfold program (Zuker, 2003)(Version 3.2). BS1 and BS2 represent the two hairpin loops formed by the CsrA-binding consensus sequences, and the two conserved motifs of ‘GGA’ within the loops are marked with arrowheads. (c) Electrophoretic mobility shift assay (EMSA). The biotin-labeled RNA probes (19 fM) were incubated with different concentrations of rCsrABb. The experiment was carried out using the wild-type probe and three mutated probes (for the mutated BS1BS2 probe, the two ‘GGA’ motifs in BS1 and BS2 were mutated to ‘AAA’; for the mutated BS1 and BS2 probes, only one ‘GGA’ motif in BS1 or BS2 was mutated to ‘AAA’).
To test if CsrABb interacts with the binding sites in the UTL, electrophoretic mobility-shift assay (EMSA) was carried out using the recombinant CsrABb protein (rCsrABb) and synthetic flaB leader transcripts as RNA probes (the sequences are described in Table 1). At a low concentration (400 nM) of rCsrABb, the recombinant protein binding to the wild-type RNA probe was detected as two shifted bands (the lower band designated as Complex I and the upper band as Complex II). As the concentration of rCsrABb increased (600 and 800 nM), the level of Complex II was significantly increased (Figure 4c). The mutation of either one of these binding sites (probes BS1 and BS2) eliminated Complex II (Figure 4c), and the mutation of both binding sites (probe BS1BS2) completely abrogated the formation of the two shifted complexes (Figure 4c). The observed gel shift pattern is consistent with the model that a CsrA dimer binding to two target sites within a single transcript (Mercante et al., 2009;Yakhnin et al., 2007), i.e., Complex I being the product of rCsrABb dimer bound to a single site, and Complex II being the product of rCsrABb bound to both binding sites within the probe. Taken together, these results indicate that CsrABb specifically binds to the flaB transcript via these two conserved binding motifs.
Table 1.
Oligonucleotide primers and RNA probes used in this study
| Primers | Descriptions | Sequences |
|---|---|---|
| P1 | rCsrABb (F) | 5’-CACCATGCTAGTATTGTCAAGAAAAG-3’ |
| P2 | rCsrABb (R) | 5’-ATTTTCATTCTTGAAATAATG-3’ |
| P3 | lac promoter, flacp (F) | 5’-GGATCCGACGTCTAATACCCGAGC-3’ |
| P4 | IPTG inducible csrABb (F) | 5’-CATATGCTAGTATTGTCAAGAAAAGC-3 |
| P5 | IPTG inducible csrABb (R) | 5’-GGATCCTTATTTGTCATCGTCGTCCTTGTAGTC ATTTTCATTCTTGAAATAATG-3’ |
| P6 | qRT-PCR, eno (F) | 5’-AACAGGAATTAACGAGGCTG-3’ |
| P7 | qRT-PCR, eno (R) | 5’-AAATTGCATTAGCACCAAGC-3’ |
| P8 | qRT-PCR, flaB (F) | 5’-CATATTCAGATGCAGACAGAGG-3’ |
| P9 | qRT-PCR, flaB (R) | 5’-CCCTGAAAGTGATGCTGGTGTG-3’ |
| P10 | EMSA probe, flaBp | 5’-GCAAAAGGAUUUGCCAAAGUCAGAAUUUAA AUUUUAUCAUGGAGGAAUGA-3’ |
| P11 | EMSA probe, flaBp (BS1 mutated) |
5’-GCAAAAAAAUUUGCCAAAGUCAGAAUUUAA AUUUUAUCAUGGAGGAAUGA-3’ |
| P12 | EMSA probe, flaBp (BS2 mutated) |
5’-GCAAAAGGAUUUGCCAAAGUCAGAAUUUAA AUUUUAUCAUAAAGGAAUGA-3’ |
| P13 | EMSA probe, flaBp (BS1 BS2 mutated) |
5’-GCAAAAAAAUUUGCCAAAGUCAGAAUUUAA AUUUUAUCAUAAAGGAAUGA-3’ |
The underlined sequences are the engineered restriction cut sites for DNA cloning. The bold sequences in EMSA probes are the mutated CsrA binding sequence, altered from GGA to AAA.
CsrABb inhibits the translation of flaB transcript
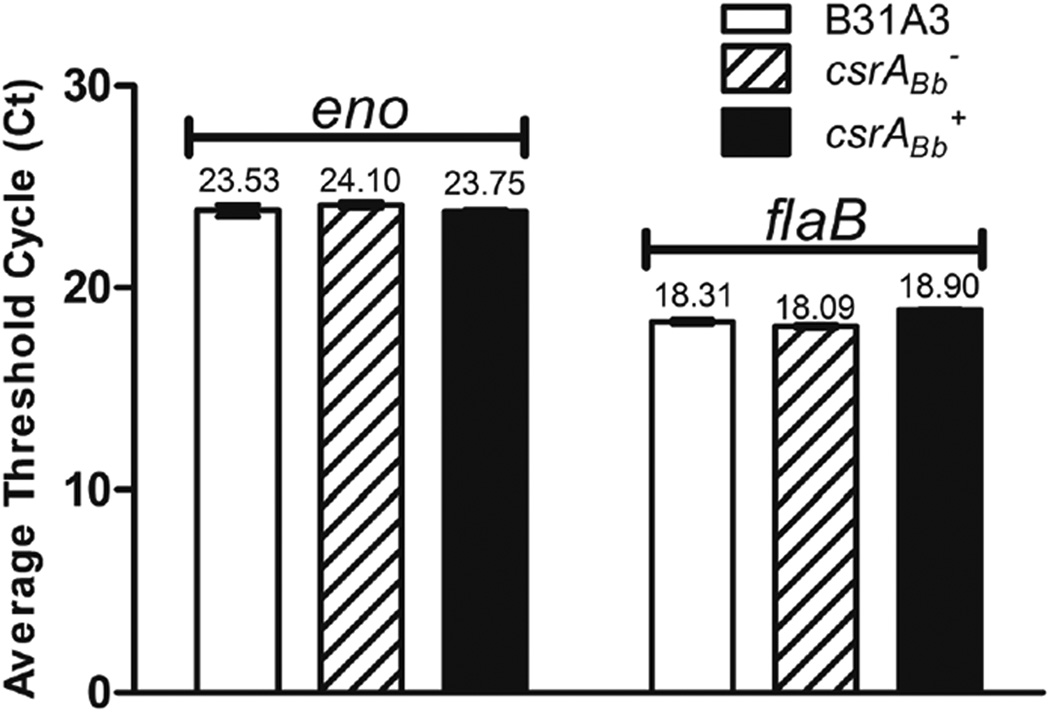
The binding of CsrABb to the flaB transcript can either reduce the stability of the transcript or block the translation initiation. To rule out one of these two possibilities, qRT-PCR analysis was conducted to compare the level of the flaB transcript of these two mutants to that of the wild type. The results were expressed as cycle threshold value (Ct) for each individual gene transcript. As shown in Figure 5, the level of the flaB transcript was similar across all three strains, implying that the action of CsrABb on FlaB occurs at the post-transcriptional level. As one of the binding sites (BS2) overlaps the SD sequence of flaB transcript (Figure 4a, b) and the EMSA assay showed that CsrABb binds to BS2 (Figure 4c), CsrABb most likely acts by blocking ribosome binding to the flaB transcript and consequently inhibiting the translation.
