Summary
The compound eye of the fruit fly, Drosophila melanogaster, has for decades been used extensively to study a number of critical developmental processes including tissue development, pattern formation, cell fate specification and planar cell polarity. To a lesser degree it has been used to examine the cell cycle and tissue proliferation. Discovering the mechanisms that balance tissue growth and cell death in developing epithelia has traditionally been the realm of those using the wing disc. However, over the last decade a series of observations has demonstrated that the eye is a suitable and maybe even preferable tissue for studying tissue growth. This review will focus on how growth of the retina is controlled by the genes and pathways that govern the specification of tissue fate, the division of the epithelium into dorsal-ventral compartments, the initiation and progression of the morphogenetic furrow and the second mitotic wave.
Keywords: Drosophila, retina, cell proliferation, growth
1. Introduction
Growth of the Drosophila retina occurs in two waves during development. The first mitotic wave, as the initial burst of growth is called, is a continuous process that begins prior to the initiation of the morphogenetic furrow and continues in regions of the disc that lie anterior to the advancing differentiating front. By contributing to the number of cells within anterior regions of the eye, this first mitotic wave essentially sets the upper limit on how many ommatidia can be generated. Several developmental events such as tissue specification, midline formation and movement of the furrow itself will contribute to the overall impact that the first mitotic wave has on tissue growth. The second epoch of proliferation occurs posterior to the furrow and is called the second mitotic wave. This tight band of cell division is responsible for generating a subset of photoreceptor neurons as well as all accessory cone and pigment cells. The second mitotic wave gives birth to the majority of cells that comprise the eye and thus is key to setting the final overall size of the retina.
Studying tissue growth in the eye, particularly that of the first mitotic wave, presents a challenge since the cellular events that promote growth are not always cleanly separated in developmental time or geographical space. For example, cells within the first and second instar eye disc are simultaneously adopting a retinal fate while being sorted into dorsal and ventral compartments. As we will see later, both of these processes are important influences on the first mitotic wave. An equally important consideration is that the factors identified as promoting tissue identity are the very same ones establishing the midline. This can sometimes make it difficult to unambiguously attribute an increase or decrease in growth rates to either developmental process. Examinations of cell proliferation within the second mitotic wave presents similar challenges as several pathways that promote cell proliferation are also important for cell fate decisions in adjacent cells. How alterations in cell fates affect cell proliferation in surrounding cells is a difficult question to answer with absolute confidence.
Interestingly though, the very issues that make studying cell proliferation in the retina a challenge are what make the developing fly eye an attractive system for studying tissue growth. We can take advantage of the integrative nature of eye development to uncover the mechanisms by which populations of cells simultaneously receive, sort, interpret and execute multiple growth signals. The asynchronous development of the eye also provides us with an opportunity to analyze the mechanisms by which a tissue recalibrates its rate of growth in real time. In this review I will attempt to document mechanisms by which retinal growth is promoted by the first and second mitotic waves.
2. Growth of Imaginal Discs: Unequal Beginnings
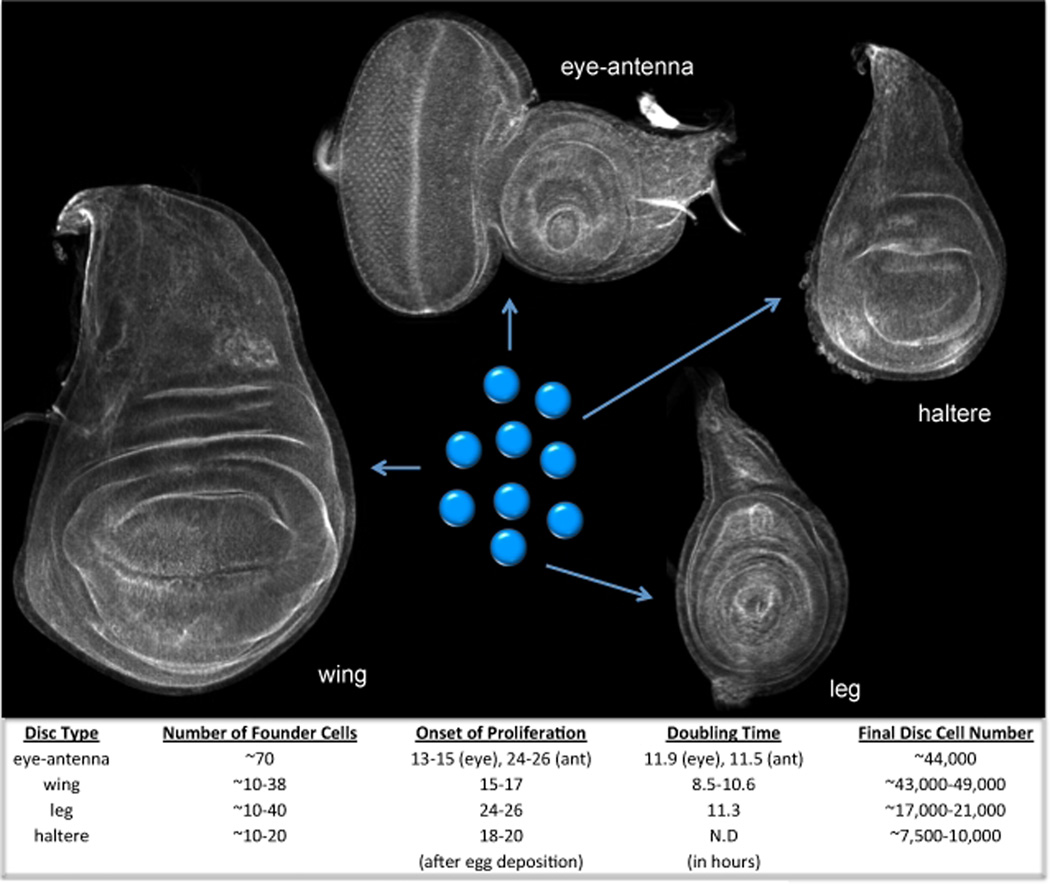
Adult compound eyes, like all other adult Drosophila structures, are derived from monolayer epithelia called imaginal discs. These epithelial sheets arise during embryogenesis when small numbers of cells are set aside from the rest of the developing embryo and begin developing asynchronously from the rest of the organism (Miall and Hammond, 1892; Kellog, 1902; Essa, 1953; Anderson, 1972a,b; Crick and Lawrence, 1975; Cohen, 1993; Held, 2002). Reflecting the differing sizes of adult appendages, each type of mature imaginal disc also contains distinct numbers of cells (Becker, 1957; Garcia-Bellido et al., 1970; Garcia-Bellido and Merriam, 1971; Morata and Garcia-Bellido, 1976; Steiner, 1975; Steiner, 1976; Martin, 1982). The different final organ sizes are achieved in part through three initial inequalities in growth control (Figure 1). First, distinct numbers of cells are set-aside during embryogenesis for each type of imaginal disc (Bryant and Schneiderman, 1969; Postlethwait and Schneiderman, 1971; Wieschaus and Gehring, 1976; Lawrence and Morata, 1977; Madhaven and Schneiderman, 1977). Second, the onset of the cell proliferative period differs for each type of disc (Madhaven and Schneiderman, 1977). And third, the cell-doubling rate is different for each kind of disc (Patterson, 1929; Bryant, 1970; Garcia-Bellido and Merriam, 1971; Postlethwait and Schneiderman, 1977; Haynie, 1975; Morata and Garcia-Bellido, 1976; Steiner, 1976; Madhaven and Schneiderman, 1977). Together, the inequality in starting cell numbers and the unique starting time and rate of proliferation allows for the eye disc to achieve a final size that is sufficient for the formation of approximately 800 ommatidia and accessory mechanosensory bristle complexes (Figure 1).
Figure 1. Tissue Growth in the Imaginal Discs of Drosophila.
This figure includes confocal images of the eye-antennal, wing, haltere and leg discs of Drosophila. The discs have been stained with phalloidin, which marks F-actin, so that the outlines of each disc can be seen. Below are the starting and final number of cells of each disc as well as the doubling times and onset of cell proliferation times.
3. Retinal Determination Genes as Regulators of Growth
Tissue selector genes commit the small clusters of cells that are set-aside during embryogenesis to adopting distinct organ fates. Once the initial fate decisions are made, then growth control mechanisms take over and generate the appropriate numbers of cells. For example, during the final larval instar stage, the eye-antennal, wing, haltere and leg discs contain approximately 44,000, 49,000, 10,000 and 21,000 cells respectively (Becker, 1957; Garcia-Bellido et al., 1970; Garcia-Bellido and Merriam, 1971; Morata and Garcia-Bellido, 1976; Steiner, 1975; Steiner, 1976; Martin, 1982). Interestingly, mutations that alter the fate of the entire organ or tissue also reset cell proliferation targets. For example, gain-of-function Antp mutations and loss-of-function Ubx alleles are characterized by the conversion of antenna and halteres to legs and wings respectively (Lewis, 1978; Schneuwly et al., 1986; Schneuwly et al., 1987a,b). The ectopic organs are indistinguishable in size when compared to normal wings and legs. Thus, the initial act of committing tissues to defined fates sets in motion a series of developmental steps that ensure a degree of growth that is commensurate with the assigned tissue fate.
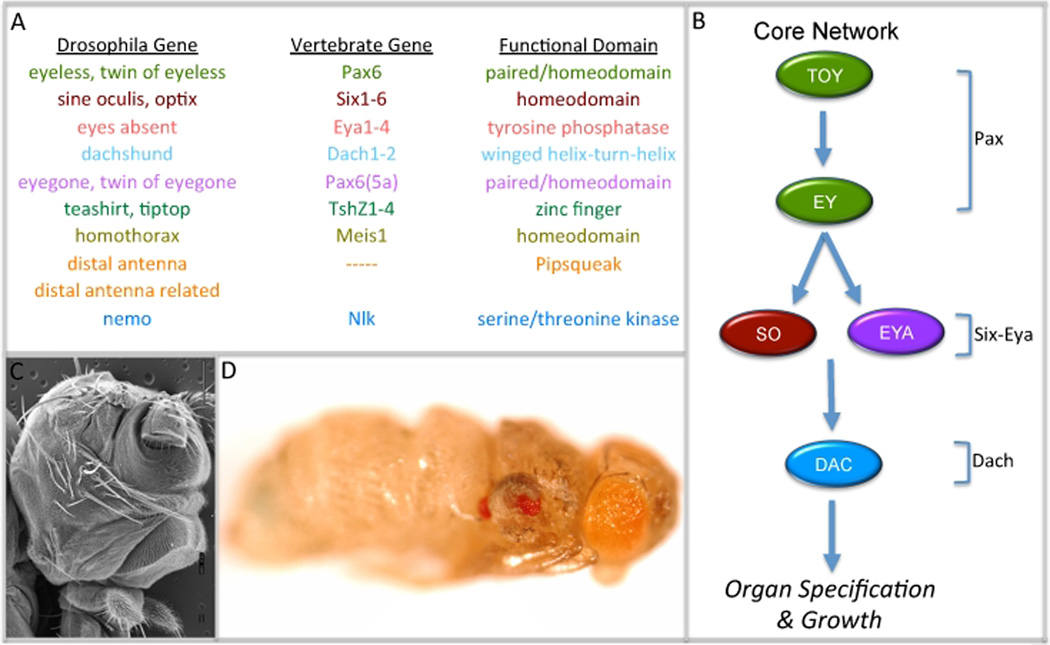
In the eye primordium, tissue specification is governed by a set of interlocking selector genes collectively termed the eye specification or retinal determination network (Figure 2A). Included within the network are the Pax6 genes eyeless (ey: Quiring et al., 1994) and twin of eyeless (toy: Czerny et al., 1999), the Pax6-like factors eyegone (eyg: Jang et al., 2003) and twin of eyegone (toe: Aldaz et al., 2003), the Six family members sine oculis (so: Cheyette et al., 1994; Serikaku and O’Tousa 1994) and optix (Seimiya and Gehring, 2000), the zinc finger transcription factors teashirt (tsh: Pan and Rubin, 1998) and tiptop (tio: Bessa et al., 2009; Datta et al., 2009), the pipsqueak DNA binding proteins distal antenna (dan: Curtiss et al., 2007) and distal antenna related (danr: Curtiss et al., 2007), the transcriptional co-activator and protein phosphatase eyes absent (eya: Bonini et al., 1993), a homolog of the Ski/Sno oncogenes dachshund (dac: Mardon et al., 1994), and the Nlk homolog nemo (Braid and Verheyen, 2008). A large body of experimental evidence has demonstrated that these genes regulate eye development by functioning through a labyrinth of complex genetic, molecular and biochemical interactions (Figure 2B).
