Abstract
Lack of stable three-dimensional structure, or intrinsic disorder, is a common phenomenon in proteins. Naturally unstructured regions are proven to be essential for carrying function by many proteins and therefore identification of such regions is an important issue. CASP has been assessing the state of the art in predicting disorder regions from amino acid sequence since 2002. Here we present the results of the evaluation of the disorder predictions submitted to CASP9. The assessment is based on the evaluation measures and procedures used in previous CASPs. The balanced accuracy and the Matthews correlation coefficient were chosen as basic measures for evaluating the correctness of binary classifications. The area under the receiving operating characteristic curve was the measure of choice for evaluating probability-based predictions of disorder. The CASP9 methods are shown to perform slightly better than the CASP7 methods but not better than the methods in CASP8. It was also shown that capability of most CASP9 methods to predict disorder decreases with increasing minimum disorder segment length.
Keywords: CASP, intrinsically disordered proteins, unstructured proteins, rediction of disordered regions, assessment of disorder prediction
INTRODUCTION
It has been widely accepted that that the ability of proteins to perform specific cellular functions is directly associated with their unique spatial structure1. Numerous experiments have shown that proteins lose their activity upon loss of ordered structure due to exposure to non-physiological environments such as high temperature, urea or acid. It was also shown that a denatured protein can regain practically all of its original activity by recovering its structure upon restoration of physiological conditions2. Based on these observations, the concept that proteins achieve their biological function upon folding into unique structural conformations became widely accepted. In the latest two decades ample information has been collected in evidence of proteins that do not follow this general rule3–6. These so-called naturally unstructured or intrinsically disordered proteins (IDPs) lack stable structures under physiological conditions but are nevertheless biologically active. Many other proteins contain structured regions alongside extended intrinsically disordered regions (IDRs) that often play an important functional role (e.g., BRCA1, a breast cancer susceptibility protein contains approximately 1,500 unstructured non-termini residues participating in repairing damaged DNA7). The IDPs and proteins including IDRs are highly abundant in both eukaryotes and prokaryotes8–10 and tend to be enriched for regulatory functions related to molecular recognition and signal transduction11–14. The number of experimentally verified IDPs and IDRs is rapidly rising15: the DisProt database16 currently contains annotations for more than 640 proteins with disordered regions, and recent reviews on this topic6,11,12 cite hundreds of papers. The association of several IDPs with human disease, such as cancer, cardiovascular disease, amyloidoses, diabetes, neurodegenerative diseases, and others, has triggered additional research on the subject17–19.
With the high level of interest in disordered proteins, a substantial effort was placed to develop experimental and computational methods to study this phenomenon12,20. Computational methods have quickly become a particularly valuable tool, in part because of their ability to keep pace with the large-scale genome sequencing projects. These techniques are based on the premise that the amino acid sequence encodes protein non-folding similarly to protein folding. Indeed, comparison of composition and complexity of protein sequences in ordered and disordered regions shows that they are statistically different12,21. Based on this observation, many methods were built to predict the IDRs through recognition of amino acid motifs characteristic of disorder.
The first formal method for computational protein disorder prediction was published in 199722, and, since then, more than fifty methods to identify disorder have been developed23–25. In a recent review, He et al24 provide a historical perspective of progress in this field, pointing out also the important role that the CASP experiments have played in these advancements since 200226.
The present paper analyzes the results obtained by the thirty-two disorder prediction groups participating in CASP9. While the initial round of disorder prediction (2002) was assessed by the organizers, the following three rounds were evaluated by independent assessors (2004, 2006, 2008). Since the methods to evaluate disorder prediction in CASP have developed to the point where assessments are relatively straightforward and can be made fully automatically, in CASP9 the evaluations were again performed by the organizers.
MATERIALS AND METHODS
Targets and definition of disorder
One hundred and twenty nine targets were released for modeling in CASP9 and all of them were made available for disorder prediction. The structures of twelve targets were not solved in time for prediction assessment and thus were canceled27, leaving 117 targets (98 X-ray and 19 NMR structures) for assessment*. Structure data for five of these targets (T0533, T0536, T0600, T0612, T0637) were compromised in the period between the corresponding server and human prediction deadlines27 and therefore only the server predictions were evaluated on these targets.
Disorder regions for each target were defined based on the best structure determination available at the time of the assessment and using the sequence released for prediction (which sometimes was slightly different than the sequence later deposited in the PDB). In cases where both the NMR and X-ray structures were available (T0551), the X-ray structure was used.
Disorder in CASP9 was defined similarly to the previous CASPs28–31. A residue was considered to be in a disordered state if it appeared in the protein’s amino acid sequence but either (1) lacked the spatial coordinates or (2) showed a high conformational variability across different X-ray chains or NMR models. We have defined “high variability” as cases where distances between positions of the same residue in any pair of models in the NMR ensemble or in any pair of X-ray chains in the asymmetric unit exceeded 3.5Å in the optimal LGA32 superposition†. In all other cases, the residue was assumed to be ordered. This is an oversimplification as residues may be disordered under physiological conditions but forced into the “ordered” state by crystallization. Also, long disordered regions often contain “dual personality” fragments33 that become structured when binding to a partner. These segments are often predicted to be ordered even though they are disordered in the absence of their partner34. However, such transitions are impossible to detect given only the crystal structure of the isolated protein.
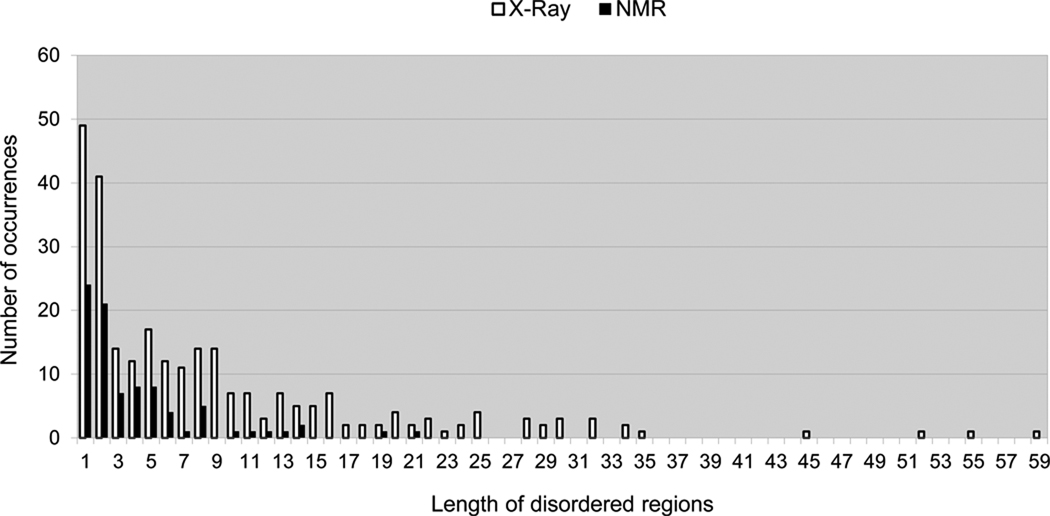
Overall, 2,677 residues (or 10.2% of residues in all CASP9 targets) were classified as disordered, including 403 residues in NMR structures (or 19.9% of all residues in NMR targets) and 2,274 residues (9.4%) in X-ray targets. Thus, percent-wise, CASP9 NMR structures contain approximately twice as many disordered residues as X-ray structures. At the target level, the fraction of disordered residues is approximately the same for both types of targets, varying from 0 to 55% in X-ray structures and from 2 to 53% in NMR structures. Two targets at the high end of this range were T0603 (X-ray, 305 residues) containing 6 separate unstructured regions summing up to 55% of its length, and T0590 (NMR, 137 residues) containing two long disordered segments covering 54% of its sequence. The statistics on the number and length of the IDRs in the CASP9 targets are shown in Figure 1. Short disordered regions are much more common than the long ones. To reduce noise due to experimental uncertainty, segments consisting of less than four consecutive residues of the same order/disorder type were not considered in the assessment. After eliminating short segments, the assessment was performed on a set of 26,075 residues, including 2,417 classified as disordered. We also assessed the ability of methods to identify longer disordered regions by setting the minimum length of a disordered region to 20, 30 and 40 residues.