Figure 5.
qRT-PCR analysis of the csrABb− mutant and the IPTG-induced csrABb+ strain. The levels of flaB transcript were measured by qRT-PCR as previously described (Sal et al., 2008). The transcript of the enolase gene (eno) was used as an internal control to normalize the qPCR data. The results were expressed as the mean threshold cycles (CT) of triplicate samples.
Modulation of CsrABb expression influences filament length and ribbon organization
Western blot analysis indicated that the level of FlaB was significantly altered in the csrABb+ and csrABb− mutants (Figure 3). We determined if the altered amounts of FlaB in these mutants influence the structure of the PFs in situ using cryo-ET. Using low magnification snapshots along a single cell, we analyzed both cell poles along with the central region of an individual cell (Figure 6). We also determined how the flagellar filaments were organized. In the wild-type cells, the PFs were clearly visible (Figure 6a), and these organelles formed a tightly packed ribbon that wrapped around the body axis in a right-handed sense as previously described (Charon et al., 2009). The angle subtended by the ribbon was 1.13 radians, which is identical to results previously obtained by cryo-ET (Charon et al., 2009). The ribbons extended from the subterminal region of the cell pole to the central region (Figure 6a). The mean number of PFs observed at each cell end was 7.9 ± 1 (range from 7 to 11, n = 11 cells); at the central region, the mean number was 7.8 ± 3.4 (range from 3 to 13, n= 11 cells). The 3D reconstruction revealed that the PFs originated from opposite cell ends and overlapped at the cell center, however, they were not localized at the same face of the cells (Figure 6a and Movie S1).
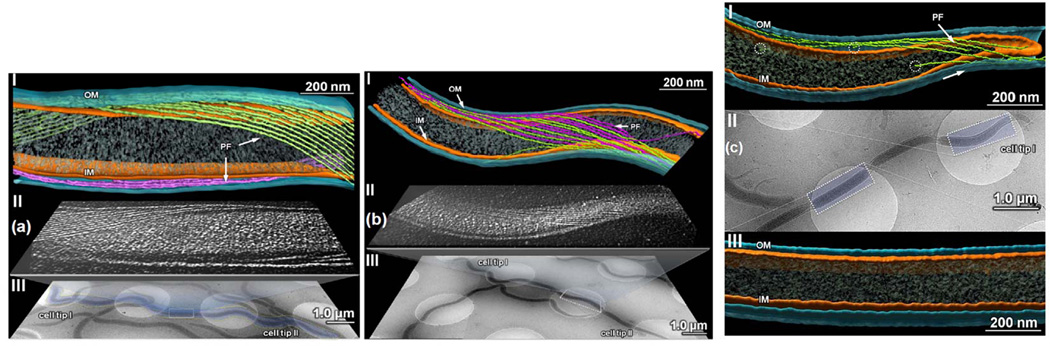
Figure 6.
Cryo-ET analysis of flagellar filaments in the wild-type, csrABb− and the IPTG-induced csrABb+ strain. (a) & (b) show the flagellar filament arrangements in the middle portions of the wild-type and the csrABb− cells, respectively. Three stages of images were used to reveal the differences between the wild-type and the mutant cells: the low-magnification images showing the overview of the cells (III), which was created by montaging several 0° tilt snapshots; the highlighted boxes showing the middle sections of the cells where high-magnification tomograms were collected (II); and above it is the 3D reconstruction produced from the data in the tomogram (I). (c) Cryo-ET analysis of the IPTG-induced csrABb+ cells: (I) 3D segmentation near the cell end, which shows the presence of the flagellar filaments (some truncated flagellar filaments are circled); (II) A low-magnification image of an IPTG-induced csrABb+ cell. High-magnification tomograms were taken of the boxed regions to produce the 3D segmentations; (III) 3D reconstruction near the middle portion of the cell, showing that there are no longer any flagellar filaments in this region of the cell.
The motility and fine structure of the csrABb− mutant was similar in some respects to specific parameters of the wild type, but different in others. Velocity measurements of swimming cells indicated that the motility of csrABb− (10.2 ± 4.5 µm/sec) was similar to the wild type (11.5 ± 0.4 µm/sec). Thus, overproducing FlaB in csrABb− did not have a noticeable effect on motility. Cryo-ET revealed some structural differences between this mutant and the wild type. The mean number of PFs at the cell poles of csrABb− was 7 ± 1.5 (range from 6 to 9, n= 9 cells), and these organelles also formed the right-handed flat ribbons (Movie S2), which is similar to that of the wild type. However, these ribbons wrapped around the cell cylinder with a smaller helix pitch than that of the wild type, as the angle subtended by the ribbon was 1.08 radians. In addition, these ribbons extensively overlapped in the central region of the cells and interdigitated with those from the opposite end (Figure 6b and Movie S2). The number of PFs observed at the cell center was increased to 12 ± 1.5 (range from 8 to 14, n = 9 cells). Thus, the increase of FlaB in the csrABb− mutant results in longer flagellar filaments that wind around the cell cylinder tighter than those of the wild type. In addition, in contrast to the wild type, these long filaments interdigitate with the filaments originating from the opposite end.