Figure 2. The Retinal Determination Network: Genes, Pathway, Loss and Gain-of-Function Phenotypes.
(A) List of retinal determination genes in flies and vertebrates. (B). Schematic of the core retinal determination pathway. (C) Scanning electron micrograph of an eya2 adult mutant head. (D) Light microscope image of an adult fly in which eyeless has been mis-expressed within the halteres and wings. Note the red ectopic eyes.
These retinal determination genes share two unique and special characteristics. First, loss-of-function mutations often lead to a near complete loss of retinal tissue (Figure 2C; Bonini et al., 1993; Cheyette et al., 1994; Mardon et al., 1994; Quiring et al., 1994; Serikaku and O’Tousa, 1994). The loss of retinal identity is accompanied by a severe inhibition of tissue growth and a dramatic increase in programmed cell death. (Bonini et al., 1993; Cheyette et al., 1994; Mardon et al., 1994; Quiring et al., 1994). As a consequence, the larval disc is significantly smaller than wild type and the morphogenetic furrow fails to initiate thereby giving rise to an adult that is missing its compound eyes. As we will see below, the reduction in size is not a non-specific effect. Several retinal determination genes are tasked primarily with promoting cell proliferation within the eye disc (Singh et al., 2002; Chao et al., 2004; Dominguez et al., 2004; Singh et al., 2004; Lopes and Cesaras, 2009; Peng et al., 2009). A few are even involved in the direct regulation of cell cycle checkpoints (Jemc and Rebay, 2007).
The genes that comprise this network are also considered special because forced expression in non-retinal tissues is sufficient to coax populations of cells into forgoing their initial fate and into adopting an eye fate (Figure 2D; Halder et al., 1995; Bonini et al., 1997; Shen and Mardon 1999; Cznery et al., 1999, Seimiya and Gehring, 2000; Jang et al., 2003; Curtiss et al., 2007; Weasner et al., 2007; Braid and Verheyen, 2008; Yao et al., 2008; Bessa et al., 2009; Datta et al., 2009). Unlike homeotic genes, expression of retinal determination genes is not capable of transforming an entire tissue. Only small numbers of cell populations within imaginal discs have enough developmental plasticity to adopt an eye fate (Salzer et al., 2010). Thus, in these forced expression assays, the retinal determination genes can only influence growth locally.
While the network is organized into a loose hierarchy, expression of genes at the top of the network is often dependent upon factors that lie at lower levels (reviewed in Kumar, 2010). Thus, the small disc phenotype that is seen in loss-of-function mutants does not necessarily mean that every member of the retinal determination network promotes cell proliferation. ey is a case in point. It is one of only four network genes that are expressed during the early embryonic growth phase of the eye primordium. Loss of this expression leads to severe reductions in the size of the eye disc (Quiring et al., 1994). Forced expression of ey can also partially rescue the growth defects of eyg mutants (Jang et al., 2003). Together these observations initially suggested that ey promotes growth of the retina. However, an insightful paper by Dominguez and colleagues revealed that eyg and to a lesser degree its paralog toe are the true growth promoters in the retina (Dominguez et al., 2004). It had been previously established that the Notch pathway is a major inducer of cell proliferation in the retina (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). The authors demonstrated that only forced expression of eyg, toe and their mammalian homolog Pax6(5a) were capable of rescuing the growth defects of Notch (N) mutants, In contrast, expression of ey and its paralog toy failed to restore growth the mutant discs (Dominguez et al., 2004).
Over the years several retinal determination genes have been implicated in directly regulating growth. These include so (Cheyette et al., 1994; Serikaku and O’Tousa, 1994), eya (Bonini et al., 1993), hth (Pai et al., 1997), tsh (Pan and Rubin, 1998) as well as eyg (Jun et al., 1998; Jang et al., 2003). The role of eyg in promoting growth appears to be related to midline formation and will be discussed below. The part that the remaining genes play in inducing cell proliferation appears tied to their roles in tissue specification and will be discussed here.
3.1 Teashirt and Homothorax: The RD Network Meets the Hippo Pathway
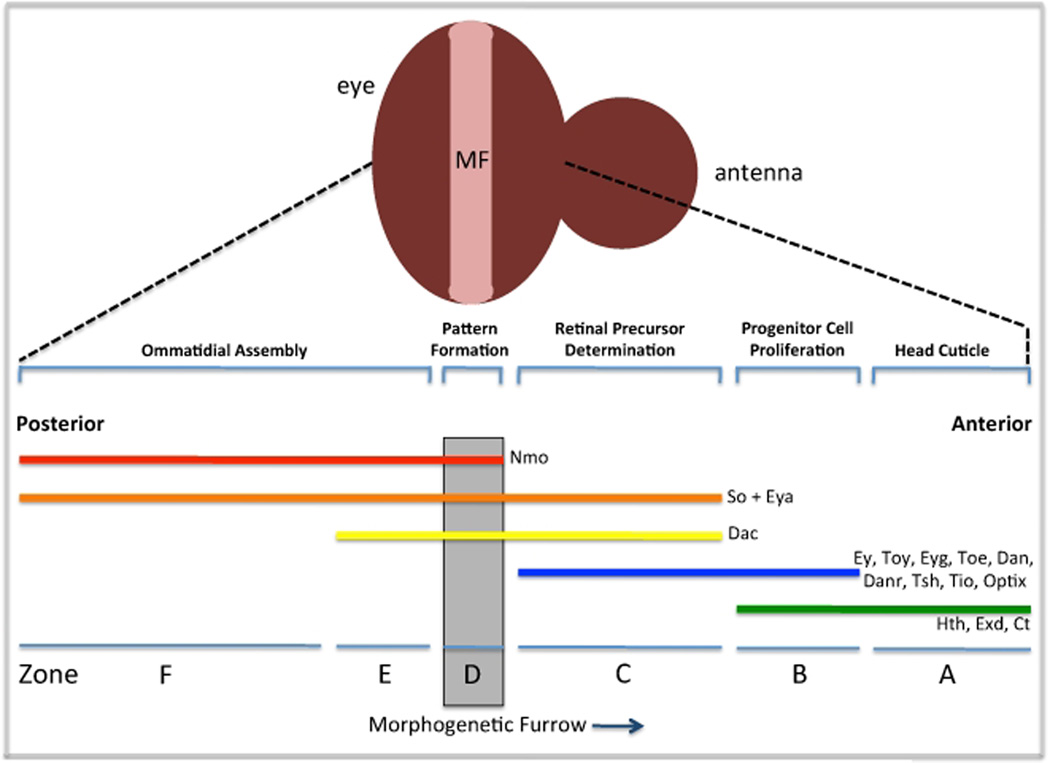
Several lines of evidence support roles for both hth and tsh in promoting growth in the developing eye. First, cells mutant for hth rarely survive in the anterior eye (Pichaud and Casares, 2000; Bessa et al., 2002) while cells over-expressing tsh overgrow (Singh et al., 2002). The genes are functionally dependent upon each other as expression of tsh leads to the induction of hth transcription (Singh et al., 2002) and as Hth and Tsh physically interact with each other (along with Ey) to repress the expression of downstream transcriptional targets (Bessa et al., 2002). Insight into potential mechanisms for tsh and hth in regulating growth came initially from an analysis of the expression patterns of each retinal determination network member (Bessa et al., 2002). Based on distinct transcriptional profiles the eye field can be divided into several regions (zone A–F) with the anterior portion of the eye occupying zones A–C (Figure 3; reviewed in Kumar, 2010).
Figure 3. Expression Patterns of the Retinal Determination Genes in the Developing Fly Eye.
Schematic diagram depicting the expression patterns of the retinal determination genes. Anterior is to the right.
Along the eye/antenna border (zone A) the only retinal determination gene that is expressed is hth along with its co-factor extradenticle (exd: Pai et al., 1997; Pichaud and Casares, 2000; Bessa et al., 2002). These cells are fated to adopt a head capsule fate and do not contribute to growth of the eye (Haynie and Bryant, 1986). Indeed, loss of either hth or exd within these cells leads to the formation of ectopic eyes with little effect on growth (Rauskolb et al., 1995; Pai et al., 1997; Gonzales-Crespo et al., 1995).