Figure 1.
Length distribution of disordered regions in CASP9 target proteins.
Participating groups and prediction format
Thirty two groups participated in prediction of disordered regions in CASP9, including 22 servers and 10 human-expert groups. These groups could submit up to five DR predictions (here called models) per target, but only models identified by the predictors as number “1” were evaluated. The overwhelming majority of groups submitted predictions on all or almost all of the targets (see Table I). The two exceptions were human-expert groups G147 and G462, which submitted predictions on 53 and 57 out of the 112 targets, respectively. We assessed the performance of these two groups but did not include them in the final rankings.
Table I.
Summary of assessment scores for disorder prediction groups in CASP9
| ID | Group Name | Targ. | TP | FP | TN | FN | Acc, CI 95% | MCC, CI 95% | AUC, CI 95% | Ranks | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | MCC | AUC | ||||||||||
| 291 | prdos2 | 117 | 1468 | 2340 | 21318 | 949 | 0.754 [0.738;0.774] | 0.418 [0.391;0.444] | 0.855 [0.846;0.863] | 1 | 3 | 1 |
| 015 | DisoPred3C | 117 | 839 | 180 | 23478 | 1578 | 0.670 [0.653;0.687] | 0.508 [0.477;0.537] | 0.854 [0.845;0.864] | 21 | 1 | 2 |
| 490 | Multicom | 110 | 953 | 934 | 21695 | 1310 | 0.69 [0.672;0.709] | 0.413 [0.379;0.448] | 0.853 [0.845;0.861] | 15 | 5 | 3 |
| 474 | Zhou-Spine-D | 117 | 1399 | 2774 | 20884 | 1018 | 0.731 [0.714;0.752] | 0.365 [0.341;0.393] | 0.832 [0.823;0.841] | 6 | 9 | 4 |
| 468 | CBRC_Poodle | 117 | 1078 | 1365 | 22293 | 1339 | 0.694 [0.677;0.713] | 0.386 [0.356;0.418] | 0.831 [0.822;0.840] | 13 | 6 | 5 |
| 119 | Multicom-refine | 117 | 1570 | 3545 | 20113 | 847 | 0.75 [0.733;0.770] | 0.365 [0.342;0.387] | 0.822 [0.812;0.832] | 2 | 8 | 6 |
| 030 | biomine_dr_pdb_c | 117 | 809 | 295 | 23363 | 1608 | 0.661 [0.646;0.679] | 0.464 [0.433;0.497] | 0.821 [0.811;0.831] | 22 | 2 | 7 |
| 351 | biomine_dr_pdb | 117 | 1444 | 2723 | 20935 | 973 | 0.741 [0.723;0.765] | 0.382 [0.357;0.407] | 0.821 [0.811;0.831] | 3 | 7 | 8 |
| 094 | McGuffin | 112 | 1078 | 1947 | 20957 | 1238 | 0.69 [0.672;0.711] | 0.338 [0.310;0.369] | 0.821 [0.812;0.830] | 14 | 12 | 9 |
| 374 | GSmetaDisorderMD | 117 | 1578 | 4213 | 19445 | 839 | 0.737 [0.720;0.759] | 0.331 [0.308;0.353] | 0.818 [0.809;0.827] | 4 | 14 | 10 |
| 165 | GSmetaserver | 117 | 1380 | 3441 | 20217 | 1037 | 0.713 [0.696;0.733] | 0.318 [0.293;0.342] | 0.815 [0.806;0.823] | 8 | 19 | 11 |
| 177 | Distill-Punch1 | 106 | 1265 | 2510 | 18649 | 953 | 0.726 [0.709;0.747] | 0.36 [0.335;0.387] | 0.8 [0.790;0.810] | 7 | 10 | 12 |
| 491 | IntFOLD-DR | 117 | 1162 | 2610 | 21048 | 1255 | 0.685 [0.668;0.706] | 0.305 [0.278;0.335] | 0.797 [0.788;0.807] | 16 | 22 | 13 |
| 010 | GSmetaDisorder | 117 | 1242 | 2943 | 20715 | 1175 | 0.695 [0.677;0.717] | 0.308 [0.281;0.335] | 0.795 [0.786;0.805] | 12 | 21 | 14 |
| 062 | Zhou-Spine-DM | 117 | 1293 | 2863 | 20795 | 1124 | 0.707 [0.691;0.725] | 0.328 [0.304;0.353] | 0.792 [0.782;0.802] | 9 | 15 | 15 |
| 421 | GSmetaDisorder3D | 117 | 964 | 1353 | 22305 | 1453 | 0.671 [0.650;0.692] | 0.348 [0.311;0.400] | 0.784 [0.773;0.794] | 20 | 11 | 16 |
| 407 | United3D | 112 | 1230 | 2814 | 20090 | 1086 | 0.704 [0.687;0.726] | 0.321 [0.293;0.354] | 0.777 [0.766;0.788] | 11 | 17 | 17 |
| 293 | biomine_dr_mixed_c | 117 | 370 | 191 | 23467 | 2047 | 0.573 [0.560;0.583] | 0.29 [0.254;0.319] | 0.771 [0.761;0.781] | 29 | 23 | 18 |
| 492 | biomine_dr_mixed | 117 | 1211 | 3116 | 20542 | 1206 | 0.685 [0.666;0.704] | 0.288 [0.261;0.34] | 0.771 [0.761;0.781] | 17 | 24 | 19 |
| 143 | OnD-CRF | 112 | 1444 | 4870 | 17502 | 850 | 0.706 [0.688;0.726] | 0.274 [0.251;0.296] | 0.761 [0.751;0.771] | 10 | 26 | 20 |
| 327 | Spritz3 | 116 | 1097 | 2061 | 21528 | 1308 | 0.684 [0.668;0.705] | 0.327 [0.302;0.355] | 0.753 [0.742;0.765] | 18 | 16 | 21 |
| 193 | Mason | 116 | 1331 | 1893 | 21697 | 1078 | 0.736 [0.719;0.758] | 0.415 [0.390;0.441] | 0.744 [0.733;0.754] | 5 | 4 | 22 |
| 392 | IsUnstruct | 112 | 1073 | 2681 | 20223 | 1243 | 0.673 [0.656;0.691] | 0.281 [0.254;0.309] | 0.735 [0.723;0.747] | 19 | 25 | 23 |
| 364 | Owls | 111 | 857 | 1369 | 21423 | 1429 | 0.657 [0.643;0.675] | 0.319 [0.292;0.33] | 0.733 [0.721;0.745] | 24 | 18 | 24 |
| 378 | ULg-GIGA | 115 | 447 | 267 | 23116 | 1917 | 0.589 [0.577;0.603] | 0.312 [0.276;0.358] | 0.721 [0.709;0.733] | 27 | 20 | 25 |
| 188 | OnD-CRF-pruned | 117 | 1271 | 4822 | 18836 | 1146 | 0.661 [0.646;0.677] | 0.221 [0.199;0.242] | 0.709 [0.697;0.721] | 23 | 28 | 26 |
| 214 | Distill | 117 | 1211 | 4566 | 19092 | 1206 | 0.654 [0.637;0.673] | 0.215 [0.193;0.237] | 0.695 [0.683;0.707] | 25 | 29 | 27 |
| 155 | MeDor | 112 | 686 | 3195 | 19709 | 1630 | 0.578 [0.561;0.594] | 0.125 [0.097;0.150] | 0.675 [0.663;0.687] | 28 | 30 | 28 |
| 114 | Lee | 111 | 469 | 237 | 22399 | 1838 | 0.596 [0.584;0.609] | 0.337 [0.306;0.365] | 0.59 [0.581;0.599] | 26 | 13 | 29 |
| 067 | HIT_Dict | 112 | 252 | 139 | 22765 | 2064 | 0.551 [0.541;0.561] | 0.24 [0.208;0.270] | 0.56 [0.548;0.571] | 30 | 27 | 30 |
| 147 | GeneSilico | 53 | 534 | 1475 | 9222 | 282 | 0.758 [0.733;0.791] | 0.349 [0.315;0.383] | 0.841 [0.827;0.855] | |||
| 462 | HEU_DisIP | 57 | 497 | 5350 | 5780 | 646 | 0.477 [0.448;0.499] | −0.03 [−0.060;−0.001] | 0.587 [0.573;0.602] | |||
The groups are sorted according to the AUC score. The two groups at the bottom of the table (in grey) are not included in the rankings as they submitted predictions on less than half of the targets.