The IPTG-induced csrABb+ strain was analyzed in detail. Before the induction, the mean length of PFs was 6.9 ± 3.0 µm (range from 5 to 10 µm, n = 11 PFs out of 6 cells examined). In contrast, the PFs in the IPTG-induced csrABb+ were notably truncated (the mean length of PFs was 1.63 ± 0.72 µm, ranging from 0.5 to 3 µm, n = 15 PFs out of three cells) (Figure 6c and movie S3). No filaments were detected in the central region of the cells (Figure 6c and movie S4). The mean number of PFs observed at the cell poles was 4 ± 1.5 (range from 2 to 6, n = 5 cells), which is less than that of the wild type. In contrast, the number of flagellar basal bodies observed at the cell poles (the mean number was 7 ± 2, n= 5 cells) was similar to that of the wild type. These observations are consistent with the above western blot results whereby only the major filament protein, FlaB, was influenced by CsrABb (Figure 3). Collectively, these results indicate that the changes in the level of CsrABb in the csrABb− mutant and the IPTG-induced csrABb+ strain specifically influenced the flagellar filaments, but not the flagellar basal bodies and hooks.
Modulation of CsrABb influences cell morphology
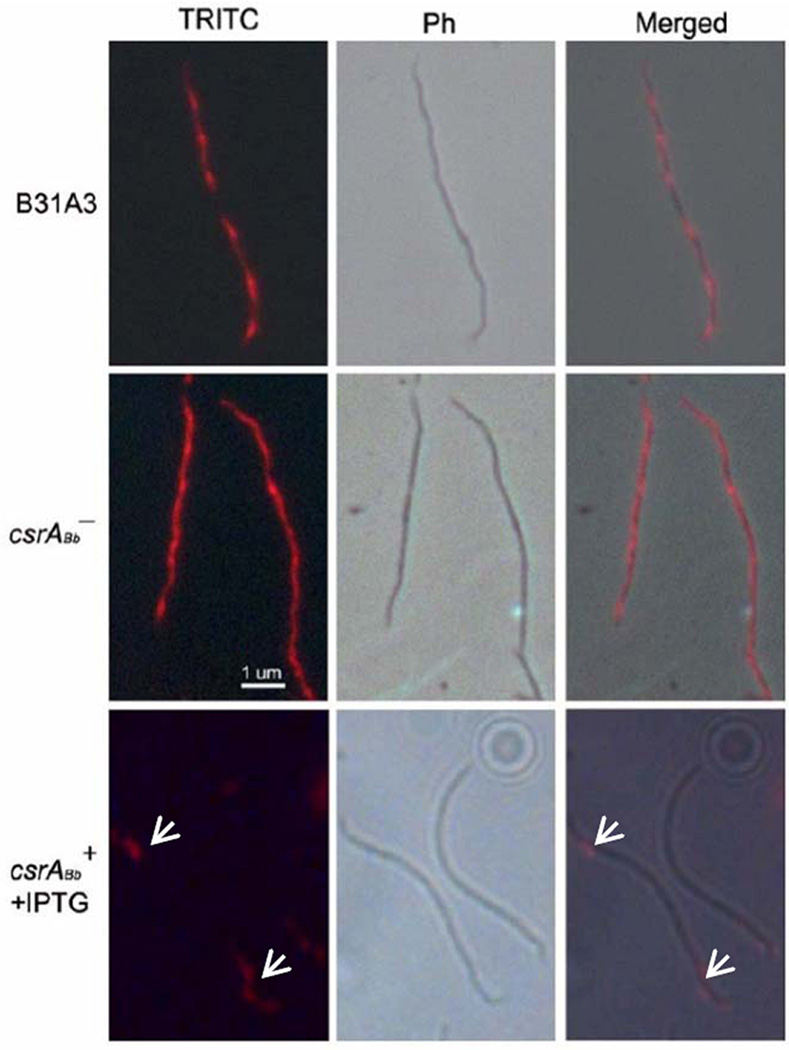
Previous studies indicate that FlaB has a skeletal function – a flaB− mutant is rod-shaped instead of being a flat wave (Motaleb et al., 2000). To determine whether the modulation of FlaB in the IPTG-induced csrABb+ strain influences the cell morphology, as well as to further examine the above cryo-ET results, an immunofluorescence assay (IFA) using antibody against FlaB was carried out. We found that B31A3 (Figure 7) and the un-induced csrABb+ cells (data not shown) had fluorescence extending along the length of the cells with a periodic intensity of fluorescence that varied every 2.98 ± 0.33 µm (range from 2.51 to 3.83 µm, n=7 cells), which is almost identical to the helix-pitch of the PFs previously measured by high-voltage electron microscopy (2.97 µm) (Goldstein et al., 1996). After induction for 5 days, the fluorescence could only be detected at the cell poles of the csrABb+ strain (Figure 7), further confirming that the PFs were truncated. These results are consistent with those obtained by cryo-ET that over-expression of CsrABb resulted in shortened PFs.
Figure 7.
Immunofluorescence assay (IFA) of the wild-type, csrABb−, and the IPTG-induced csrABb+ strain using anti-FlaB antibody. The cells were fixed with methanol, stained with the anti-FlaB antibody, and counterstained with anti-mouse Texas red antibody. The micrographs were taken under DIC and TRITC (1000 x), and the resulting images were then merged. Arrows represent the location of FlaB in the IPTG-induced csrABb+ strain.