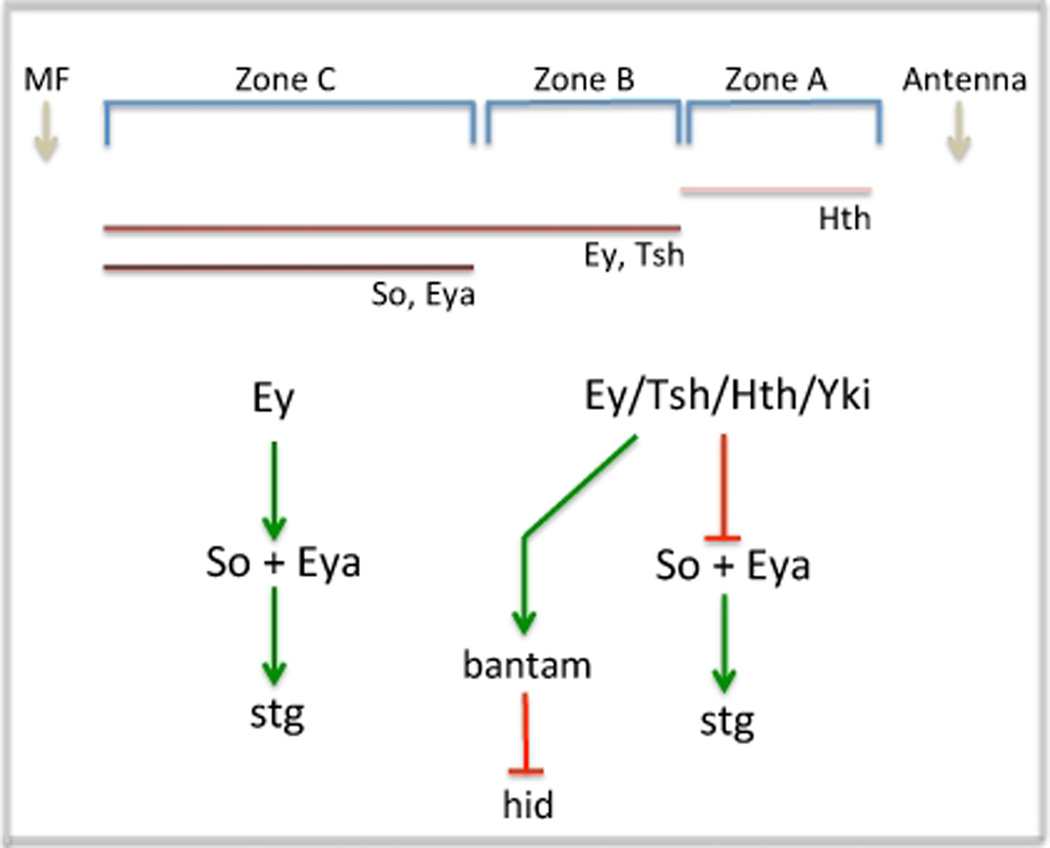
Just adjacent lies a narrow swathe of cells (Zone B) that are locked into a highly proliferative state. Several retinal determination genes including ey/toy (Quiring et al., 1994; Czerny et al., 1999), eyg/toe (Aldez et al., 2003; Jang et al., 2003), hth (Bessa et al., 2002), tsh/tio (Pan and Rubin 1998; Bessa et al., 2009; Datta et al., 2009) and dan/danr (Curtiss et al., 2007) are expressed within this tract of cells. One key to maintaining cells within this zone in a proliferative state is the repression of three retinal determination genes: so, eya and dac, a task that appears to be accomplished by the Tsh-Hth-Ey complex described above (Bessa et al., 2002). A consequence of this repression in Zone B that is relevant to tissue growth is down-regulation of string (stg), the fly homolog of yeast cdc25 and regulator of the G2/M transition. In zones A and B where hth is expressed stg expression is absent. In contrast, as hth expression is eliminated in cells just ahead of the furrow (zone C) stg expression is activated. A genetic relationship between these two genes exists as clones lacking hth show ectopic stg expression (Lopes and Casares, 2009). But hth does not appear to promote tissue growth through direct regulation of stg. As we will see below that appears to be the task of the so and eya genes. In cells closest to the morphogenetic furrow (Zone C) hth is lost and as a result the repression of so, eya and dac is relieved. The initiation of so and eya transcription will lead to the activation of stg (Jemc and Rebay, 2007) which is a key step in preparing cells for entrance into the furrow where they will undergo a genomic switch that will transition them to a state of terminal differentiation (Figure 4; Jasper et al., 2002).
Figure 4. Model for RD Network Control of the Retinal Cell Cycle.
In this model cell proliferation in the most anterior segments of the eye field is achieved by the activation of bantam, a microRNA that blocks translation of hid RNA, and transcriptional repression of both so and eya, two genes required for the activation of stg. In zone C cells, the So-Eya complex activates stg thereby moving cells through the cell cycle so that they can be arrested in G1 within the furrow.
In addition to the regulation of the cell cycle via stg, a recent study demonstrated that tsh and hth promote tissue growth through interactions with the Hippo tumor suppressor pathway (Peng et al., 2009). Hippo signaling is essential for correct growth control in both Drosophila and mammals (Dong et al., 2007; Pan, 2007; Halder and Johnson, 2011). The target of this pathway is yorkie (yki), a transcriptional co-activator (Huang et al., 2005; Dong et al., 2007, Oh and Irvine, 2008; Zhang et al., 2008). In its unphosphorylated state, Yki is localized to the nucleus and participates in the activation of growth promoting genes. One well-established binding partner is the TEAD/TEF transcription factor Scalloped (Sd: Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008). As loss of sd has little to no effect in the retina, the proliferative effects that Yki has in the eye must be mediated through other DNA binding proteins. The identity of that binding partner came to light when it was discovered that tissue overgrowths induced by simultaneous expression of tsh and hth are mediated by the Hippo pathway. Hth does indeed form a biochemical complex with Yki. The Hth-Yki transcription factor binds to an enhancer within the bantam (ban) microRNA locus thereby activating its expression (Peng et al., 2009). ban is known to block cell death by binding to the 3` UTR of head involution defective (hid) mRNA thereby inhibiting its translation (Brennecke et al., 2003). ban has been previously identified as a genetic target of the Hippo pathway in the wing (Nolo et al., 2006; Thompson and Cohen, 2006). It now is considered a molecular target of both Hippo and the RD network in the eye (Figure 4).
3.2 The So-Eya Complex: Talking to the Cell Cycle
The loss of either so or eya, the founding members of the Six and Eya protein families, in the Drosophila retina is characterized by a dramatic loss of the adult compound eyes. This loss in tissue identity is accompanied by a massive increase in programmed cell death thereby resulting in an eye disc that is only a fraction of normal size (Milani 1941; Milani 1951; Bonini et al., 1993; Cheyette et al., 1994; Serikaku and O’Tousa, 1994). However, simple over-expression of so or its paralogs, DSix4 and optix, does not lead to tumorigenesis in flies. This is in contrast to the dramatic effects on tissue growth that are seen when members of the Six or Eya families are mis-regulated in vertebrates. For example, over-expression of Six3, a vertebrate homolog of so, induces enlargement of the zebrafish rostral forebrain (Kobayashi et al., 1998). And inappropriate levels of several Six and Eya family members are observed in a multitude of human cancers (reviewed in Christensen et al., 2008). In total, these results suggest that Six and Eya family members function as cell survival factors as well as growth promoters.
So is a homeodomain-containing DNA binding protein (Cheyette et al., 1994; Serikaku and O’Tousa, 1994) while Eya is a transcriptional co-activator and protein tyrosine phosphatase (Pignoni et al., 1997; Xu et al., 1997; Ohto et al., 1999; Li et al., 2003; Silver et al., 2003; Rayapureddi et al., 2003; Tootle et al., 2003). Through direct physical interactions these proteins form a composite transcription factor (Pignoni et al., 1997) which initiates the expression of other retinal determination network members, genes that are involved in cell fate specification, as well as factors that control the movement of the morphogenetic furrow (Suzuki and Saigo, 2000; Yan et al., 2003; Pappu et al., 2005; Pauli et al., 2005; Zhang et al., 2006; Tanaka-Matakatsu and Du, 2008). Evidence from both flies and vertebrates indicates that the So-Eya complex influences cell proliferation by directly regulating several cell cycle components. In human cells, Six1 is known to regulate c-myc, cyclin D1, cyclin A1 (Li et al., 2003; Coletta et al., 2004; Yu et al., 2006). In the fly retina the So-Eya complex has been shown to bind to an enhancer element within the stg locus (Edgar and O’Farrell, 1989; Jemc and Rebay, 2007). stg is expressed in a tight band just ahead of the morphogenetic furrow and is thought to shepherd the asynchronously dividing cells ahead of the furrow through G2 and into mitosis (Thomas et al., 1994). This is thought to be important for synchronizing cells within the furrow where they are arrested in G1 (Ready et al., 1976; Horsfield et al., 1998; Escudero and Freeman, 2007; Firth et al., 2010). The loss of stg arrests cells in G2 and prevents them from entering mitosis and results in the over-recruitment of photoreceptor and cone cells (Mozer and Easwarachandran, 1999). Thus, one function of the So-Eya complex may be to synchronize cells at the G1 phase by activating and maintaining cdc25/stg expression ahead of the furrow (Figure 4).
4. The Midline: Growth from the Center
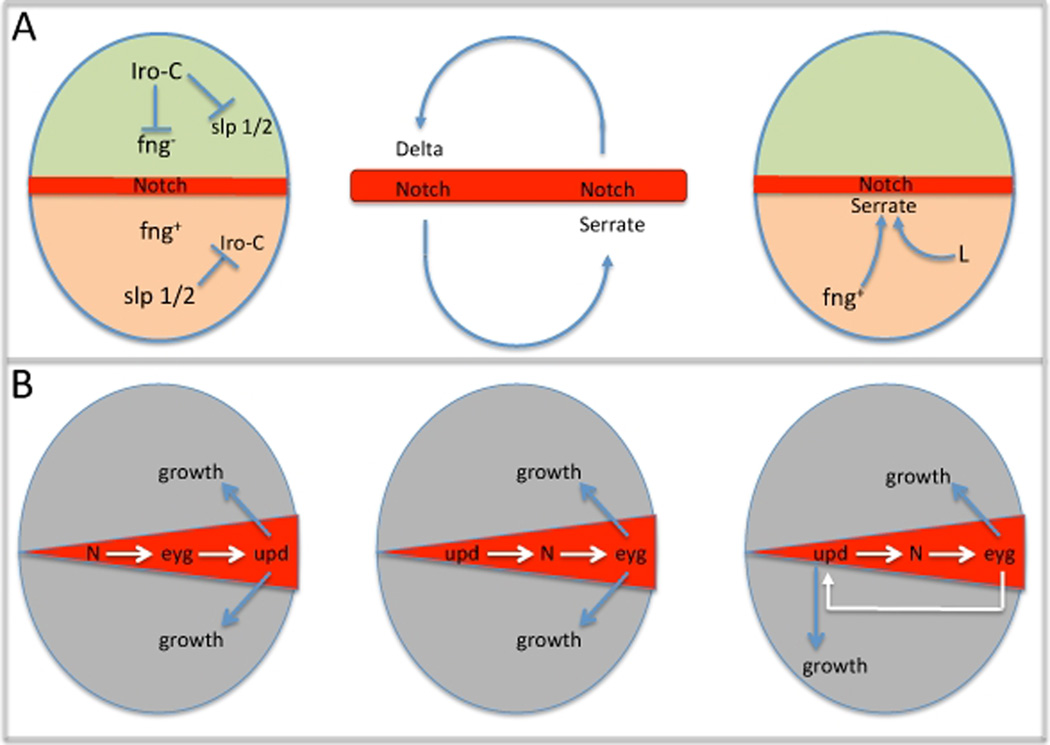
The division of a homogeneous tissue into broad compartmental domains is one of the first steps towards generating a system with multiple cell types. The Drosophila eye is initially borne with only a ventral address (Singh et al., 2003). This homogeneity is broken during the first larval instar when a dorsal fate is imposed on a subset of the eye field thereby creating two distinct retinal zip codes: ventral and dorsal (Maurel-Zaffran and Treisman, 2000; Singh et al., 2005). At the molecular level, the dorsal and ventral fates arise by the juxtaposition of selector genes (Iro-C and Slp1/2) whose expression patterns meet at but never cross the midline (Figure 5A, left panel; McNeill et al., 1997; Sato and Tomlinson, 2007). As a consequence, ventral cells express fringe (fng) while dorsal cells do not (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). The confrontation of fng+ and fng− cells is conserved although the orientation in the eye is inverted when compared to other tissues such as the wing (Irvine and Wieschaus, 1994). As a result, cells at the midline undergo Notch dependent tissue growth. Ubiquitous expression of fng throughout the eye, or the loss of Notch signaling blocks growth and the resulting eye discs are nearly indistinguishable from those that harbor mutations within the retinal determination network (Cagan and Ready, 1989; Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998). In contrast, activation of Notch signaling leads to tissue over-growth (de Celis et al., 1998; Go et al., 1998; Reynolds-Kenneally and Mlodzik, 2005).
Figure 5. Notch Signaling at the Midline Promotes Growth.
(A) Schematics depicting the activation of Notch at the midline, the amplification of the Notch signal and the activation of the Ser ligand within the ventral half of the eye. (B) Models depicting the genetic hierarchy of Notch mediated growth in the eye. These models include the activities of Notch, eyegone and JAK/STAT signaling.