The format of the predictions in the DR category has not changed since CASP5. The predictors were asked to identify the IDRs by assigning to each residue a binary classifier of order or disorder (“O” for the ordered state and “D” for the disordered), and a probability of belonging to a disordered region (a real number in the [0;1] range). The detailed description of the DR format can be found at the Prediction Center website http://predictioncenter.org/casp9/index.cgi?page=format#DR. Learning from the lessons of CASP8, in CASP9 we required that all the residues that were assigned a binary disordered/ordered tag were also assigned probability values above/below 0.5, respectively. The value of 0.5 was reserved for residues where predictors were undecided.
Evaluation criteria
Disorder predictions in CASP9 were evaluated with the MCC, Acc, and AUC measures also used in previous CASPs29–31. The Sw measure was dropped from the assessment as it was shown35 to be equivalent to the Acc, when calculated with the weights used in CASPs6–8. The statistical significance of the differences in group performance was assessed using procedures adopted in CASP730, i.e. the bootstrap confidence interval method36,37 and the DeLong tests38.
Measures for evaluating binary order/disorder predictions
In CASP, the ability to correctly assign the order/disorder tags to residues in a target has been evaluated with several measures28–31: sensitivity and specificity (used in CASP5–8), statistical accuracy Q2 (CASP6), Matthews correlation coefficient MCC (CASP6), the weighted score Sw (CASP6–8), and the balanced accuracy Acc (CASP7–8).
The disorder prediction data are characterized by a large class imbalance: in the latest five CASPs ordered residues outnumbered disordered ones 9 to 1 or higher. As disordered residues are relatively rare and therefore harder to predict, their correct prediction should be rewarded more generously than the prediction of ordered residues, and vice versa – the incorrect prediction of disordered regions should be penalized less severely than the incorrect prediction of ordered residues. Not all measures are equally effective in handling these tasks. Below, we briefly discuss the relative strengths and weaknesses of the aforementioned evaluation measures for the disorder assessment.
Sensitivity and specificity
are the two statistical measures routinely used for evaluating the accuracy of a two-class binary predictor. In prediction of disorder, TP (true positives) and TN (true negatives) are the numbers of correctly predicted disordered and ordered residues, respectively; FP (false positives) and FN (false negatives) are the numbers of misclassified ordered and disordered residues, and Nd and No are the total numbers of disordered and ordered residues in all targets predicted by a particular group. Specificity determines the fraction of negative examples (ordered residues) correctly identified in a prediction. For datasets dominated by negative examples, specificity is high for practically all predictors and therefore is not a discriminative measure of prediction quality. Sensitivity represents the fraction of positive examples (disordered residues) correctly identified in a prediction and has a better discriminative power but at the same time is completely insensitive to negative examples (see the corresponding formula). Predictors can increase the sensitivity or specificity of their classifiers by deliberately predicting more residues as disordered or ordered, respectively. There is a tradeoff between these two measures and increasing one of them usually leads to decreasing the other. A prediction method can be considered to perform well only if it scores high in both sensitivity and specificity; neither of these two measures is a good estimator of methods’ strength when used alone. The one-sided nature of sensitivity and specificity can be overcome by employing measures that use all four parameters of prediction quality (TP, FP, TN and FN).
The statistical accuracy (used under the name of Q2 in previous CASP disorder prediction assessments) is calculated according to
| (1) |
and accounts for all four components of prediction quality. Nevertheless, it strongly favors conservative classifications (i.e. predicting more residues as ordered)29,30 and therefore is not well suited for disorder assessment.
The balanced accuracy Acc
| (2) |
is a much better measure as it does not reward over-prediction of the ordered state. On contrary, it has a desired feature of rewarding prediction of disordered state more generously than the prediction of the ordered, but it is also known to strongly favor greedy classifications (i.e. predicting more residues as disordered)29.
The Matthews correlation coefficient (MCC)
| (3) |
does not favor over-prediction of any of the prediction classes and had been recommended for handling cases with skewed class frequencies39,40. MCC varies between −1 and 1 with a random prediction scoring zero. It was noticed, though, that MCC can yield unreasonably high scores in cases where prediction algorithms assign very few or no false positives and at the same time very few true positives41. As this situation can happen in DR prediction (over-prediction of ordered residues), we have conducted a numerical experiment to estimate the scale of possible discrepancies. We have run these calculations on artificial datasets with the TP, TN, FP and FN values varying in the ranges typical of the CASP9 data. The MCC appeared to yield reasonable and consistent scores for all combinations of prediction characteristics, leading to a conclusion that in general it does not overinflate scores for over-prediction of ordered residues in our data.