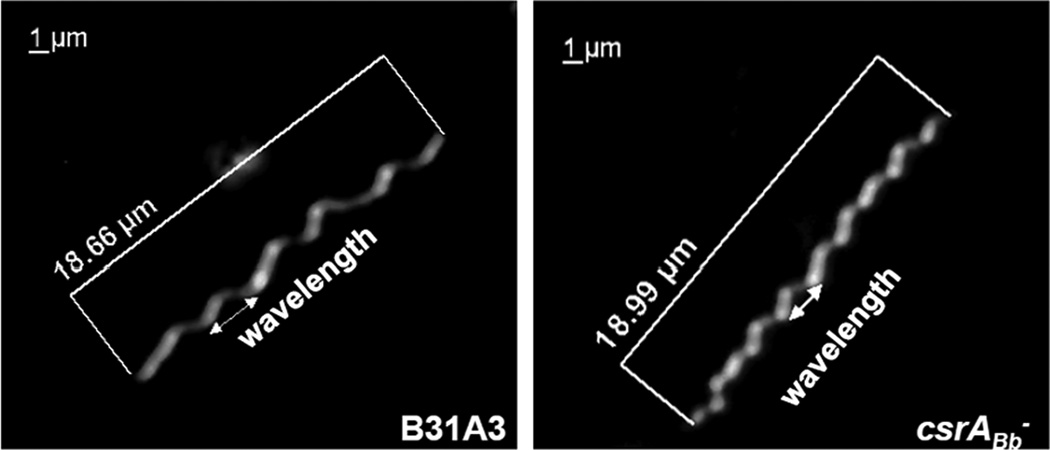
The increase in FlaB synthesis in the csrABb− mutant influenced cell morphology in an opposite manner than that of the IPTG-induced csrABb+ cells. As noted above, the IFA results revealed that the flagellar filaments in the wild type had a periodic intensity of fluorescence. However, this pattern was different in the csrABb− mutant, as the fluorescence appeared more heterogeneous and less periodic (Figure 7). We measured the wavelengths of the cells by dark-field microscopy, as previous studies have shown that the helix-pitch of the PFs in situ and the wavelengths of the cells are equivalent (Goldstein et al., 1996). We found that for the wild type, there were approximately 6 waves per cell (range from 5 to 6 waves out of 10 cells examined), with a mean wavelength of 3.07 ± 0.55 µm (range from 2.52 to 3.62 µm) (Figure 8), which is similar to the previous measurements by both dark-field light microscopy (3.19 µm) and high voltage electron microscopy (2.97 µm) (Goldstein et al., 1994;Goldstein et al., 1996). In contrast, the csrABb− mutant cells had more waves (the mean number was 8, ranging from 7 to 9 out of 10 cells examined), and the waves were shorter (the mean wavelength was 2.16 ± 0.25 µm, range from 1.91 to 2.41µm). These results suggest that the longer flagellar filaments in the csrABb− mutant not only influenced how the bundles interacted with one another within the cells by interdigitating, but also impacted the intricacies of the flat wave periodicity of the cells. The finding of the shorter cellular wavelength in the mutant is consistent with the results from cryo-ET (Figure 6b), as the latter indicated that the PFs wrapped around the cell cylinders in the csrABb− mutant more tightly than in the wild type.
Figure 8.
Dark-field microscopic analysis of the csrABb− mutant. Cells were visualized under dark-field illumination at 1000 x magnification using a Zeiss Axiostar plus microscope. The cell length and the wavelength are labeled.
DISCUSSION
CsrA regulates its targeted genes at the post-transcriptional level by affecting the stability or the translation of a given transcript (Romeo, 1998;Wang et al., 2005;Wei et al., 2001). In E. coli, CsrA positively regulates flagellar synthesis by increasing the stability of the flhDC transcripts (Wei et al., 2001). In contrast to E. coli, the CsrA of B. subtilis negatively regulates Hag, the major flagellin protein, by blocking the translation of the hag transcript (Yakhnin et al., 2007). The evidence presented in this report indicates that CsrABb negatively regulates the major flagellin protein FlaB of B. burgdorferi at the translational level. First, the over-expression of CsrABb substantially repressed the level of FlaB. After the induction of CsrABb with IPTG, the amount of FlaB in the csrABb+ strain was decreased by approximately 75% (Figure 3a). Second, the absence of CsrABb increased the level of FlaB by 4-fold (in the csrABb− mutant) over the wild type (Figure 3b). Third, the transcriptional analysis showed that the regulatory role of CsrABb on FlaB occurs at the post-transcriptional level, i.e., qRT-PCR analysis showed that the flaB transcript level remained unchanged in the csrABb− mutant and the csrABb+ strain induced with IPTG (Figure 5). Finally, EMSA assays of the wild type and mutated leader sequences of flaB transcript indicated that CsrABb specifically binds to the two predicted CsrA binding sites located within the UTL region of flaB transcripts (Figure 4). Taken together, these results indicate that CsrABb is a repressor of FlaB, and it most likely acts by binding to the leader sequence and consequently blocking the translation of the flaB transcript. The identified mechanism is very similar to that proposed in B. subtilis (Yakhnin et al., 2007). It is noteworthy to point out that the flaB transcript is often used as an internal control to evaluate the expressional levels of other genes, as it is often stated, based on transcript analysis, that flaB is constitutively expressed during the growth and the life cycle of B. burgdorferi (Revel et al., 2002;Gilmore, Jr. et al., 2001; Magnarelli et al., 1992). However, this conclusion needs to be tempered, as the studies reported here reveal that the translation of flaB message is under the control of CsrABb and the amount of flaB message being synthesized does not necessarily correlate with the amount of FlaB protein being made.
To determine the overall impact of CsrABb on the other flagellar proteins, the levels of FliG2 and FlgE, which represent the flagellar basal body C-ring complex and flagellar hook (Sal et al., 2008; Li et al., 2010b), respectively, were measured by western blots. The results showed that the amounts of FliG2 and FlgE remained unchanged in both the csrABb− mutant and the IPTG-induced csrABb+ strain (Figure 3). Furthermore, cryo-ET analysis indicated that the changes of CsrABb specifically influenced the length of the flagellar filaments in the csrABb− mutant and the IPTG-induced csrABb+ strain, but not the number of basal bodies per cell (Figure 6). Taken together, both western blots and structural analyses indicate that CsrABb specifically impacts PF synthesis by regulating FlaB synthesis and filament length. In the enteric bacteria, the regulation of flagellar filament length is proposed to be measured based on the rate of the secretion of FlgM (Keener, 2006). During the early synthesis of flagellar filament, the secretion of FlgM through the completed basal body-hook complex is rapid. Low intracellular concentration of FlgM allows the production of flagellin proteins from the σ28 promoter, which leads to the elongation of flagellar filament. As the flagellar filament length increases, the secretion rate of FlgM decelerates and once again results in intracellular accumulation of FlgM. This accumulation inhibits the σ28 activity, shutting off the production of the flagellin. However, such regulatory mechanism does not exist in B. burgdorferi (Charon and Goldstein, 2002;Fraser et al., 1997). The studies in this report suggest that CsrABb may play a similar role as FlgM --acting as a negative regulator of FlaB which turns off filament synthesis when flagellar assembly is completed. The synthesis of FlaB can be energetically costly, as FlaB constitutes about 10–14% of the total cellular protein of B. burgdorferi (Motaleb et al., 2004). The identified mechanism may help the spirochete to conserve energy by preventing excess FlaB protein production during the flagellar synthesis and assembly.