4.1 Induction and Maintenance of Notch Signaling at the Midline
In Drosophila, the ligands for the N receptor are encoded by Delta (Dl) and Serrate (Ser: Dexter, 1914; Fehon et al., 1990; Fleming et al., 1990; Rebay et al., 1991; Thomas et al., 1991; Lieber et al., 1992; de Celis et al., 1993; Kooh et al., 1993). In the wing, where Notch signaling has been extensively studied, Dl and Ser are expressed in complementary patterns with the former restricted to the ventral compartment and the latter confined to dorsal territories (Doherty et al., 1996). In the eye the situation is reversed with Dl being present in the dorsal domain and Ser present in the ventral compartment (Cho and Choi, 1998; Cho et al., 2000). In both tissues the expression and activation of the N receptor is at its highest along the dorso-ventral boundary (Huppert et al., 1997). The juxtaposition of ligands on either side of the midline creates a positive-feedback loop that maintains Notch signaling across the midline and as a consequence promotes growth in the retina (Figure 5A, middle panel).
During development the patterns of N, Dl and Ser expression can vary considerably. At certain stages, N expression will be coincident with Dl and Ser while at other stages the receptor will be expressed in a complementary pattern to its ligands (Kooh et al., 1993). In the eye, cells along the dorsal side of the midline co-express N and Dl while their counterparts along the ventral side of the midline co-express N and Ser. Similar receptor-ligand relationships are also seen in the developing wing and are thought to mediate a positive-feedback loop between cells that straddle the D/V border (Diaz-Benjumea, 1995; Huppert et al., 1997). In the eye the model is that Ser expression in the ventral compartment signals across the midline to induce Notch-dependent activation of Dl transcription. Dl, in the dorsal compartment, in turn signals back across the midline to activate Ser expression via N. As Dl and Ser are short range signaling molecules this reinforcing feedback mechanism has the effect of increasing N signaling just at the midline (Figure 5A, middle panel).
A role for the Ser ligand in the activation of N-dependent growth came from demonstrations that loss-of-function Ser mutants had small eyes (Speicher et al., 1994; Go et al., 1998). There was immediate interest in identifying the mechanisms that activated Ser expression. Its restriction to the ventral compartment suggested the upstream regulators would themselves either be expressed or function only within the ventral half of the eye. Earlier reports had proposed that, in the wing, fng could function as an upstream positive regulator of Ser (Diaz-Benjumea and Cohen, 1993; Kim et al., 1995). This also turned out to be case in the eye as loss of fng within the ventral compartment results in a down regulation of Ser expression (Cho and Choi, 1998). Another regulator of Ser expression turned out to be the product of the Lobe (L) gene. L mutants were first identified by the presence of a small eye (Morgan et al., 1925). And although L is expressed in both dorsal and ventral compartments the tissue that is lost is exclusively of ventral identity. In clones that lack L, Ser expression is also down regulated (Chern and Choi, 2002). Taken together, the evidence so far indicates that the selective induction of Ser in the ventral half of the eye is due to the activities of fng and L (Figure 5A. right panel).
4.2 Eyegone: At the Intersection of Growth Control and the RD Network
Pax genes encode transcription factors that are defined by the presence of two DNA binding domains: a Paired domain and a Homeodomain. The N-terminal (PAI) and C-terminal (RED) segments of the Paired domain can each interact with DNA when expressed separately but in the context of an intact Paired domain binding will occur pre-dominantly through the PAI domain. The RED domain becomes the predominant DNA binding domain only when the PAI domain is either removed or its function blocked (Czerny et al., 1993; Epstein et al., 1994; Kozmik et al., 1997). In flies, this situation manifests itself in the form of two Pax6-like proteins, Eyg and Toe, both of which contain only an intact RED domain (Jones et al., 1998; Jun et al., 1998; Jang et al., 2003; Aldaz et al., 2003). Similar situations exist in vertebrates as alternate splicing events in the Pax3, Pax6 and Pax8 genes generate protein isoforms that all contain a disrupted and inactive PAI domain (Cnerny et al., 1993; Epstein et al., 1994; Kozmik et al., 1997; Chi and Epstein, 2002).
Adult flies mutant for eyg display retinal phenotypes that range from having small to no compound eyes. This is reflected in the below average size of third instar larval eye disc (Jang et al., 2003). Forced over-expression of either eyg or toe induces tissue proliferation in areas of the surrounding head capsule that already expresses retinal determination factors but cannot induce ectopic eye formation in tissues such as the wing, leg and halteres (Jang et al., 2003; Yao et al., 2008). Taken together these results suggest that Eyg and Toe may play significant roles in promoting growth in the eye.
A link between eyg and Notch-dependent growth was first intimated by a comparison of expression patterns. At the midline eyg expression overlaps that of Notch signal activation (Jang et al., 2003). The same is true of toe, whose expression is identical to eyg (Yao et al., 2008). A functional connection between both Pax6-like proteins and Notch activation came from a pair of manuscripts showing that forced expression of either eyg or toe is sufficient to rescue the growth defects associated with loss of N. Similarly, loss of either Pax6-like gene could suppress the increased growth that results from hyper-activation of the Notch pathway (Chao et al., 2004; Dominguez et al., 2004). These reports also showed that Notch signaling induced expression of eyg. Of all the retinal determination genes, only eyg and toe mediate Notch-dependent induction of cell proliferation in the retina. Despite the small disc phenotype seen in ey mutants, forced expression of ey (and its paralog toy) fail to rescue the growth defects of N mutants (Dominguez et al., 2004). As ey lies genetically and molecularly upstream of both so, eya and dac it is unlikely that any one of these factors would rescue N mutants. Thus the available data so far suggests that Notch signaling promotes growth in the retina via the transcriptional activity of the Pax6-like factors Eyg and Toe (Figure 5B, left panel).
4.3 From Notch to the JAK/STAT Pathway
The Janus Kinase (JAK)/Signal Transducers and Activators of Transcription (STAT) signaling pathway is thought to be a global regulator of tissue growth. Eye discs devoid of the unpaired (upd) ligand have small eyes while forced expression leads to increased cell proliferation (Chen et al., 2002; Bach et al., 2003; Chen et al., 2003; Tsai and Sun, 2004). Like N and eyg, upd is expressed at the midline although its transcription is restricted to its intersection with the posterior margin (Tsai and Sun, 2004). This localized area is called “the firing point” because it is from here that the morphogenetic furrow initiates. One school of thought suggests that N/eyg-dependent growth is mediated by JAK/STAT signaling while another supports a model in which the JAK/STAT pathway sits at the top of the growth control circuit and promotes growth in the eye through Notch/eyg. Several lines of evidence support the first model. First, loss of either N or eyg leads to a reduction in upd transcription while forced expression of either component activates upd expression (Chao et al., 2004; Reynold-Kenneally and Mlodzik, 2005). Second, N and/or eyg-dependent growth can be blocked by the loss of upd and several other JAK/STAT components (Reynold-Kenneally and Mlodzik, 2005). Together, these data and those linking Notch signaling to eyg suggest a model in which localized activation of N at the midline initiates eyg expression which in turn leads to activation of the JAK/STAT pathway resulting in non-autonomous long-range growth of the entire eye disc (Figure 5B, left panel).
The second model, in which JAK/STAT signaling lies upstream of and promotes growth through N, has been recently proposed as an alternative (Figure 5B, middle panel: Gutierriz-Avino et al., 2009). In this paper the authors demonstrate that the growth defects associated with expression of Protein Inhibitor of STAT (PIAS) can be rescued by the activation of the Notch pathway. They also demonstrated that the tissue overgrowth phenotype which is observed when upd is over-expressed can be suppressed if the fng+/fng− border (and thus Notch activation) is disturbed (this was achieved by ubiquitous expression of Iro-C genes). At the moment it is not clear if these models are mutually exclusive of one another. There is room for both modes of growth control to exist if one were to incorporate a positive feedback loop between Notch signaling and the JAK/STAT pathway. This could act to continually promote localized Notch signaling at the midline while promoting long-range growth throughout the disc via JAK/STAT Figure 5B, right panel).
5. The Morphogenetic Furrow: How Point of Origin and Pace can Influence Organ Size
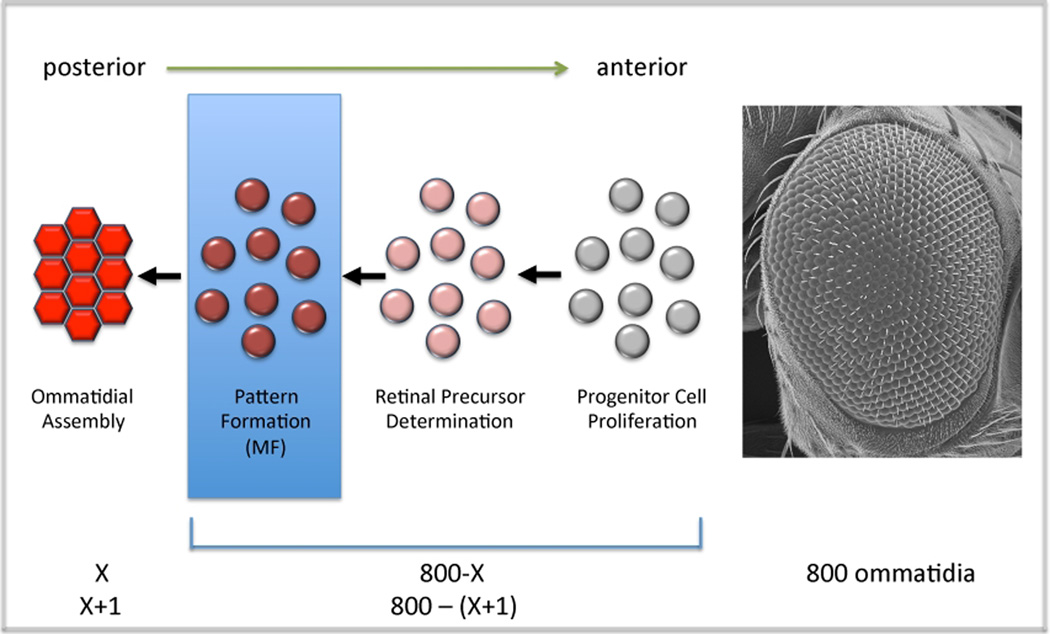
During the third and final instar a wave of morphogenesis initiates at the posterior margin of the disc and sweeps across the eye field until it reaches the eye/antennal border which lies at the far anterior edge of the eye disc (Weismann, 1864, Melamed and Trujillo-Cenoz, 1975; Ready et al., 1976). The leading edge of this wave front can be visualized by a dorso-ventral groove in the epithelium and is called the morphogenetic furrow. As it traverses the eye field it transforms a sea of undifferentiated cells into regularly spaced unit eyes or ommatidia (Krafka, 1924, Medvedev, 1935, Steinberg, 1943; Waddington and Perry, 1960; Ready et al., 1976; Tomlinson and Ready, 1987a,b; Wolff and Ready, 1991). Since the adult compound eye will ultimately contain approximately 800 such units, a continual re-adjustment of growth rates ahead of the furrow is predicted to accompany the addition of each new ommatidial row behind the furrow (Figure 6). The overall size of the adult eye can be reduced by either altering the point along the retinal periphery at which the furrow initiates or by accelerating the pace at which the furrow races across the epithelium (Ma and Moses, 1995; Treisman and Rubin, 1995; Brown et al., 1995; Zelhof et al., 1997; Bhattacharaya and Baker, 2009). The reduction in tissue size is likely due to an imbalance between the rates of patterning and growth with d[P]/d[G] ratio being significantly greater than 1.