The general conclusions on the effectiveness of measures (1)–(3) hold true for the CASP9 data. First, the tendency of Q2 to unreasonably favor conservative predictions can be illustrated by an example of two CASP9 groups: G291 and G067. Group G291 is ranked high according to all three measures used in our evaluation (Table I), while group G067 is at the very bottom of the table. Surprisingly, G067 outscores G291 0.91 to 0.87 according to Q2 (data not shown). This result can be directly attributed to more conservative predictions submitted by G067 (only 391 residues predicted as disordered; the remaining 98.5% of residues predicted as ordered, the highest figure in CASP9). Second, both Acc and MCC, reproduce the overall trends in prediction quality fairly well but emphasize numerical contributions from TP, TN, FP and FN differently (on CASP data, the Spearman ranked correlation coefficient between these two measures is only ρ=0.56). The balanced accuracy (Acc) generously rewards correct prediction of disordered regions and mildly penalizes their incorrect prediction, encouraging development of riskier methods tuned to identify large numbers of disordered residues. The MCC is more balanced, it does not reward “greedy” predictions as strongly as the Acc does, but instead rewards classifiers with higher predictive precision
| (4) |
The difference between the Acc and MCC can be illustrated by the example of groups G119 and G015 (Table I). Group G119 is one of the most “greedy” CASP9 classifiers. It has predicted 5115 residues as disordered, but only 1570 of these classifications had been correct (31%). Group G015 predicted only 1019 residues as disordered, of which 839 were correct (82%). The Acc score favors G119 as able to identify almost twice as many disordered residues as G015. At the same time the MCC favors G015 for obtaining a much higher level of precision, while still predicting a relatively high number of disordered residues. The decision of which of these two measures should be used in assessments is to some degree subjective and therefore we present the results of both, noting the better balance of the MCC.
In previous three CASPs, also the Sw score was extensively used in assessments
This measure was specifically designed29 to address the imbalance in the ratio of ordered and disordered residues through adjustable weights wo and wd. It was recently shown35 that for the weights used in CASP6–8
this score is equivalent to the Acc as there is a linear relationship between the two:
Therefore we kept only one of these measures (Acc) in our analysis.
In addition to the scores used in previous CASPs we have tested other evaluation measures. One such measure is the F-score, which, similarly to the MCC, had been recommended to handle skewed data42,43. Our calculations on CASP data have shown a high correlation of this measure with the MCC (Spearman’s ρ=0.9) and therefore these results are not shown.
Measures for evaluating probability-based predictions of disorder
The ability to identify the IDRs through assigning per residue disorder confidence scores [0;1] was assessed with the receiver operating characteristic (ROC) analysis. This method is frequently used to assess the accuracy of a classifier, and has been previously used in the assessment of protein disorder predictions (both in CASP and elsewhere)29–31,44.
In essence, a ROC curve illustrates the correspondence between the true positive rate of a predictor (Sensitivity) and its false positive rate (FPR = FP/(TN + FP) 1 − Specificity) for a set of probability thresholds (from 0 to 1 in our case). For each threshold, a residue is considered as a positive example (disordered) if its predicted probability is equal to or greater than the threshold value. The area under a ROC curve (AUC) is indicative of the classifier accuracy. An AUC of 1 identifies a perfect predictor, while an AUC of 0.5 corresponds to a random classifier. We have computed the AUC scores using the trapezoid integration rule with a threshold increment of 0.01.
Statistical significance of differences in group performance
Performance of groups as binary order/disorder classifiers was statistically compared using the re-sampling procedure. For each group, 80% of targets were randomly drawn from the list of targets predicted by that group and the evaluation scores were re-calculated on that subset. The procedure was repeated 1000 times, and a discrete distribution of the two-class classifications was learned for every group. Based on these distributions, we have calculated the 95% confidence intervals for each assessment measure using the two-tailed bootstrap percentile method36,37. Statistical significance of the differences in group performance was inferred based on the comparison of the confidence intervals obtained for each group45.
Performance of groups as predictors of the per-residue disorder probabilities was compared using the DeLong non-parametric tests38, designed to assess the statistical significance of the differences between the AUC scores in the ROC analysis. The evaluation was performed using the statistical package R46 and the pROC library47.
RESULTS
Performance of disorder prediction methods
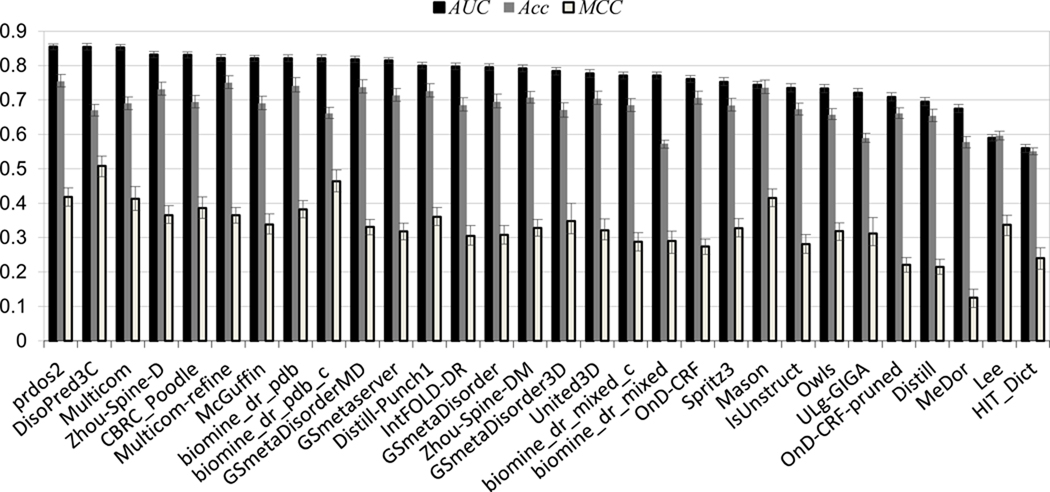
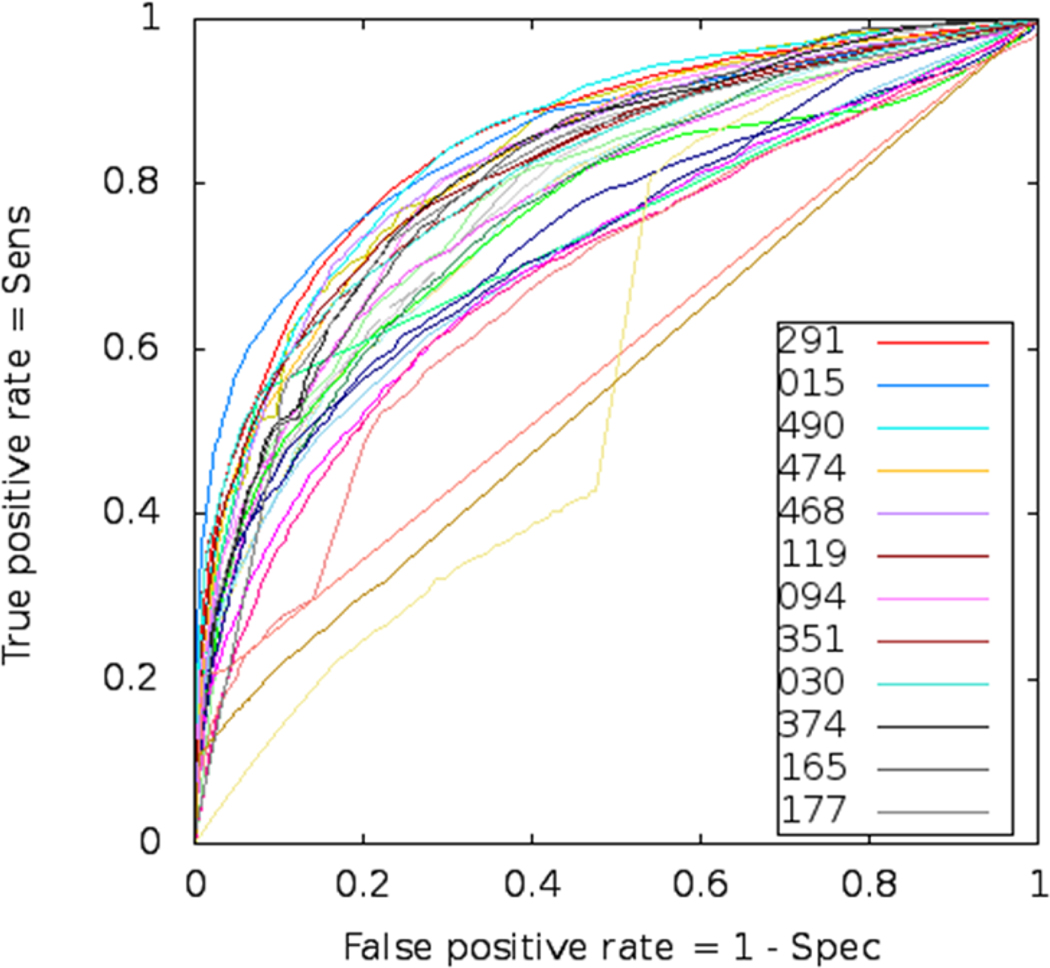
Numerical evaluations of DR predictions for all groups participating in CASP9 are summarized in Table I and illustrated in Figure 2. Scores from the main three evaluation measures used in our assessment (Acc, MCC and AUC) are provided together with the ranges of the corresponding 95% confidence intervals and the group ranks. The ROC curves based on the continuous-scale disorder predictions are plotted in Figure 3. Note that the uneven distribution of the assigned probability scores can affect the smoothness of ROC curves, which is imperative for an accurate calculation of the AUC scores. In CASP9, all top-ranked groups have assigned sufficiently distinct probabilities to enable an accurate calculation of the AUC scores. The only exception is group G193, which submitted predictions yielding good scores according to the binary-classification measures Acc and MCC but poor AUC scores in the probability-based analysis. This was due to uniformly assigning a value of zero to all residues predicted as ordered.