The change of cell morphology in the csrABb− mutant is quite intriguing. IFA results showed that the flagella helically wrapped around the wild-type cell bodies (Figure 7). A similar pattern was seen in the csrABb− mutant but with a smaller helical pitch, suggesting that the filaments in the mutant may overlap more extensively and lead to a higher number of waves with smaller amplitude than seen in the wild type. These observations were further substantiated with the cryo-ET analysis whereby the PFs wrapped around the body axis in a right-handed sense, and that the helix pitch was smaller in the csrABb− mutant than the wild type. The observed phenotype can be explained by the model first proposed by Goldstein et al. and detailed by Dombrowski et al. (Goldstein et al., 1994;Dombrowski et al., 2009). In this model, the spirochete cell is considered as an elastic object in which the PFs and the cell cylinder exert forces onto each other and cause them to deform. In the csrABb− mutant, the length of PFs is increased whereas the cell length remains unchanged (Figure 8). Thus in order to accommodate the increase length of PFs, the filaments in the mutant may change the normal ratio between the bending properties of the PFs and the cell cylinder, leading to the formation of increase wave number per cell and decrease wavelength and amplitude in the mutant.
B. burgdorferi is maintained through a complex enzootic cycle involving tick and mammalian hosts. To adapt to these different hosts, the spirochete has evolved a complex regulatory network to robustly alter its gene expression, including several hallmark virulence factors [for recent reviews, see (Rosa et al., 2005;Samuels, 2011) ]. The results reported here show that CsrABb acts as a negative regulator for flagellar assembly and motility, whereas recent studies described that CsrABb positively regulates several virulence factors of B. burgdorferi and it is required for the infectivity (Rajasekhar Karna et al., 2010;Sanjuan et al., 2009;Sze and Li, 2010). Thus, CsrABb can be an important regulator coordinating the relationship between motility and other virulence factor expression during the infection. For example, under the fed-tick condition, the levels of OspC and several other virulence factors are robustly increased (Schwan, 2003;Yang et al., 2000). In order to preferentially express these factors that are required for the spirochete to adapt to a new host, B. burgdorferi may need to transiently decrease the synthesis of FlaB. Along with this proposition, a condition mimicking tick-feeding (37°C/pH 6.8) increases the expression of CsrABb (Sanjuan et al., 2009). In addition, a recent report describes that B. burgdorferi appears to be nonmotile within the lumens of the fed ticks (Dunham-Ems et al., 2009). It is possible that CsrABb may be a factor responsible for the observed phenotype (e.g., the blood meal and/or unknown factors present in the milieu of tick midguts may trigger the expression of CsrABb, which in turn decreases the level of FlaB and consequently results in a nonmotile phenotype). We are currently investigating the role of CsrABb in tick infection and the transmission between the tick vector and the mammalian hosts. The results from these studies will further elucidate the role of CsrABb in the processes of the disease.
As a global regulator, CsrA is orchestrated with other regulators, such as alternative sigma factor RpoS, RNA chaperone Hfq, (3’-5’)-cyclic-diguanosine monophosphate (c-di-GMP) signaling system as well as bacterial two-component systems (TCS) (Babitzke and Romeo, 2007; Martinez et al., 2011;Jonas et al., 2010;Yakhnin et al., 2011). These regulators have been recently identified in B. burgdorferi and their roles in the physiology and virulence of the spirochete are beginning to be established [for recent review, see (Samuels, 2011)]. Recent studies have shown that CsrABb positively regulates RpoS (Sze and Li, 2010;Rajasekhar Karna et al., 2010), a global regulator of B. burgdorferi (Caimano et al., 2007). RpoS has been shown to interplay with several other important regulators of B. burgdorferi, including RpoN (σ54, Hubner et al., 2001;Fisher et al., 2005), Rrp2 (a two-component regulator, Yang et al., 2003), BosR (an oxidative stress regulator, Ouyang et al., 2011;Ouyang et al., 2009;Hyde et al., 2010; Boylan et al., 2003), DsrABb (a small noncoding RNA, Lybecker and Samuels, 2007) as well as HfqBb (a RNA chaperone, Lybecker et al., 2010). Thus it is very important and intriguing to further explore if the similar scenario found in E. coli and S. enterica also exists in B. burgdorferi.