Figure 6. Progression of the Furrow and the Constant Recalculation of Growth Rates.
This model describes the readjustment of growth rates that is likely to occur as new rows of ommatida are added to the developing eye. It also briefly describes the cellular steps that cells ahead of the furrow take on the way to ommatidial assembly.
5.1 Wingless Signaling Inhibits the Initiation of Ectopic Furrows
Having a single firing center is important for maintaining a balance between patterning and growth rates. The creation of ectopic firing points directs the birth of new morphogenetic furrows (Ma and Moses, 1995, Chanut and Heberlein, 1997; Pignoni and Zipursky, 1997; Kumar and Moses, 2001). In these situations cell proliferation rates ahead of the furrow do not increase therefore endogenous and ectopic furrows together quickly pattern all available cells before the number of cells needed to support an 800 unit display have been generated resulting in eyes that are small and disorganized.
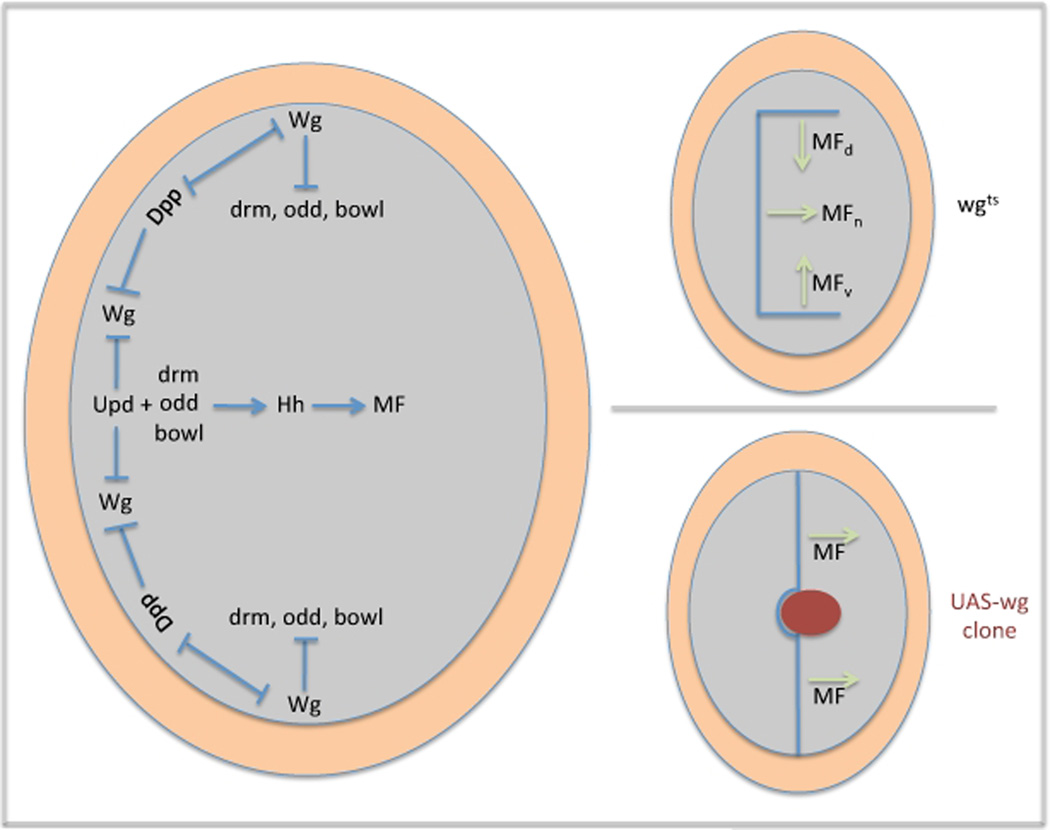
Limiting the eye to a single firing center is a task that falls largely to the Wingless (Wg) signaling cascade. In late second/early third instar eye discs wg expression is found along the dorsal and ventral margins ahead of the advancing morphogenetic furrow. Its loss at the edges of the eye field leads to the birth of ectopic furrows (Figure 7, upper right panel; Ma and Moses, 1995). Together, the three furrows quickly overwhelm and pattern all available precursor cells before the full complement of retinal progenitors are produced. Wg signaling alone is sufficient to block pattern formation as ubiquitous wg expression throughout the eye field can inhibit the launching of the endogenous furrow from the firing center and its progression across the eye field (Figure 7, lower right panel; Treisman and Rubin, 1995).
Figure 7. Initiation of the Morphogenetic Furrow and the Firing Center.
The left panel describes the mechanisms by which Dpp, JAK/STAT and Wg restrict the firing center to the intersection of the posterior margin and the midline. The top right panel describes the initiation of ectopic furrows in response to the loss of wg expression. The bottom right panel describes a furrow stop phenotype that results from ectopic wg expression within the eye field.
Prior to the initiation of the morphogenetic furrow wg expression and activity is down-regulated at the posterior margin of the eye disc by JAK/STAT and Decapentaplegic (Dpp) signaling (Figure 7, left panel; Heberlein et al., 1993; Wiersdorff et al., 1996; Chanut and Heberlein, 1997; Hazlett et al., 1998; Dominguez and Hafen, 1997; Ekas et al., 2006; Tsai et al., 2007). Loss of dpp expression is not accompanied by an increase in wg transcription suggesting that the functional antagonism between these two pathways is at the post-transcriptional level (Treisman and Rubin, 1995). In contrast, loss of JAK/STAT signaling leads to a cell autonomous increase in wg expression. However, it appears that JAK/STAT regulation of wg is indirect as STAT92E binding sites are not present within the STAT responsive wg 3` eye enhancer (Ekas et al., 2006; Tsai et al., 2007). As expected, forced activation of either pathway down-regulates wg transcription and leads to the formation of ectopic furrows from the lateral and anterior margins of the disc (Chanut and Heberlein, 1997; Pignoni and Zipursky, 1997; Ekas et al., 2006; Tsai et al., 2007). Together, these results suggest that the competition between Wg, Dpp and JAK/STAT signals limits the eye to one firing point at the intersection of the posterior margin and the D/V midline and that this is a key step in preserving proper growth of the eye field (Figure 7, left panel).
The competing activities of the of JAK/STAT, Dpp and Wg signaling cascades results in the activation of Hh signaling at the intersection of the posterior margin and the midline. It is from this point that the morphogenetic furrow initiates and begins its journey across the eye field (Ma et al., 1993; Dominguez and Hafen, 1997). The mechanism by which hh transcription was activated at the firing center remained elusive for many years. But recently, three zinc finger transcription factors, Odd skipped (Odd), Brother of odd with entrails limited (Bowl) and Drumstick (Drm) have been shown to be expressed along the posterior margin and are sufficient to initiate hh transcription and retinogenesis (Figure 7; Bras-Pereira et al., 2006). The results in this paper also provided a mechanistic link between Wg and Hh signaling. The authors demonstrated that Wg inhibits drm transcription at the margins anterior to the furrow. Thus, it seems that Wg can prevent ectopic furrow formation by blocking the activation of hh by odd/drm/bowl. Similar linkages between Hh and Wg pathways using odd class genes are preserved in other imaginal discs as well as the insect gut and embryonic epidermis (Hatini et al., 2005).
5.2 Extramacrochaetae Regulates the Velocity of the Morphogenetic Furrow
An equally important factor in maintaining balance between patterning and growth rates is the speed at which the furrow navigates across the eye disc. Several indirect attempts to clock the furrow have been attempted over the years with contradicting results. An initial attempt registered a biphasic rate in which a new row was laid down once every 100 minutes in the posterior half of the eye and once every 70 minutes in the anterior half (Campos-Ortega and Hofbauer, 1977). A later measurement, one that is more widely accepted, suggests that the furrow creates a new ommatidial row every 120 minutes across the entire disc (Basler and Hafen, 1989). Whichever measurement is correct, it is imperative that the furrow does not move too rapidly across the eye field. If the furrow did move too rapidly then the pool of retinal precursors would be quickly transformed into photoreceptors and their capacity to generate the requisite number of cells needed for a fully formed adult eye would be exhausted.
One gene that appears to regulate the velocity of the furrow is extramacrochaetae (emc), which encodes a helix-loop-helix (HLH) protein (Botas et al., 1982). Emc and its vertebrate homologs, the Inhibitors of Differentiation (Id1-4) belong to a unique subclass of basic helix-loop-helix (bHLH) transcription factors. bHLH proteins form either homo or heterodimers through the HLH domain and can either activate or repress transcription of downstream target genes through binding of the basic domain dimer to the N or E box (Murre et al., 1989a,b; Bertrand et al., 2002). Emc works by forming inactive heterodimers with bHLH proteins. Since Emc lacks a basic domain (Ellis et al., 1990; Garrell and Modolell, 1990) the HLH-bHLH heterodimer is unable to bind nucleic acids. Thus Emc affects transcription through protein sequestration (van Doren et al., 1991). At the writing of this review very few binding partners have been identified in vitro and even fewer in the developing eye.
During eye development emc is expressed in a dorso-ventral stripe ahead of the advancing morphogenetic furrow (Brown et al., 1995). Simultaneous reductions in both emc and hairy (h) levels lead to an acceleration of the furrow and a dramatic decrease in the overall size of the eye disc (Brown et al., 1995; Bhattacharya and Baker, 2009). Initial reports put forth a model in which H, a bHLH transcription factor and Emc functioned together to slow the furrow (Brown et al., 1995), but more recent results have shown that the loss of emc by itself is all that is required for the furrow to accelerate across the disc (Bhattacharya et al., 2009). Reduction in the size of eyes that completely lack emc must be analyzed with some caution. Clones of emc null alleles do not survive unless they are generated in a genetic background that is heterozygous for dominant mutations within ribosomal proteins (referred to as Minute mutations). In this genetic background cells that lack emc have a competitive advantage over the surrounding tissue. Therefore the small eye phenotype may be due to a combination of phenotypes: the acceleration of the furrow across the eye field as well as potential regulation of the cell cycle. Indeed, several human cancers are associated with elevated levels of Id1-4 (Desprez et al., 1998; Parrinello et al., 2001; Dong et al., 2003; Itahana et al., 2003; Han et al., 2004; Kebebew et al., 2000; Kebebew et al., 2003; Ling et al., 2004; Chaudhary et al., 2005; Tsuchiya et al., 2005; Yuen et al., 2006; Tam et al., 2008; Zhao et al., 2008; Tang et al., 2009).