Figure 2.
Performance of DR groups according to three evaluation scores: AUC (black bars), Acc (grey bars) and MCC (light grey bars). The groups are sorted according to decreasing AUC score. The error bars on the plot indicate boundaries of the 95% confidence intervals for each measure.
Figure 3.
ROC curves of disordered region predictions for all CASP9 groups. Legends are shown for the best 12 groups according to the AUC. There are four non-regular ROCs corresponding to poorly performing groups, two of which misinterpreted DR format (G193 used only a single value for ordered residues and G114 did not use continuous scale but rather 5 different numbers).
Table I shows that prdos2 is the only group to rank among the top three prediction groups according to all three evaluation measures. In addition to this group, there are three other groups (Zhou-Spine-D, Multicom-refine and biomine_dr_pdb) to rank among the best 10 groups according to all three measures.
Figure 2 shows that there are several groups that perform equally well according to the Acc measure (grey bars). However, as we have discussed in Materials and Methods, some high Acc scores may be an artifact due to over-prediction of disordered residues. As the scores for top groups are very close, the statistical significance of the differences between them could not be established by the comparison of the confidence intervals.
Group DisoPred3C has obtained a relatively low Acc score but, at the same time, the best and the second best MCC and AUC scores, respectively. The high MCC score can be attributed to the high (highest in CASP9) precision (4) of classifications submitted by this group. The low Acc score is most likely due to the relatively low levels of disorder prediction. Based on the comparison of confidence intervals for the MCC scores, results of DisoPred3C are statistically better than those of all other groups, except for biomine_dr_pdb_c, which is second best according to the MCC.
Groups prdos2, DisoPred3C and Multicom are the best performing groups according to the probability-based assessment. These groups are statistically indistinguishable from each other by the AUC score and better than all other groups according to the results of the DeLong tests (see Table II). This conclusion is also confirmed by the comparison of the AUC confidence intervals (Table I).
Table II.
Statistical comparison of the best 12 groups according to the AUC score
| 291 | 015 | 490 | 474 | 468 | 119 | 094 | 351 | 030 | 374 | 165 | 177 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 291 | X | |||||||||||
| 015 | 0.97 | X | ||||||||||
| 490 | 0.79 | 0.83 | X | |||||||||
| 474 | <0.01 | <0.01 | <0.01 | X | ||||||||
| 468 | <0.01 | <0.01 | <0.01 | 0.92 | X | |||||||
| 119 | <0.01 | <0.01 | <0.01 | 0.14 | 0.18 | X | ||||||
| 094 | <0.01 | <0.01 | <0.01 | 0.10 | 0.14 | 0.93 | X | |||||
| 351 | <0.01 | <0.01 | <0.01 | 0.10 | 0.13 | 0.87 | 0.93 | X | ||||
| 030 | <0.01 | <0.01 | <0.01 | 0.10 | 0.13 | 0.86 | 0.92 | 0.99 | X | |||
| 374 | <0.01 | <0.01 | <0.01 | 0.04 | 0.05 | 0.61 | 0.66 | 0.74 | 0.75 | X | ||
| 165 | <0.01 | <0.01 | <0.01 | <0.01 | 0.01 | 0.28 | 0.30 | 0.37 | 0.38 | 0.56 | X | |
| 177 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.03 | X |
Results of the non-parametric DeLong tests on the AUC scores. Shaded cells highlight pairs of groups that are statistically indistinguishable at the 0.05 significance level.
Evaluation of results for longer disorder regions
As noted earlier, short disordered regions prevail in the CASP data set (see Figure 1). While such regions may sometimes consist of chain termini or short loops without any obvious functional role, they are often of functional importance (for example, flaps over enzyme active sites, pieces of chain that order into DNA grooves, or loops that become ordered in protein-protein interfaces), so their inclusion in the methods testing is important. At the same time, long disorder regions require separate attention as they are found in abundance in the human disease-associated proteins48 and their properties and functional roles are likely different from those of short disorder regions (for example, ordering of complete domains upon complex formation). The issue of the different length of disordered regions has been taken into consideration in several disorder predictions methods49–53. To address this issue in the assessment, we additionally evaluated the predictions taking into account only segments longer than a specified length cutoff.