In summary, although accumulating evidence suggests that CsrABb could be an important regulator of B. burgdorferi (Sze and Li, 2010;Rajasekhar Karna et al., 2010;Sanjuan et al., 2009), understanding its role and potential mechanisms involved is still in its infancy and several key questions are needed to be further addressed before reaching more comprehensive conclusions, e.g., Does CsrABb function as a global regulator as its counterpart of E. coli? Is CsrABb orchestrated with other regulatory networks? How is CsrABb activated during the enzootic cycle of B. burgdorferi? Addressing these questions can help to further enlighten our current understanding about gene regulation in B. burgdorferi.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
B. burgdorferi sensu stricto wild type B31A3 strain (a low passage virulent clone) and A3-LS (a clone derived from B31A3 carrying a lacI gene) were used in this study (Elias et al., 2002;Gilbert et al., 2007). Cells were maintained at 34°C in Barbour-Stoenner-Kelly (BSK-II) medium in the presence of 3.4% carbon dioxide. The strains were grown in the appropriate antibiotic for selective pressure as needed: kanamycin (300 µg/ml), gentamicin (40 µg/ml), and streptomycin (50 µg/ml). For the over-expression of CsrABb, 105 of stationary phase csrABb+ cells were inoculated into 10 ml of BSK II medium containing the appropriate antibiotics in the presence of a final concentration of 1 mM IPTG and cultivated until the cell density reached 108cells/ml. The E. coli TOP10 strain (Invitrogen, Carlsbad, CA) was used for DNA cloning, and the BL21 CodonPlus strain (Stratagene, La Jolla, CA) was used for the expression of the recombinant protein. The E. coli strains were cultured in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Constructing plasmids for the over-expression of csrABb
To conditionally over-express CsrABb, bb0184 was PCR amplified using primer P4/P5. A DNA fragment encoding the FLAG-tag (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) was added to the 3’ end of csrABb to assist further characterization (e.g., identifying protein /mRNA that potentially interacts with CsrABb). The obtained PCR fragment was fused downstream of the flac promoter in the vector of pTA-flacp (kindly provided by Dr. Samuels) at the engineered NdeI and BamHI restriction cut sites. The resulting flacp-csrABb fragment was further PCR amplified using primers P3/P5. The obtained PCR product was cloned into the pGEM-T-easy vector (Promega, Madison, WI) and finally cloned into the B. burgdorferi shuttle vector pBSV2G (Tilly et al., 2009) at the BamHI cut site. The resulting plasmid was named pflacpCsrA (Figure 1a) and was used to construct a strain that over-expresses CsrABb under the control of the flac promoter.
Immunoblot analysis
B. burgdorferi cells were harvested at approximately 108 cells/ml. For the western blots, the same amount of whole cell lysates (10∼50 µg) was separated on SDS-PAGE gel and transferred to PVDF membrane (Bio-Rad Laboratories, Hercules, CA). The immunoblots were probed with specific antibodies against various flagellar proteins, and DnaK was used as an internal control. Monoclonal antibodies to FlaB, FlaA, and DnaK were provided by A. Barbour (University of California, Irvine), B. Johnson (Center for Disease Control and Prevention, Atlanta) and J. Benach (State University of New York, Stony Brook), respectively. Polyclonal antibodies to FlgE and FliG2 were described in previous publications (Sal et al., 2008;Li et al., 2010b). A monoclonal antibody against the FLAG epitope (Sigma Aldrich, St. Louis, MO) was used to confirm the level of CsrABb. Immunoblots were developed using horseradish peroxidase labeled secondary antibody with an ECL luminol assay as previously described (Li et al., 2010b). Signals were quantified using the Molecular Imager ChemiDoc XRS System with the Image Lab software (Bio-Rad Laboratories).
Bacteria swarm plate assay and motion analysis
Swarm plate analysis was conducted as previously described (Li et al., 2002;Li et al., 2010b;Motaleb et al., 2000). Briefly, 5 µl of cultures (108cells/ml) was spotted onto 0.35% agarose containing BSK-II medium diluted 1:10 with Dulbecco’s PBS without divalent cations. Plates were incubated for 3–4 days at 34°C with 3.4% CO2. The diameter of the swarm ring was measured and recorded in millimeters. The wild- type B31A3 strain was used as a positive control, and a previously constructed non-motile flaB− mutant was used as a negative control to determine the initial inoculum size (Motaleb et al., 2000). The velocity of spirochetal cells was measured using a computer-based bacteria tracking system as previously described (Bakker et al., 2007). The data were statistically analyzed by one way ANOVA followed by Turkey’s multiple comparison at P< 0.01.
RNA preparation and quantitative reverse transcription PCR (qRT-PCR)
RNA samples were prepared as previously described (Sal et al., 2008;Brooks et al., 2003). Briefly, 50 ml of B. burgdorferi mid-log phase (∼ 5 × 107cells/ml) cultures were harvested, and total RNA was extracted using TRI reagent (Sigma-Aldrich) following the manufacturer’s instructions. The resultant samples were treated with Turbo DNase (Ambion, Austin, TX) at 37°C for 2 hours to eliminate genomic DNA contamination. The resultant RNA samples were re-extracted using acid-phenol:chloroform (Ambion). This was followed by precipitation in isopropanol and washing with 70% ethanol. The RNA pellets were resuspended in RNase-free water. To produce cDNA, 1 µg of RNA was reverse transcribed using AMV reverse transcriptase (Promega). The qPCR was performed using iQ SYBR Green Supermix and a MyiQ Thermal Cycler (Bio-Rad Laboratories). The transcript of the enolase gene (eno, BB0337) was used as an internal control to normalize the qPCR data as described before (Sal et al., 2008). The results were expressed as threshold cycle (CT) value. The primers used for the qRT-PCR are listed in Table 1.
Cryo-electron tomography analysis
Viable bacterial cultures were centrifuged to increase the concentration to ∼2×109 cells/ml. A 4 µl sample was deposited onto freshly glow-discharged holey carbon grids for 1 min. The grids were blotted with filter paper and rapidly frozen in liquid ethane using a gravity-driven plunger apparatus as previously described (Liu et al., 2009). The resulting frozen-hydrated specimens were imaged at −170 °C using a Polara G2 electron microscope (FEI Company, Hillsboro, OR) equipped with a field emission gun and a 4K × 4K CCD (16 megapixel) camera (TVIPS; GMBH, Germany). The microscope was operated at 300 kV with a magnification of 4,700× for taking low-magnification images along the length of individual cells. The montage of those images gave an overview of each organism. Two or three regions along the same organism were selected for further tomographic analysis at a magnification of 31,000×, resulting in an effective pixel size of 5.6 Å after 2×2 binning. The FEI “batch tomography” program was utilized to effectively collect low-dose single-axis tilt series at −8 µm defocus with a cumulative dose of ∼100 e-/Å2 distributed over 65 images, covering an angular range from −64° to +64°, with an angular increment of 2°.
3D reconstruction and visualization
Tilted images were aligned and reconstructed using package IMOD (Kremer et al., 1996). The location of each tomogram was recognized from the overview picture of each organism at low magnification. In total, 35 tomograms were generated from 9 cells of WT, 18 tomograms from 9 cells of the csrABb− mutant, and 10 tomograms from 5 cells of the IPTG-induced csrABb+ strain. These tomograms were visualized in IMOD, and then segmented manually with Amira (MCS). A sigmoid filter was applied to highlight the membranes and filaments, which were segmented and generated into non-manifold triangulated surfaces.