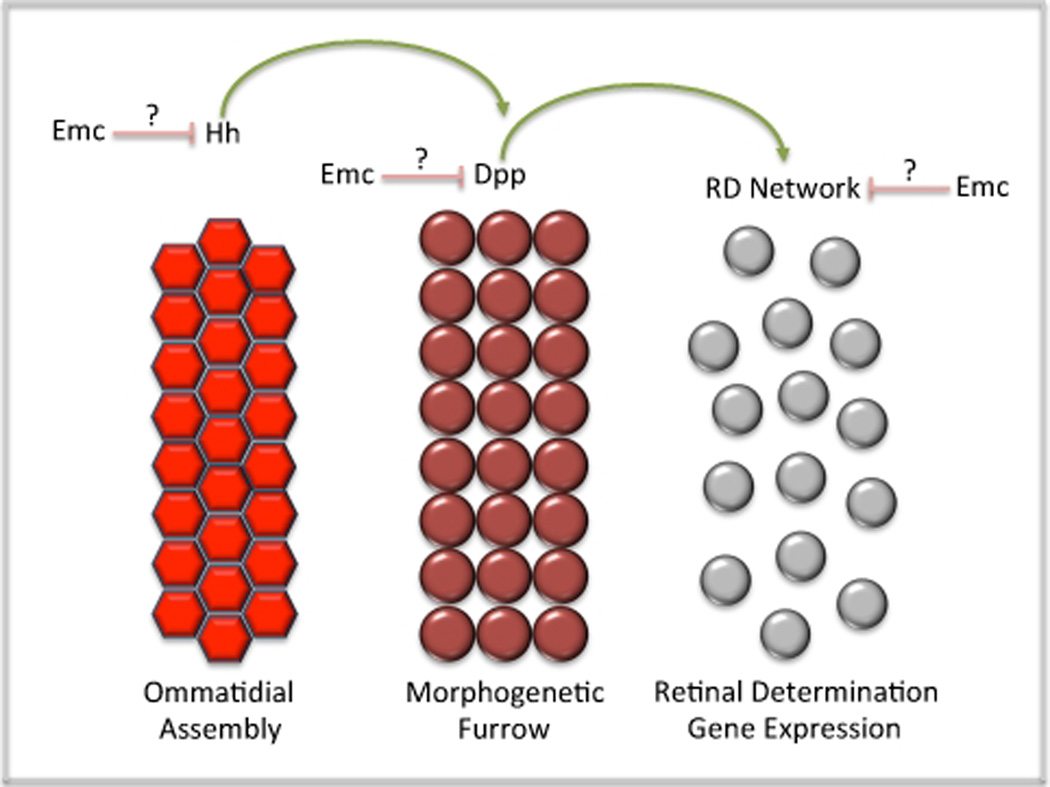
The mechanism by which Emc regulates the pace of furrow movement is yet to be resolved although several clear possibilities exist. Emc could potentially function to regulate the breadth and intensity of the Hedgehog signal, the transmission and execution of instructions from the Dpp pathway or the activity of the retinal determination network ahead of the advancing morphogenetic furrow. Equally likely is the potential for emc to simultaneously regulate multiple signaling events within and ahead of the furrow (Figure 8). Irrespective of which pathway is being regulated by Emc, a key to truly understanding the role of insect and vertebrate HLH proteins in growth control will require the identification of tissue specific binding partners and an understanding of how their sequestration affects regulation of the cell cycle.
Figure 8. Regulation of Furrow Movement and Velocity by Extramacrochaetae.
Hh and Dpp signaling as well as members of the RD network are known to regulate the progression of the morphogenetic furrow. Emc is known to slow the rate of furrow progression. This figure describes the points of potential regulation for Emc.
6. The Second Mitotic Wave: Where Size Really Matters
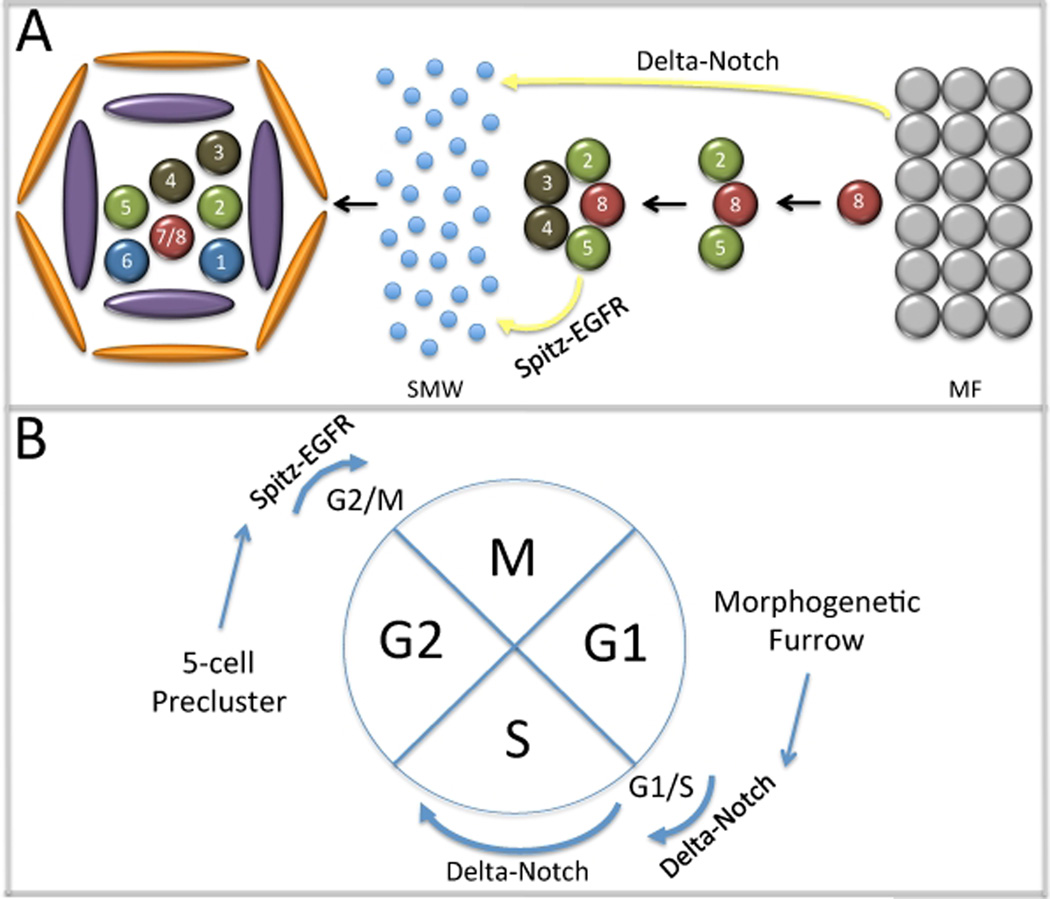
As cells pass through the morphogenetic furrow a subset immediately exits the cell cycle and adopts a photoreceptor fate. These cells (R8, R2/5, R3/4) form the first five cells of the ommatidium and are collectively referred to as the 5-cell precluster (Ready et al., 1976). The remaining cells will undergo a final synchronized round of mitosis before exiting the cell cycle themselves. This second mitotic wave will give rise to three photoreceptor subtypes (R1, R6 and R7), the four cone cells, all thee pigment cell subtypes and the four cells that comprise the bristle complex (Ready et al., 1976; Tomlinson, 1985). Correct cell fate specification occurs in the absence of the second mitotic wave although not enough cells are generated to produce 800 complete ommatidia. Instead unit eyes within the anterior half of the retina contain the full balance of photoreceptor, cone, pigment and bristle cells while those within the posterior half are depleted for the later borne cells (de Nooij and Hariharan, 1995). This result indicates that the sole role of the second mitotic wave is to generate large quantities of cells. And since the majority of cells that comprise each ommatidium is generated in this round of cell division, the second mitotic wave can also be thought of as being the major determinant of the overall size of the adult retina (Figure 9).
Figure 9. Notch and EGF Receptor Regulation of Second Mitotic Wave Checkpoints.
(A) Schematic diagram of ommatidial assembly. Delta-Notch signaling from the furrow and Spitz-EGFR from the precluster regulate progression of cells through the second mitotic wave. (B) Notch signaling regulates entry into S phase and the length of time that cells remain in S phase. EGF Receptor signaling promotes the transition of cells into mitosis.
6.1 A Passage to the Second Mitotic Wave: Notch Controls the G1/S Checkpoint
In order for cells, which are arrested in G1 within the morphogenetic furrow, to enter the second mitotic wave they must first negotiate the G1/S cell cycle checkpoint. This is accomplished through Notch signaling emanating from cells within the furrow (Figure 9). Cells within the furrow express and secrete the Dl ligand while non-precluster cells express the N receptor. Upon activation of the N receptor, cells enter the second mitotic wave by navigating the G1/S transition through modulations of RBF and E2F activity, two well-known regulators of G1 and S cell cycle phases (Baonza and Freeman, 2005; Firth and Baker, 2005). Other reports have extended these results by showing that Notch dependent entry into the second mitotic wave requires the CyclinE/Cdk2 complex, which is known to directly regulate Rb and E2F activity (Sukhanova and Du, 2008). Similarly, the CyclinD/Cdk4 complex also regulates both Rb and E2F activity (Mittnacht, 1998; Lundberg and Weinberg, 1998; Xin et al., 2002). Previous studies in vertebrate tissue culture systems had shown that expression of a mutant form of Rb lacking cdk phosphorylation sites can induce G1 arrest (Brown et al., 1999). However, at least within the second mitotic wave both CyclinE/Cdk2 and CyclinD/Cdk4 complexes may actually be required for the proper completion of S phase rather than regulating the initial entry into S phase. Despite forced expression of mutant Rb proteins in all cells behind the furrow, cells enter the second mitotic wave normally but do not exit S phase on time (Xin et al., 2002). The implication is that Notch signaling may actually regulate two steps: the initial entry into the second mitotic wave and the length of time that cells reside within S phase (Figure 9A).
6.2 The G2/M Checkpoint: EGF Receptor Signaling from the Pre-cluster
Entry into the second mitotic wave is not the only point at which non-autonomous signals from cells outside the wave are required. A second step, the transition into M phase from G2, also requires a long-range signal. In contrast to the G1/S transition, this step appears to be controlled by the EGF Receptor pathway and the signal emanates not from the furrow but rather from the five-cell pre-cluster (Figure 9). Initial insights into a potential role for EGF Receptor signaling in retinal growth control came from a study that demonstrated a requirement for this pathway in cell proliferation (Xu and Rubin, 1993). Other reports demonstrated that the ligand Spitz (Spi) and the two processing/presenting factors Star (S) and Rhomboid (Rho) are distributed within cells of the pre-cluster but not the furrow (Freeman et al., 1992; Heberlein et al., 1993; Tio and Moses, 1997). Similarly, loss of the EGF Receptor itself results in the loss of all photoreceptors that are normally borne in the second mitotic wave (Freeman, 1996; Kumar et al., 1998).
Removal of spi from the pre-cluster or of the EGF receptor from cells within the second mitotic wave does not affect the level of BrdU incorporation, suggesting that passage from the G1 to S phase does not require EGF Receptor signaling. In contrast, Cyclin B accumulation is higher than normal indicating that cells within the second mitotic wave are arrested in G2. In complementary experiments, hyper-activation of EGF Receptor signaling results in the elimination of Cyclin B and an increase in the number of cells undergoing mitosis. These experiments suggested that EGF Receptor signaling is necessary and sufficient for cells to transition through the G2/M checkpoint within the second mitotic wave (Baker and Yu, 2001). A subsequent paper demonstrated that the effect of the EGF Receptor pathway on the G2/M checkpoint was genetically dependent upon canonical Ras/MAPK signaling and the Ets transcription factor Pointed (Pnt), a downstream target of the cascade (Brunner et al., 1994; O’Neill et al., 1994; Yang and Baker, 2003).