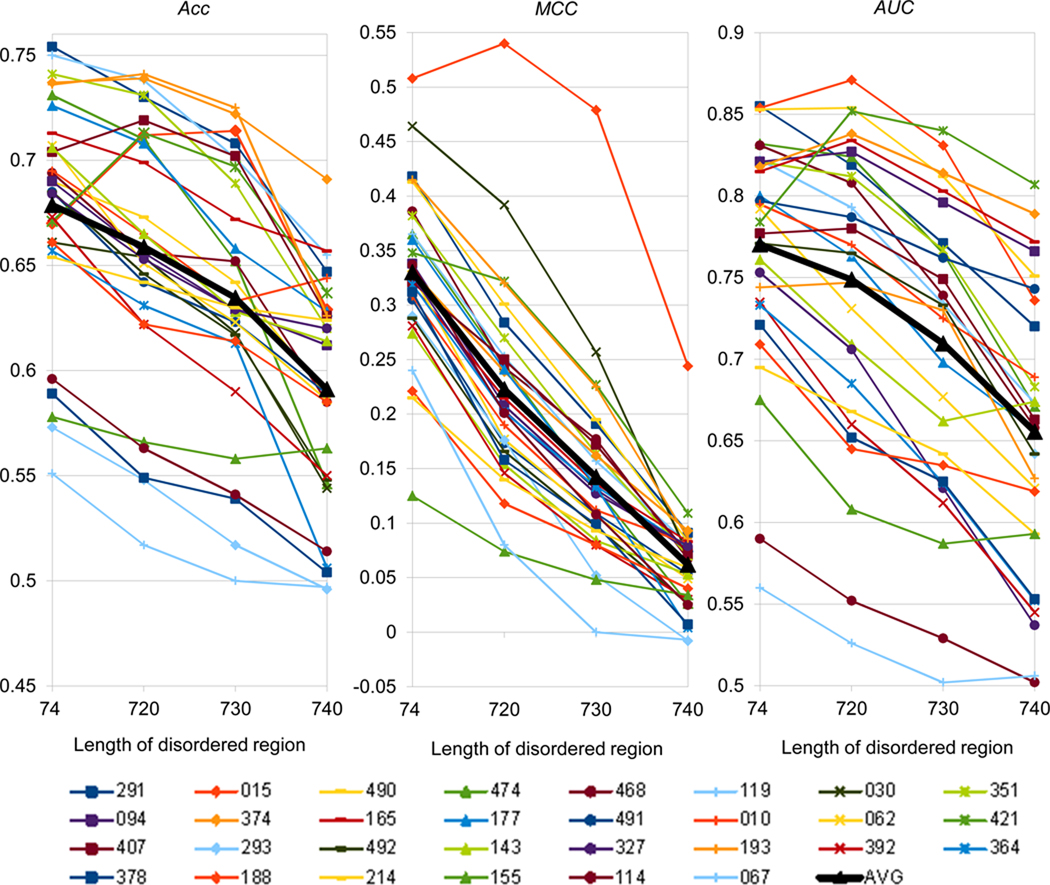
Figure 4 compares results of CASP9 methods for four minimum length thresholds: 4, 20, 30 and 40 residues. The “average group” splines (‘AVG’, thicker line) in all three panels of the graph show that the discriminatory power of the methods tends to decrease with the increase of the minimum disorder segment length. The average drop in performance is moderate according to the Acc and AUC scores and more pronounced according to the MCC score. The MCC panel suggests that an average CASP9 method can identify 40+ residue long disorder segments just slightly better than a random predictor (MCC=0). It should be mentioned, though, that the results for disorder regions spanning 40 residues or more should be interpreted with caution as there were only four qualified segments constituting to only 0.8% of all residues in CASP9 targets.
Figure 4.
Comparison of prediction performance across four different minimum disorder segment length thresholds. Different panels show scores for different evaluation measures (Acc, MCC and AUC). Each group is marked with a different color; groups in the legend are sorted according to the AUC score (across and then down); the artificial average group (‘AVG’, black thicker line) is added to the graph as a point of reference.
Curves for the vast majority of participating groups follow the average trend to decrease, resulting in high correlation (0.85 – 0.98) between the scores of the same evaluation measure at neighboring length thresholds. The lowest (even though still high in absolute value: 0.85) correlation between the 4+ and 20+ Acc score sets reflects the fact that this score was the most prone to the shifting of ranks. While the majority of groups performed worse in identifying 20+ residue long disorder segments (compared to 4+ segments), there were five groups that performed somewhat better, with two of them - DisoPred3C (G015) and GSMetaDisorder3D (G421) - improving their Acc scores significantly (by more than 6%) and consequently raising their ranks by 13 positions (to #8 and #7). DisoPred3C is the only group that demonstrated an ability to better discriminate 20+ residue long disorder regions according to all three evaluation scores, and is the best group in this length range according to the MCC and AUC scores. This group also has quite high scores for the 30+ residue-long regions, comparable to those they obtained for the 4+ ones. GSMetaDisorder3D also proved to be successful in identifying longer disorder segments, consistently placing in the best three according to the MCC and AUC scores at all longer disorder length levels.
Comparison between recent CASPs
To compare the accuracy of disorder predictions across all CASPs, we have re-evaluated predictions in previous CASPs using exactly the same disorder definitions and evaluation measures as in CASP9. Even so, it is hard to ensure full objectivity of such a comparison as the targets, methods and databases change in time.
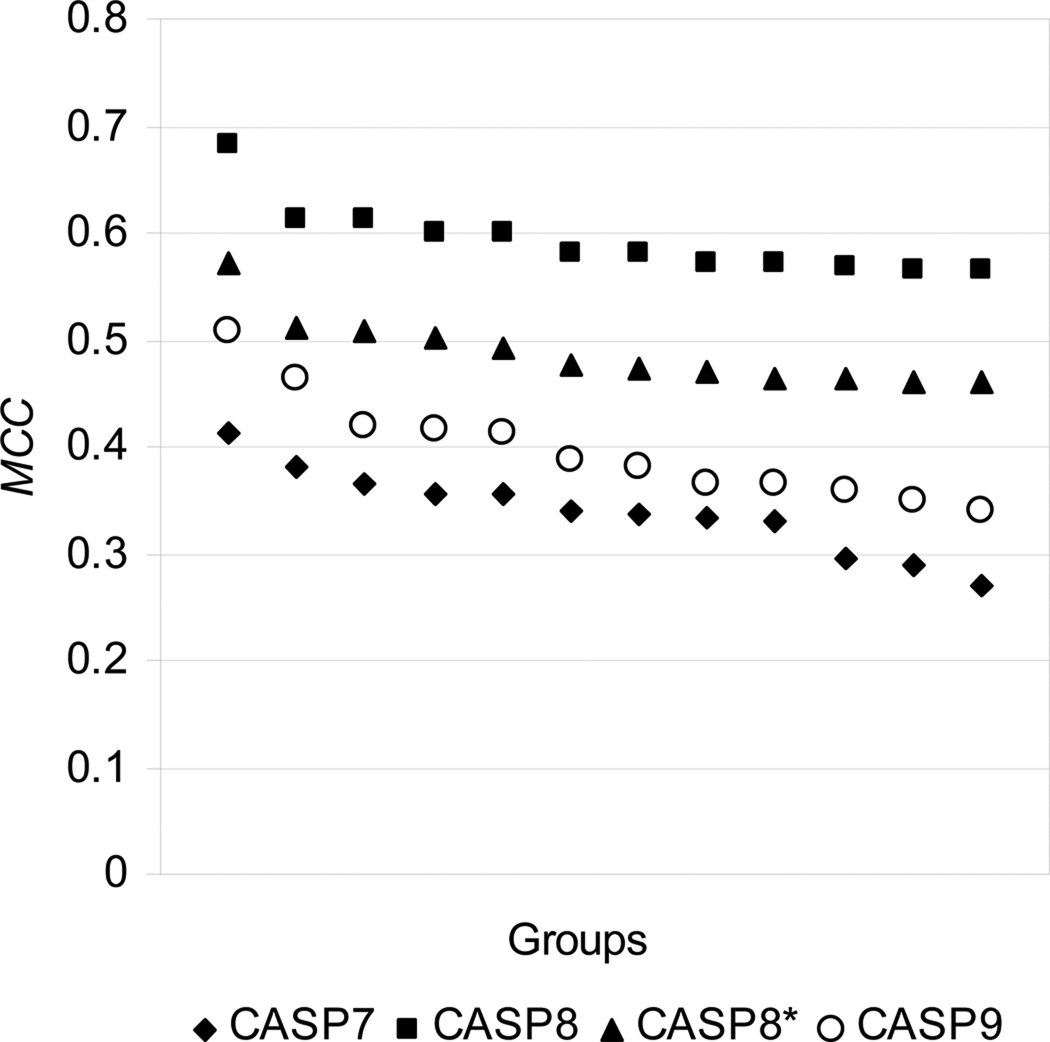
Figure 5 shows the results of a comparison of the MCC scores for the twelve best performing groups in the latest three CASPs. The CASP9 scores are higher than those in CASP7 but lower than in CASP8. This tendency holds when these methods are compared using other scores (see Figure S1 in Supplementary Material). As the majority of the top performing CASP8 methods were among the best also in CASP9, the drop in performance is most likely due to a greater difficulty of the CASP9 targets54.