Immunofluorescence assay (IFA) and dark-field microscopy analysis
IFA was conducted to localize FlaB as previously described (Li et al., 2010b). Briefly, 1.5 ml cultures of wild-type B31A3, csrABb− and csrABb+ +IPTG were harvested, washed twice with PBS buffer (phosphate-buffered saline, pH 7.5), and then treated with methanol at −20°C for 1 hour. The collected cells were then treated with lysozyme (1 mg/ml) in GTE buffer (50 mM glucose, 25 mM Tris, and 1 mM EDTA, pH 7.5) for 1 hour at room temperature. Cells were then placed on poly-L-lysine-coated cover slips, and allowed to fully air dry. The obtained cover slips were first incubated in a blocking solution (2% BSA in PBS, pH 7.5) for 1 hour, followed by incubation in the blocking solution containing 1:500 diluted FlaB antibody for 1 hour at room temperature. Finally, the cover slips were washed five times with PBS, incubated with the secondary goat anti-mouse Texas red antibody (Invitrogen) for 1 hour at room temperature, washed with PBS, and mounted in 40% glycerol for image processing as described previously (Li et al., 2010b).
For the dark-field microscopy analysis, 100 µl of stationary phase (∼108 cells/ml) wild type B31A3 and csrABb− mutant cultures were harvested, washed twice with PBS buffer, and then resuspended in PBS buffer. Cells were then visualized under dark-field illumination at 100x magnification. Cell images were taken using a Zeiss Axiostar plus microscope and processed using AxioVision software (Zeiss, Germany).
Purification of recombinant CsrABb protein (rCsrABb)
Recombinant CsrABb protein was purified as previously described (Sze and Li, 2010). Briefly, the entire BB0184 gene was PCR amplified using primers P1/P2 (Table 1) and ligated to the pET101/D expression vector (Invitrogen). The resulting plasmid was then transformed into BL21 CodonPlus cells (Stratagene). The expression of the recombinant CsrABb was induced using 1 mM IPTG. The recombinant protein (rCsrABb) was purified at 4°C using HisTrap HP columns (GE Healthcare, Piscataway, NJ) as previously described (Yang et al., 2010). The final purified protein was dialyzed in a buffer (20mM Tris-HCl, 100mM KCl, 10mM MgCl2, and 25% glycerol, pH7.5) at 4°C overnight. The purified rCsrABb was used for either immunization or the electrophoretic mobility shift assay as previously described (Sze and Li, 2010).
Electrophoretic mobility shift assay (EMSA)
RNA probes were commercially synthesized (Integrated DNA Technologies, Coralville, IA) and labeled using Ambion’s BrightStar Psoralen-Biotin Nonisotopic Labeling Kit following the manufacturer’s instruction. EMSA was carried out as previously described with minor modifications (Yang et al., 2010;Dubey et al., 2005;Brencic and Lory, 2009;Baker et al., 2007). Briefly, rCsrABb, biotin-labeled RNA (19fM), 10U RNasin ribonuclease inhibitor (Promega), and 32.5 ng yeast RNA were included in 10 µl of reaction buffer (10 mM Tris-HCl, pH7.5, 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, and 10% glycerol). The reactions were incubated at 37°C for 30 min to allow the rCsrABb-RNA complex formation. Reactions were separated on 15% native polyacrylamide gels and signals were developed using Ambion’s BrightStar Nonisotopic Detection System following the manufacturer’s instruction.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Samuels for providing the plasmids and A3-LS strain, A. Barbour and J. Benach for providing antibodies, and T. Romeo for providing the E. coli csrA− mutant strain (TRMG1655) and thoughtful discussions. We also thank C. Wolgemuth for helpful discussions. This research was supported by Public Health Service grants (AI073354 and AI078958) and American Heart Association grants to C. Li; NIAID grant (AI29743) to N. Charon; NIAID grant (AI087946) and Welch Foundation grant (AU-1714) to J. Liu.
REFERENCE
- 1.Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 2.Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol. 2009;63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakker RG, Li C, Miller MR, Cunningham C, Charon NW. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl Environ Microbiol. 2007;73:1180–1188. doi: 10.1128/AEM.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci U S A. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 11.Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, et al. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombrowski C, Kan W, Motaleb MA, Charon NW, Goldstein RE, Wolgemuth CW. The elastic basis for the shape of Borrelia burgdorferi. Biophys J. 2009;96:4409–4417. doi: 10.1016/j.bpj.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, Radolf JD. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest. 2009;119:3652–3665. doi: 10.1172/JCI39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 19.Ge Y, Charon NW. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 20.Ge Y, Li C, Corum L, Slaughter CA, Charon NW. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J Bacteriol. 1998;180:2418–2425. doi: 10.1128/jb.180.9.2418-2425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge Y, Old IG, Girons IS, Charon NW. The flgK motility operon of Borrelia burgdorferi is initiated by a sigma70-like promoter. Microbiology. 1997;143(Pt 5):1681–1690. doi: 10.1099/00221287-143-5-1681. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert MA, Morton EA, Bundle SF, Samuels DS. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol Microbiol. 2007;63:1259–1273. doi: 10.1111/j.1365-2958.2007.05593.x. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore RD, Jr, Mbow ML, Stevenson B. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 2001;3:799–808. doi: 10.1016/s1286-4579(01)01435-6. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein SF, Buttle KF, Charon NW. Structural analysis of the Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J Bacteriol. 1996;178:6539–6545. doi: 10.1128/jb.178.22.6539-6545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein SF, Charon NW, Kreiling JA. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc Natl Acad Sci U S A. 1994;91:3433–3437. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmann JD. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991;5:2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- 27.Hovind-Hougen K. Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J Biol Med. 1984;57:543–548. [PMC free article] [PubMed] [Google Scholar]
- 28.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 30.Hyde JA, Shaw DK, Smith R, III, Trzeciakowski JP, Skare JT. Characterization of a conditional bosR mutant in Borrelia burgdorferi. Infect Immun. 2010;78:265–274. doi: 10.1128/IAI.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La VC, et al. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella typhimurium. Environ Microbiol. 2010;12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keener JP. How Salmonella typhimurium measures the length of flagellar filaments. Bull Math Biol. 2006;68:1761–1778. doi: 10.1007/s11538-005-9033-0. [DOI] [PubMed] [Google Scholar]