A direct link between EGF Receptor signaling and the G2/M checkpoint was established by an analysis of several transcription factors that are known downstream components of the EGF Receptor pathway (Baonza et al., 2002). These include pnt (see above), yan (Rebay and Rubin, 1995) and tramtrack (ttk: Read, 1992; Lai et al., 1996; Tang et al., 1997). Removal of Pnt, a transcriptional activator, results in a severe reduction in the number of mitotic figures (Baonza et al., 2002; Yang and Baker, 2003). In contrast, an increase in the mitotic index is seen in cells that lack Ttk, a transcriptional repressor. (Baonza et al., 2002). Furthermore, both Pnt and one isoform of Ttk (Ttk69) were shown to directly bind to regulatory sequences within an eye specific enhancer element of stg/cdc25, the universal regulator of the G2/M checkpoint.
Is stg the only target of EGF Receptor signaling within the second mitotic wave? A recent paper suggests that the answer is no. The authors used a microarray-based approach to compare the expression profile of wild type eye discs to those that lack the second mitotic wave due to manipulations in EGF signaling. A number of new genes were identified and may play roles at the G2/M checkpoint (Firth and Baker, 2007)
7. Concluding Remarks
Despite the increased awareness of the Drosophila retina as a model system for studying growth control mechanisms, our understanding of how cell proliferation is regulated in the eye is still in its infancy. In fact, in terms of identifying the full complement of genes, pathways and networks that regulate the size of the adult eye, we have only scratched the surface. In this review, I have briefly discussed the contributions that the retinal determination network, the dorsal-ventral midline, the morphogenetic furrow and the second mitotic wave make to the growth of the Drosophila retina. These components are just a few inputs into the overall growth control circuit. Additional inputs that are known to play important roles in tissue growth (but are not discussed here) include, but are not limited to, tumor suppressor pathways, apical-basal polarity regulators and programmed cell death cascades. How all of these influences are integrated into the central growth pacemaker is an open question and certainly one of fundamental importance. As increasing numbers of studies continue to use the eye to study growth control, future authors of similar reviews will be able to provide a significantly more complete picture of how cell proliferation levels are modulated during development to produce adult structures of the appropriate size and shape: not too big, not too small.
Acknowledgements
I would like to thank Bonnie Weasner, Brandon Weasner, Abigail Anderson and Carrie Spratford for critical comments on this manuscript. I would also like to apologize to anyone whose work was not cited here. J.P. Kumar is supported by a grant from the National Institutes of Health (2R01 EY014863).
References
- Aldaz S, Morata G, Azpiazu N. The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development. 2003;130:4473–4482. doi: 10.1242/dev.00643. [DOI] [PubMed] [Google Scholar]
- Anderson DT. The development of hemimetabolous insects. In: Counce S, Waddington CH, editors. Developmental Systems: Insects. New York: Academic Press; 1972a. pp. 96–163. [Google Scholar]
- Anderson DT. The development of hemimetabolous insects. In: Counce S, Waddington CH, editors. Developmental Systems: Insects. New York: Academic Press; 1972b. pp. 165–242. [Google Scholar]
- Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–1166. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell. 2005;8:529–539. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Baonza A, Murawsky CM, Travers AA, Freeman M. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol. 2002;4:976–980. doi: 10.1038/ncb887. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development. 1989;107:723–731. doi: 10.1242/dev.107.4.723. [DOI] [PubMed] [Google Scholar]
- Becker HJ. Uber Rontgenmossaikflecken und Defektmutationen am Auge von Drosophila und die Entwicklungsphysiologie des Auges. Z Induk Abst Vererb Lehre. 1957;88:333–373. [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bessa J, Carmona L, Casares F. Zinc-finger paralogues tsh and tio are functionally equivalent during imaginal development in Drosophila and maintain their expression levels through auto- and cross-negative feedback loops. Dev Dyn. 2009;238:19–28. doi: 10.1002/dvdy.21808. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev Biol. 2009;327:288–300. doi: 10.1016/j.ydbio.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Botas J, Moscoso del Prado J, Garcia-Bellido A. Gene-dose titration analysis in the search of trans-regulatory genes in Drosophila. Embo J. 1982;1:307–310. doi: 10.1002/j.1460-2075.1982.tb01165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braid LR, Verheyen EM. Drosophila nemo promotes eye specification directed by the retinal determination gene network. Genetics. 2008;180:283–299. doi: 10.1534/genetics.108.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bras-Pereira C, Bessa J, Casares F. Odd-skipped genes specify the signaling center that triggers retinogenesis in Drosophila. Development. 2006;133:4145–4149. doi: 10.1242/dev.02593. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB. Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell. 1995;80:879–887. doi: 10.1016/0092-8674(95)90291-0. [DOI] [PubMed] [Google Scholar]
- Brown VD, Phillips RA, Gallie BL. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol. 1999;19:3246–3256. doi: 10.1128/mcb.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Bryant PJ. Cell lineage relationships in the imaginal wing disc of Drosophila melanogaster. Dev Biol. 1970;22:389–411. doi: 10.1016/0012-1606(70)90160-0. [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Schneiderman HA. Cell lineage, growth, and determination in the imaginal leg discs of Drosophila melanogaster. Dev Biol. 1969;20:263–290. doi: 10.1016/0012-1606(69)90015-3. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes & Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hofbauer A. Cell clones and pattern formation on the lineage of photoreceptor cells in the compound eye of Drosophila. Wilhelm's Roux's Archives. 1977;181:227–245. doi: 10.1007/BF00848423. [DOI] [PubMed] [Google Scholar]
- Chanut F, Heberlein U. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development. 1997;124:559–567. doi: 10.1242/dev.124.2.559. [DOI] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Schmidt M, Sadler-Riggleman I. Negative acting HLH proteins Id 1, Id 2, Id 3, and Id 4 are expressed in prostate epithelial cells. The Prostate. 2005;64:253–264. doi: 10.1002/pros.20238. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Oh SW, Zheng Z, Chen HW, Shin HH, Hou SX. Cyclin D-Cdk4 and cyclin E-Cdk2 regulate the Jak/STAT signal transduction pathway in Drosophila. Dev Cell. 2003;4:179–190. doi: 10.1016/s1534-5807(03)00024-8. [DOI] [PubMed] [Google Scholar]
- Chern JJ, Choi KW. Lobe mediates Notch signaling to control domain-specific growth in the Drosophila eye disc. Development. 2002;129:4005–4013. doi: 10.1242/dev.129.17.4005. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Cho KO, Chern J, Izaddoost S, Choi KW. Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell. 2000;103:331–342. doi: 10.1016/s0092-8674(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396:272–276. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Cohen SM. In: Imaginal disc development. Bate M, Martinez Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 747–841. [Google Scholar]
- Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Muller-Tidow C, Golub TR, Kawakami K, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH, Lawrence PA. Compartments and polyclones in insect development. Science. 1975;189:340–347. doi: 10.1126/science.806966. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Burnett M, Mlodzik M. distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev Biol. 2007;306:685–702. doi: 10.1016/j.ydbio.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- Datta RR, Lurye JM, Kumar JP. Restriction of ectopic eye formation by Drosophila teashirt and tiptop to the developing antenna. Dev Dyn. 2009 doi: 10.1002/dvdy.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, del Arco A, Garcia-Bellido A. Genetic and molecular characterization of a Notch mutation in its Delta- and Serrate-binding domain in Drosophila. Proc Natl Acad Sci U S A. 1993;90:4037–4041. doi: 10.1073/pnas.90.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Tyler DM, de Celis J, Bray SJ. Notch signalling mediates segmentation of the Drosophila leg. Development. 1998;125:4617–4626. doi: 10.1242/dev.125.23.4617. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Hariharan IK. Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- Desprez PY, Lin CQ, Thomasset N, Sympson CJ, Bissell MJ, Campisi J. A novel pathway for mammary epithelial cell invasion induced by the helix-loop-helix protein Id-1. Mol Cell Biol. 1998;18:4577–4588. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter JS. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am Nat. 1914;48:712–758. [Google Scholar]
- Diaz-Benjumea FJ, Cohen SM. Interaction between dorsal and ventral cells in the imaginal disc directs wing development in Drosophila. Cell. 1993;75:741–752. doi: 10.1016/0092-8674(93)90494-b. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development. 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eassa YEE. The development of imaginal buds in the head of Pieris brassicae Linn. (Lepidoptera) Trans R Entomol Soc Lond. 1953;104:39–51. [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekas LA, Baeg G-H, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Spann DR, Posakony JW. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Escudero LM, Freeman M. Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC developmental biology. 2007;7:13. doi: 10.1186/1471-213X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev Biol. 2007;307:521–538. doi: 10.1016/j.ydbio.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Bhattacharya A, Baker NE. Cell cycle arrest by a gradient of Dpp signaling during Drosophila eye development. BMC developmental biology. 2010;10:28. doi: 10.1186/1471-213X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4:2188–2201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF Receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Merriam JR. Parameters of the wing imaginal disc development of Drosophila melanogaster. Dev Biol. 1971;24:61–87. doi: 10.1016/0012-1606(71)90047-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Merriam J. Developmental compartmentalization in the dorsal mesothoracic disc of Drosophila. Dev Biol. 1970;48:132–147. doi: 10.1016/0012-1606(76)90052-x. [DOI] [PubMed] [Google Scholar]
- Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Avino FJ, Ferres-Marco D, Dominguez M. The position and function of the Notch-mediated eye growth organizer: the roles of JAK/STAT and four-jointed. EMBO Rep. 2009;10:1051–1058. doi: 10.1038/embor.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Gou C, Hong L, Liu J, ZheyiHan, Liu C, Wang J, Wu K, Ding J, Fan D. Expression and significances of Id1 helix-loop-helix protein overexpression in gastric cancer. Cancer letters. 2004;216:63–71. doi: 10.1016/j.canlet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, DiNardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiaton. Genes Dev. 2005;19:709–718. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie JL. PhD Thesis. Irvine: Universitat of California; 1975. Intercalary regeneration and pattern formation in imaginal discs of Drosophila melanogaster. [Google Scholar]
- Haynie JL, Bryant PJ. Development of the eye-antennal imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J Exp Zool. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, Rubin GM. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-x. [DOI] [PubMed] [Google Scholar]
- Held LI. Developmental and Cell Biology Series. Vol. 39. Cambridge Press; 2002. Imaginal Discs: The Genetic and Cellular Logic of Pattern Formation; p. 460. [Google Scholar]
- Horsfield J, Penton A, Secombe J, Hoffman FM, Richardson H. decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development. 1998;125:5069–5078. doi: 10.1242/dev.125.24.5069. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Itahana Y, Singh J, Sumida T, Coppe JP, Parrinello S, Bennington JL, Desprez PY. Role of Id-2 in the maintenance of a differentiated and noninvasive phenotype in breast cancer cells. Cancer Res. 2003;63:7098–7105. [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jasper H, Benes V, Atzberger A, Sauer S, Ansorge W, Bohmann D. A genomic switch at the transition from cell proliferation to terminal differentiation in the Drosophila eye. Dev Cell. 2002;3:511–521. doi: 10.1016/s1534-5807(02)00297-6. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NA, Kuo YM, Sun YH, Beckendorf SK. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development. 1998;125:4163–4174. doi: 10.1242/dev.125.21.4163. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci U S A. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebebew E, Treseler PA, Duh QY, Clark OH. The helix-loop-helix transcription factor, Id-1, is overexpressed in medullary thyroid cancer. Surgery. 2000;128:952–957. doi: 10.1067/msy.2000.111082. [DOI] [PubMed] [Google Scholar]