Figure 5.
Comparison of the performance of the best 12 groups in the latest 3 CASPs. Groups in each CASP are sorted according to the MCC score. CASP8 results are evaluated for both the full set of targets and the set without target T0500, a long, completely unfolded protein considerably influencing the scores. The reduced target set is marked with an asterisk in the legend. The scores in CASP9 are higher than in CASP7 but lower than in CASP8. The CASP8–CASP9 drop in scores may be attributed to the greater difficulty of targets in CASP9.
CONCLUSIONS
The number of disorder prediction methods published in the literature is now well over fifty24 and continues to grow as new methods continue to appear55–58. This growth correlates well with the increase in the number of disorder prediction groups participating in CASP experiments. However, the increased number of participating groups does not seem to result in a better performance. Rather, our analysis show that the scores obtained in CASP9 have slightly decreased in comparison with those in CASP8 according to all three measures used in the CASP9 evaluation. As we discussed in this paper, this might be related to a higher difficulty of the CASP9 targets but perhaps also to the lack of conceptually new methods. New meta-predictors or slight modifications of established methods were not sufficient to achieve substantial progress in the field. By analyzing the submitted abstracts we could identify only one group (Zhou-Spine-D), claiming development of a conceptually new method based on neural networks. This method was assessed to be among top 10 according to all scores in CASP9, but did not outperform other already established CASP performers. A brief description of the best performing automatic methods participated in CASP9 is provided in Table III. It seems that performance of disorder prediction methods in CASP has reached a plateau and new breakthroughs are needed.
Table III.
Methods description for the best CASP9 DR servers
| CASP9 group name and HTTP address | Description |
|---|---|
| PrDOS259 http://prdos.hgc.jp/cgi-bin/top.cgi | SVM algorithm based on sequence profiles combined with a template-based predictor. |
| DisoPred3C44 http://bioinf.cs.ucl.ac.uk/disopred | SVM trained on high-resolution X-ray structures. Uses profiles from 15 positions around each residue as an input vector. |
| MULTICOM-refine55 casp.rnet.missouri.edu/predisorder.html | One-Dimensional Recursive Neural Network (1D-RNN). Predicts the disorder probability of each residue along a protein sequence taking as input the sequence profile, predicted secondary structure, and solvent accessibility. |
| Zhou-Spine-D sparks.informatics.iupui.edu/SPINE-D | Two-layered Neural Network followed by a filter. The input features include residue-level and window-level information calculated from amino acid sequence, seven representative physical parameters60, PSI-BLAST profile, predicted secondary structure61 and solvent accessibility torsion-angle fluctuation62. |
| Zhou-Spine-DM sparks.informatics.iupui.edu/SPINE-DM | Meta approach that employs a two-layer Neural Network with a filter. Combines input from six disorder predictors: VSL249, DISOPred244, DisPro1.063, IUPred64 and SPINE-D (above). |
| CBRC_Poodle51 http://mbs.cbrc.jp/poodle/poodle-i.html | SVM integrating three own SVM predictors: Poodle-S65 and Poodle-L66 specialized in short and long disorder regions, respectively, and Poodle-W67 targeting unfolded protein prediction. |
| biomine_dr_pdb, biomine_dr_pdb_c57 biomine-ws.ece.ualberta.ca/MFDp.html | Two meta + SVM approaches. Combine predictions from DISOPred244, DISOclust68 and IUPred64, and additionally use predicted secondary structure, solvent accessibility, B-factors and backbone dihedral torsion angles in SVM learning. |
| GSmetaDisorderMD, GSmetaserver http://iimcb.genesilico.pl/metadisorder/ | Two Genetic Algorithms combining predictions from GSmetaDisorder (consensus of 13 DR servers) and GSmetaDisorder3D (consensus of missing residues in multiple sequence alignments produced by fold recognition methods). The two methods use different weight optimization scores. |
| OnD-CRF69 http://babel.ucmp.umu.se/ond-crf/ | Machine learning technique based on Conditional Random Fields. Uses only sequence and predicted secondary structure as inputs. |
| Mason70 www.cs.gmu.edu/~mlbio/svmprat | SVM method integrating flexible window-based profile kernels and predicted secondary structure. |
| McGuffin (as DISOClust68 on all CASP9 3D server models) www.reading.ac.uk/bioinf/DISOclust/ | DISOClust server was tested in CASP9 as a server group (IntFOLD-DR) and a human-expert group (McGuffin). Both groups exploited the same approach based on conservation of ordered residues within multiple structures. The McGuffin group used consensus of all 3D server models submitted to CASP, while IntFOLD-DR - only of those available from the in-house nFOLD4 method. |
Besides more effective disorder prediction methods, we also need better target sets in CASP, since the vast majority of targets are solved by X-ray crystallography and therefore typically contain only short disorder regions. This type of data likely does not fully represent the type of disorder observed in functionally relevant long disordered segments. Thus, test sets containing more targets with extended disordered regions are required for more comprehensive testing of disorder prediction methods.
For the first time, we have analyzed differences in the capability of methods to recognize disorder regions of different length. The surprising result is that, independent of the exact evaluation metrics, there is a rather dramatic fall-off in performance with disorder length increase. Perhaps this reflects a tendency for the methods used for CASP to be trained on the short disorder segments typical of the targets. Nevertheless, it is a disturbing result.
Supplementary Material
ACKNOWLEDGEMENTS
This work was partially supported by the US National Library of Medicine (NIH/NLM) – grant LM007085 to KF and by Award No. KUK-I1-012-43 made by King Abdullah University of Science and Technology (KAUST) to AT.
Abbreviations
- 3D
three-dimensional
- DR
disordered residues
- IDP (IDR)
Intrinsically Disordered Protein (Region)
- MCC
the Matthews Correlation Coefficient
- ROC
the Receiver Operating Characteristic
- AUC
Area Under the ROC Curve
Footnotes
Target T0549, which was excluded from the tertiary structure assessment as lacking big parts of structure, was retained for the disorder assessment.
This definition slightly differs from the previous CASP definitions, in part due to the 3.5Å deviation criterion for X-ray structures, which classifies additional 246 residues (or 1.0% of all residues in CASP9 X-ray structures) as disordered.