- 35.Kimsey RB, Spielman A. Motility of Lyme disease spirochetes in fluids as viscous as the extracellular matrix. J Infect Dis. 1990;162:1205–1208. doi: 10.1093/infdis/162.5.1205. [DOI] [PubMed] [Google Scholar]
- 36.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 37.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol. 2010;169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 39.Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc Natl Acad Sci U S A. 2002;99:6169–6174. doi: 10.1073/pnas.092010499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Corum L, Morgan D, Rosey EL, Stanton TB, Charon NW. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J Bacteriol. 2000a;182:6698–6706. doi: 10.1128/jb.182.23.6698-6706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Motaleb A, Sal M, Goldstein SF, Charon NW. Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol. 2000b;2:345–354. [PubMed] [Google Scholar]
- 42.Li C, Sal M, Marko M, Charon NW. Differential regulation of the multiple flagellins in spirochetes. J Bacteriol. 2010a;192:2596–2603. doi: 10.1128/JB.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C, Wolgemuth CW, Marko M, Morgan DG, Charon NW. Genetic analysis of spirochete flagellin proteins and their involvement in motility, filament assembly, and flagellar morphology. J Bacteriol. 2008;190:5607–5615. doi: 10.1128/JB.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Xu H, Zhang K, Liang FT. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol Microbiol. 2010b;75:1563–1576. doi: 10.1111/j.1365-2958.2010.07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- 48.Lybecker MC, Abel CA, Feig AL, Samuels DS. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2010;78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 50.Macnab RM. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 51.Magnarelli LA, Fikrig E, Berland R, Anderson JF, Flavell RA. Comparison of whole-cell antibodies and an antigenic flagellar epitope of Borrelia burgdorferi in serologic tests for diagnosis of Lyme borreliosis. J Clin Microbiol. 1992;30:3158–3162. doi: 10.1128/jcm.30.12.3158-3162.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol. 2009;392:511–528. doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mofunanya A, Li FQ, Hsieh JC, Takemaru K. Chibby forms a homodimer through a heptad repeat of leucine residues in its C-terminal coiled-coil motif. BMC Mol Biol. 2009;10:41. doi: 10.1186/1471-2199-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriarty TJ, Norman MU, Colarusso P, Bankhead T, Kubes P, Chaconas G. Real-time high resolution 3D imaging of the lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS Pathog. 2008;4:e1000090. doi: 10.1371/journal.ppat.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Motaleb MA, Sal MS, Charon NW. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J Bacteriol. 2004;186:3703–3711. doi: 10.1128/JB.186.12.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motaleb MA, Sultan SZ, Miller MR, Li C, Charon NW. CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. J Bacteriol. 2011;193:3332–3341. doi: 10.1128/JB.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol. 2008;67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norris SJ, Charon NW, Cook RG, Fuentes MD, Limberger RJ. Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J Bacteriol. 1988;170:4072–4082. doi: 10.1128/jb.170.9.4072-4082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang Z, Deka RK, Norgard MV. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 2011;7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol. 2009;74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajasekhar Karna SL, Sanjuan E, Esteve-Gassent MD, Miller CL, Maruskova M, Seshu J. CsrABb modulates levels of lipoproteins and key regulators of gene expression (RpoS and BosR) critical for pathogenic mechanisms of Borrelia burgdorferi. Infect Immun. 2010;79:732–744. doi: 10.1128/IAI.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 66.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosa PA, Tilly K, Stewart PE. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol. 2005;3:129–143. doi: 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- 68.Sadziene A, Thomas DD, Bundoc VG, Holt SC, Barbour AG. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J Clin Invest. 1991;88:82–92. doi: 10.1172/JCI115308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sal MS, Li C, Motalab MA, Shibata S, Aizawa S, Charon NW. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J Bacteriol. 2008;190:1912–1921. doi: 10.1128/JB.01421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samuels DS. Gene Regulation in Borrelia burgdorferi. Annu Rev Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 71.Sanjuan E, Esteve-Gassent MD, Maruskova M, Seshu J. Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi. Infect Immun. 2009;77:5149–5162. doi: 10.1128/IAI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwan TG. Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi. Biochem Soc Trans. 2003;31:108–112. doi: 10.1042/bst0310108. [DOI] [PubMed] [Google Scholar]
- 73.Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol. 2010;77:128–142. doi: 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szczepanski A, Furie MB, Benach JL, Lane BP, Fleit HB. Interaction between Borrelia burgdorferi and endothelium in vitro. J Clin Invest. 1990;85:1637–1647. doi: 10.1172/JCI114615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sze C, Li C. Inactivation of bb0184 that encodes a carbon storage regulator A represses the infectivity of Borrelia burgdorferi. Infect Immun. 2010;79:1270–1279. doi: 10.1128/IAI.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 78.Tilly K, Bestor A, Dulebohn DP, Rosa PA. OspC-independent infection and dissemination by host-adapted Borrelia burgdorferi. Infect Immun. 2009;77:2672–2682. doi: 10.1128/IAI.01193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Timmermans J, Van ML. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 81.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 82.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol. 2007;64:1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 83.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, Babitzke P. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by Esigma(70) and Esigma(S) , and indirect transcriptional activation by CsrA. Mol Microbiol. 2011;81:689–704. doi: 10.1111/j.1365-2958.2011.07723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang TY, Sung YM, Lei GS, Romeo T, Chak KF. Posttranscriptional repression of the cel gene of the ColE7 operon by the RNA-binding protein CsrA of Escherichia coli. Nucleic Acids Res. 2010;38:3936–3951. doi: 10.1093/nar/gkq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 86.Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, Li C. Transcription and genetic analyses of a putative N-acetyl-muramyl-L-alanine amidase in Borrelia burgdorferi. FEMS Microbiol Lett. 2009;290:164–173. doi: 10.1111/j.1574-6968.2008.01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.