- Kebebew E, Treseler PA, Duh QY, Clark OH. The helix-loop-helix protein, Id-1, is overexpressed and regulates growth in papillary thyroid cancer. Surgery. 2003;134:235–241. doi: 10.1067/msy.2003.227. [DOI] [PubMed] [Google Scholar]
- Kellog VL. The development and homologies of the mouth parts of insects. Am Nat. 1902;36:683–706. [Google Scholar]
- Kim J, Irvine KD, Carroll SB. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Toyama R, Takeda H, Dawid IB, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- Kooh PJ, Fehon RG, Muskavitch MA. Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development. 1993;117:493–507. doi: 10.1242/dev.117.2.493. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. Embo J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafka J. Development of the compound eye of Drosophila melogaster and its bar-eyed mutant. Biol Bull. 1924;47:143–149. [Google Scholar]
- Kumar JP. Retinal determination the beginning of eye development. Curr Top Dev Biol. 2010;93:1–28. doi: 10.1016/B978-0-12-385044-7.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Moses K. The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development. 2001;128:2689–2697. doi: 10.1242/dev.128.14.2689. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo BZ, Moses K. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Harrison SD, Karim F, Li Y, Rubin GM. Loss of tramtrack gene activity results in ectopic R7 cell formation, even in a sina mutant background. Proc Natl Acad Sci. 1996;93:5025–5030. doi: 10.1073/pnas.93.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Morata G. The early development of mesothoracic compartments in Drosophila. An analysis of cell lineage and fate mapping and an assessment of methods. Dev Biol. 1977;56:40–51. doi: 10.1016/0012-1606(77)90153-1. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Lieber T, Wesley CS, Alcamo E, Hassel B, Krane JF, Campos-Ortega JA, Young MW. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron. 1992;9:847–859. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- Ling MT, Wang X, Lee DT, Tam PC, Tsao SW, Wong YC. Id-1 expression induces androgen-independent prostate cancer cell growth through activation of epidermal growth factor receptor (EGF-R) Carcinogenesis. 2004;25:517–525. doi: 10.1093/carcin/bgh047. [DOI] [PubMed] [Google Scholar]
- Lopes CS, Casares F. hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol. 2009;339:78–88. doi: 10.1016/j.ydbio.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Moses K. wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development. 1995;121:2279–2289. doi: 10.1242/dev.121.8.2279. [DOI] [PubMed] [Google Scholar]
- Ma C, Zhou Y, Beachy PA, Moses K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75:927–938. doi: 10.1016/0092-8674(93)90536-y. [DOI] [PubMed] [Google Scholar]
- Madhaven MM, Schneiderman HA. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux Archive. 1977;183:269–305. doi: 10.1007/BF00848459. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Martin PF. Direct determination of the growth rate of Drosophila imaginal discs. J Exp Zool. 1982;222:97–102. [Google Scholar]
- Maurel-Zaffran C, Treisman JE. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development. 2000;127:1007–1016. doi: 10.1242/dev.127.5.1007. [DOI] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Medvedev NH. Genes and development of characters. I. The study of the growth of the imaginal discs of eyes of the wild type larvae and three mutants - Lobe, glass and eyeless in Drosophila melanogaster. Z Induk Abst Vererb Lehre. 1935;70:55–72. [Google Scholar]
- Melemad J, Trujilo-Cenoz O. The fine structure of the eye imaginal disc in muscoid flies. J Ultrastrcut Res. 1975;51:79–93. doi: 10.1016/s0022-5320(75)80010-4. [DOI] [PubMed] [Google Scholar]
- Miall LC, Hammond AR. The development of the head of the imago of Chironomus. Trans Linn Soc Zool. 1892;5:265–279. [Google Scholar]
- Milani R. Two new eye-shape mutant alleles in Drosophila melanogaster. D I S. 1941;14:52. [Google Scholar]
- Milani R. Linkage data: The locus so of Drosophila melanogaster. DIS. 1951;25:79. [Google Scholar]
- Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- Morata G, Garcia-Bellido A. Developmental analysis of some mutants of the bithorax system of Drosophila. Wilhelm Roux Archive. 1976;1979:125–143. doi: 10.1007/BF00848298. [DOI] [PubMed] [Google Scholar]
- Morgan LV. Polyploidy in Drosophila melanogaster with two attached X chromosomes. Genetics. 1925;10:148–178. doi: 10.1093/genetics/10.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozer BA, Easwarachandran K. Pattern formation in the absence of cell proliferation: tissue-specific regulation of cell cycle progression by string (stg) during Drosophila eye development. Dev Biol. 1999;213:54–69. doi: 10.1006/dbio.1999.9350. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989a;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989b;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1997;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci U S A. 1998;95:15508–15512. doi: 10.1073/pnas.95.26.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Lin CQ, Murata K, Itahana Y, Singh J, Krtolica A, Campisi J, Desprez PY. Id-1, ITF-2, and Id-2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. J Biol Chem. 2001;276:39213–39219. doi: 10.1074/jbc.M104473200. [DOI] [PubMed] [Google Scholar]
- Patterson JT. The production of mutations in somatic cells of Drosophila melanogaster by means of X-rays. J Exp Zool. 1929;53:327–372. [Google Scholar]
- Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005;132:2771–2782. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F, Casares F. homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev. 2000;96:15–25. doi: 10.1016/s0925-4773(00)00372-5. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Schneiderman HA. A clonal analysis of development in Drosophila melanogaster: morphogenesis, determination and growth in the wild type antenna. Dev Biol. 1971;24:477–519. doi: 10.1016/0012-1606(71)90061-3. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans [see comments] Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Smith KM, Peifer M, Wieschaus E. extradenticle determines segmental identities throughout Drosophila development. Development. 1995;121:3663–3673. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Read D, Levine M, Manley JL. Ectopic expression of the Drosophila tramtrack gene results in multiple embryonic defects, including repression of even-skipped and fushi tarazu. Mech Dev. 1992;38:183–195. doi: 10.1016/0925-4773(92)90052-l. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Salzer CL, Kumar JP. Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS One. 2010;5:e8510. doi: 10.1371/journal.pone.0008510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tomlinson A. Dorsal-ventral midline signaling in the developing Drosophila eye. Development. 2007;134:659–667. doi: 10.1242/dev.02786. [DOI] [PubMed] [Google Scholar]
- Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987a;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- Schneuwly S, Kuroiwa A, Baumgartner P, Gehring WJ. Structural organization and sequence of the homeotic gene Antennapedia of Drosophila melanogaster. Embo J. 1986;5:733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S, Kuroiwa A, Gehring WJ. Molecular analysis of the dominant homeotic Antennapedia phenotype. Embo J. 1987b;6:201–206. doi: 10.1002/j.1460-2075.1987.tb04739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya M, Gehring WJ. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development. 2000;127:1879–1886. doi: 10.1242/dev.127.9.1879. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O'Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Choi KW. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development. 2003;130:6351–6360. doi: 10.1242/dev.00864. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Choi KW, Sun YH. Dorso-ventral asymmetric functions of teashirt in Drosophila eye development depend on spatial cues provided by early DV patterning genes. Mech Dev. 2004;121:365–370. doi: 10.1016/j.mod.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Singh A, Lim J, Choi KW. Dorso-ventral boundary is required for organizing growth and planar polarity in the Drosophila eye. In: Mlodzik M, editor. Planar Cell Polarization during Development: Advances in Developmental Biology and Biochemistry. Elsevier Science and Technology Books; 2005. pp. 59–91. [Google Scholar]
- Speicher SA, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 1994;120:535–544. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- Steinberg AG. The development of the wild type and bar eyes of Drosophila melanogaster. Can J Res. 1943;21:277–283. [Google Scholar]
- Steiner E. PhD Thesis. Universitat Zurich; 1975. Establishment of compartments in the developing leg imaginal discs of Drosophila melanogaster. [DOI] [PubMed] [Google Scholar]
- Steiner E. Establishment of compartments in the developing leg imaginal discs of Drosophila melanogaster. Wilhelm Roux Archive. 1976;180:9–30. doi: 10.1007/BF00848882. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Saigo K. Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development. 2000;127:1531–1540. doi: 10.1242/dev.127.7.1531. [DOI] [PubMed] [Google Scholar]
- Tam WF, Gu TL, Chen J, Lee BH, Bullinger L, Frohling S, Wang A, Monti S, Golub TR, Gilliland DG. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112:1981–1992. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Tang R, Hirsch P, Fava F, Lapusan S, Marzac C, Teyssandier I, Pardo J, Marie JP, Legrand O. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009 doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Gunning DA, Cho J, Zipursky L. Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell. 1994;77:1003–1014. doi: 10.1016/0092-8674(94)90440-5. [DOI] [PubMed] [Google Scholar]
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo Pathway Regulates the bantam microRNA to Control Cell Proliferation and Apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tio M, Moses K. The Drosophila TGF alpha homolog Spitz acts in photoreceptor recruitment in the developing retina. Development. 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morp. 1985;89:313–331. [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987a;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987b;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Treisman JE, Rubin GM. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Yao JG, Chen PH, Posakony JW, Barolo S, Kim J, Sun YH. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306:760–771. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Okaji Y, Tsuno NH, Sakurai D, Tsuchiya N, Kawai K, Yazawa K, Asakage M, Yamada J, Yoneyama S, et al. Targeting Id1 and Id3 inhibits peritoneal metastasis of gastric cancer. Cancer science. 2005;96:784–790. doi: 10.1111/j.1349-7006.2005.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Waddington CH, Perry MM. The ultrastructure of the developing eye of Drosophila. Proc Roy Soc Biol Sci. 1960;153:155–178. [Google Scholar]
- Weasner B, Salzer C, Kumar JP. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev Biol. 2007;303:756–771. doi: 10.1016/j.ydbio.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. Die nachembryonale entwicklung der Musciden nach beobachtungen an Musca vomitoria und Sarcophaga carnaria. Zeit Wiss Zool. 1864;14:187–336. [Google Scholar]
- Wiersdorff V, Lecuit T, Cohen SM, Mlodzik M. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development. 1996;122:2153–2162. doi: 10.1242/dev.122.7.2153. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Gehring W. Clonal analysis of primordial disc cells in the early embryo of Drosophila melanogaster. Dev Biol. 1976;50:249–263. doi: 10.1016/0012-1606(76)90150-0. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xin S, Weng L, Xu J, Du W. The role of RBF in developmentally regulated cell proliferation in the eye disc and in Cyclin D/Cdk4 induced cellular growth. Development. 2002;129:1345–1356. doi: 10.1242/dev.129.6.1345. [DOI] [PubMed] [Google Scholar]
- Xu PX, Woo I, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Yao JG, Weasner BM, Wang LH, Jang CC, Weasner B, Tang CY, Salzer CL, Chen CH, Hay B, Sun YH, et al. Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev Biol. 2008;315:535–551. doi: 10.1016/j.ydbio.2007.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66:1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Id proteins expression in prostate cancer: high-level expression of Id-4 in primary prostate cancer is associated with development of metastases. Mod Pathol. 2006;19:931–941. doi: 10.1038/modpathol.3800602. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Ghbeish N, Tsai C, Evans RM, McKeown M. A role for ultraspiracle, the Drosophila RXR, in morphogenetic furrow movement and photoreceptor cluster formation. Development. 1997;124:2499–2506. doi: 10.1242/dev.124.13.2499. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008a;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZR, Zhang ZY, Zhang H, Jiang L, Wang MW, Sun XF. Overexpression of Id-1 protein is a marker in colorectal cancer progression. Oncology reports. 2008b;19:419–424. [PubMed] [Google Scholar]