REFERENCES
- 1.Wu H. Studies on denaturation of proteins. XIII. A theory of denaturation. Chin J Physiol. 1931;1:219–234. [PubMed] [Google Scholar]
- 2.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(96):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 5.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith CH. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol. 2005;345(2):275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 8.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4(12):728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue B, Williams RW, Oldfield CJ, Dunker AK, Uversky VN. Archaic chaos: intrinsically disordered proteins in Archaea. BMC Syst Biol. 2010;4 Suppl 1:S1. doi: 10.1186/1752-0509-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogatyreva NS, Finkelstein AV, Galzitskaya OV. Trend of amino acid composition of proteins of different taxa. J Bioinform Comput Biol. 2006;4(2):597–608. doi: 10.1142/s0219720006002016. [DOI] [PubMed] [Google Scholar]
- 11.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 12.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6(5):1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9 Suppl 2:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35(Database issue):D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6(5):1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10 Suppl 1:S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raychaudhuri S, Dey S, Bhattacharyya NP, Mukhopadhyay D. The role of intrinsically unstructured proteins in neurodegenerative diseases. PLoS ONE. 2009;4(5):e5566. doi: 10.1371/journal.pone.0005566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracken C, Iakoucheva LM, Romero PR, Dunker AK. Combining prediction, computation and experiment for the characterization of protein disorder. Curr Opin Struct Biol. 2004;14(5):570–576. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disorder regions in proteins from amino acid sequence. Proc IEEE Int Conf Neural Networks. 1997;1:90–95. [Google Scholar]
- 23.Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65(1):1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 24.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19(8):929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 25.Dosztanyi Z, Meszaros B, Simon I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief Bioinform. 2010;11(2):225–243. doi: 10.1093/bib/bbp061. [DOI] [PubMed] [Google Scholar]
- 26.Moult J, Fidelis K, Zemla A, Hubbard T. Critical assessment of methods of protein structure prediction (CASP)-round V. Proteins. 2003;53 Suppl 6:334–339. doi: 10.1002/prot.10556. [DOI] [PubMed] [Google Scholar]
- 27.Kinch L, Shi S, Cheng H, Cong Q, Pei J, Schwede T, Grishin N. CASP9 target classification. Proteins. 2011 doi: 10.1002/prot.23190. (Current) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melamud E, Moult J. Evaluation of disorder predictions in CASP5. Proteins. 2003;53 Suppl 6:561–565. doi: 10.1002/prot.10533. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Dunbrack RL., Jr Assessment of disorder predictions in CASP6. Proteins. 2005;61 Suppl 7:167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- 30.Bordoli L, Kiefer F, Schwede T. Assessment of disorder predictions in CASP7. Proteins. 2007;69 Suppl 8:129–136. doi: 10.1002/prot.21671. [DOI] [PubMed] [Google Scholar]
- 31.Noivirt-Brik O, Prilusky J, Sussman JL. Assessment of disorder predictions in CASP8. Proteins. 2009;77 Suppl 9:210–216. doi: 10.1002/prot.22586. [DOI] [PubMed] [Google Scholar]
- 32.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Stec B, Godzik A. Between order and disorder in protein structures: analysis of "dual personality" fragments in proteins. Structure. 2007;15(9):1141–1147. doi: 10.1016/j.str.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobanov MY, Furletova EI, Bogatyreva NS, Roytberg MA, Galzitskaya OV. Library of disordered patterns in 3D protein structures. PLoS Comput Biol. 2010;6(10):e1000958. doi: 10.1371/journal.pcbi.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19(9):1141–1164. doi: 10.1002/(sici)1097-0258(20000515)19:9<1141::aid-sim479>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 37.Wilcox RR. Fundamentals of modern statistical methods : substantially improving power and accuracy. New York, NY: Springer; 2010. p. 249. xvi. [Google Scholar]
- 38.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 39.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 40.Carugo O. Detailed estimation of bioinformatics prediction reliability through the Fragmented Prediction Performance Plots. BMC Bioinformatics. 2007;8:380. doi: 10.1186/1471-2105-8-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16(5):412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 42.van Rijsbergen CJ. Foundation of evaluation. J of Documentation. 1974;30(4):365–373. [Google Scholar]
- 43.Sokolova M, Japkowicz N, Szpakowicz S. Beyond Accuracy, F-score and ROC: a Family of Discriminant Measures for Performance Evaluation. Lecture Notes in Comp Sci. 2006;4304:1015–1021. [Google Scholar]
- 44.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Payton ME, Greenstone MH, Schenker N. Overlapping confidence intervals or standard error intervals: what do they mean in terms of statistical significance? J Insect Sci. 2003;3:34. doi: 10.1093/jis/3.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The R development Core Team. Vienna: 2006. R: a language and environment for statistical computing. [Google Scholar]
- 47.2011 http://ca.expasy.org/tools/pROC/ [Google Scholar]
- 48.Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YY, Romero P, Cortese MS, Uversky VN, Dunker AK. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24(10):435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13(1):71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirose S, Shimizu K, Noguchi T. POODLE-I: Disordered region prediction by integrating POODLE series and structural information predictors based on a workflow approach. In Silico Biology. 2010;10(0015) doi: 10.3233/ISB-2010-0426. [DOI] [PubMed] [Google Scholar]
- 52.Han P, Zhang X, Norton RS, Feng ZP. Large-scale prediction of long disordered regions in proteins using random forests. BMC Bioinformatics. 2009;10:8. doi: 10.1186/1471-2105-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vullo A, Bortolami O, Pollastri G, Tosatto SC. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34(Web Server issue):W164–W168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kryshtafovych A, Fidelis K, Moult J. CASP9 results compared to those of previous CASP experiments. Proteins. 2011 doi: 10.1002/prot.23182. (Current) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng X, Eickholt J, Cheng J. PreDisorder: ab initio sequence-based prediction of protein disordered regions. BMC Bioinformatics. 2009;10:436. doi: 10.1186/1471-2105-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS ONE. 2009;4(2):e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizianty MJ, Stach W, Chen K, Kedarisetti KD, Disfani FM, Kurgan L. Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics. 2010;26(18):i489–i496. doi: 10.1093/bioinformatics/btq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35(Web Server issue):W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meiler J, Muller M, Zeidler A, Schmaschke F. Generation and evaluation of dimension-reduced amino acid parameter representations by artificial neural networks. J Mol Model. 2001;7(9):360–369. [Google Scholar]
- 61.Faraggi E, Yang Y, Zhang S, Zhou Y. Predicting continuous local structure and the effect of its substitution for secondary structure in fragment-free protein structure prediction. Structure. 2009;17(11):1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang T, Faraggi E, Zhou Y. Fluctuations of backbone torsion angles obtained from NMR-determined structures and their prediction. Proteins. 2010;78(16):3353–3362. doi: 10.1002/prot.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng JL, Sweredoski MJ, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Min Knowl Disc. 2005;11(3):213–222. [Google Scholar]
- 64.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 65.Shimizu K, Hirose S, Noguchi T. POODLE-S: web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics. 2007;23(17):2337–2338. doi: 10.1093/bioinformatics/btm330. [DOI] [PubMed] [Google Scholar]
- 66.Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. POODLE-L: a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics. 2007;23(16):2046–2053. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 67.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGuffin LJ. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics. 2008;24(16):1798–1804. doi: 10.1093/bioinformatics/btn326. [DOI] [PubMed] [Google Scholar]
- 69.Wang L, Sauer UH. OnD-CRF: predicting order and disorder in proteins using [corrected] conditional random fields. Bioinformatics. 2008;24(11):1401–1402. doi: 10.1093/bioinformatics/btn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rangwala H, Kauffman C, Karypis G. svmPRAT: SVM-based protein residue annotation toolkit. BMC Bioinformatics. 2009;10:439. doi: 10.1186/1471-2105-10-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.