Abstract
The synthesis and degradation of anthropogenic and natural organohalides are the basis of a global halogen cycle. Chlorinated hydroquinone metabolites (CHMs) synthesized by basidiomycete fungi and present in wetland and forest soil are constituents of that cycle. Anaerobic dehalogenating bacteria coexist with basidiomycete fungi in soils and sediments, but little is known about the fate of these halogenated fungal compounds. In sediment microcosms, the CHMs 2,3,5,6-tetrachloro-1,4-dimethoxybenzene and 2,3,5,6-tetrachloro-4-methoxyphenol (TCMP) were anaerobically demethylated to tetrachlorohydroquinone (TCHQ). Subsequently, TCHQ was converted to trichlorohydroquinone and 2,5-dichlorohydroquinone (2,5-DCHQ) in freshwater and estuarine enrichment cultures. Screening of several dehalogenating bacteria revealed that Desulfitobacterium hafniense strains DCB2 and PCP1, Desulfitobacterium chlororespirans strain Co23, and Desulfitobacterium dehalogenans JW/DU1 sequentially dechlorinate TCMP to 2,3,5-trichloro-4-methoxyphenol and 3,5-dichloro-4-methoxyphenol (3,5-DCMP). After a lag, these strains demethylate 3,5-DCMP to 2,6-DCHQ, which is then completely dechlorinated to 1,4-dihydroquinone (HQ). 2,5-DCHQ accumulated as an intermediate during the dechlorination of TCHQ to HQ by the TCMP-degrading desulfitobacteria. HQ accumulation following TCMP or TCHQ dechlorination was transient and became undetectable after 14 days, which suggests mineralization of the fungal compounds. This is the first report on the anaerobic degradation of fungal CHMs, and it establishes a fundamental role for microbial reductive degradation of natural organochlorides in the global halogen cycle.
Anthropogenic production of organohalides stands at more than 15,000 compounds (20), while an additional 3,000 are biotically or abiotically synthesized in the environment (13, 14). The natural production of halogenated organic compounds represents the synthesis portion of a global halogen cycle (see reference 25 for a recent review). These compounds were potential substrates for microbial dehalogenation long before the human introduction of additional organohalides. The microbial degradation of the halogenated aromatic compounds into nonhalogenated organic material, biomass, or CO2 is requisite for the completion of the halogen cycle. Many bacteria and fungi are capable of total or partial degradation of anthropogenic halogenated compounds such as chlorophenols, chlorobenzoates, polychlorinated biphenyls, chlorobenzenes, and chlorinated aliphatic compounds (reviewed in references 15, 17, and 48). Far less is known about the biodegradation, especially anaerobic degradation, of natural organohalides.
Basidiomycete fungi are particularly adept at synthesizing halogenated aromatic compounds, such as the chlorinated hydroquinone metabolites (CHMs) (reviewed in reference 9). In addition to being chlorinated, CHMs are symmetrically hydroxylated or methoxylated at the 1 and 4 positions. The CHM 2,3,5,6-tetrachloro-4-methoxyphenol (TCMP, also known as drosophilin A) was the first chlorinated compound to be isolated and characterized from a basidiomycete fungus (1). Ten different genera of fungi are known to produce CHMs (9, 37, 39). TCMP and 2,3,5,6-tetrachloro-1,4-dimethoxybenzene (TCDB), another CHM of fungi, can also be produced microbially by aerobic bacteria. Suzuki (36) demonstrated that a soil Mycobacterium sp. could dechlorinate by hydrolysis anthropogenic pentachlorophenol (PCP) to tetrachlorohydroquinone (TCHQ), which the bacterium then methylates. Häggblom et al. (18) showed that Rhodobacter spp. could also methylate TCHQ to TCMP and TCDB.
The fungal CHMs are environmentally significant due to their antimicrobial activity (1, 27) and their potential physiological role during lignin degradation, since they can serve as substrates for lignin peroxidases (19, 42). The fungi that synthesize CHMs are common and produce high levels of absorbable organic halogens, including CHMs, when grown on ligninocellulosic substrates (45). Specifically, TCMP has been detected in composite forest litter (9). The deposition of the CHMs into the environment in association with degrading plant material renders these compounds available to anaerobic dehalorespiring bacteria, which utilize halogenated organic compounds as terminal electron acceptors for growth. The introduction of halogenated aromatic compounds by fungi into anoxic environments such as flooded soil, sediment, and rotting logs could have driven the evolution of reductive dehalogenation in anaerobic bacteria and influenced anaerobic metabolism of aromatic compounds in general.
Other than reports on the anaerobic biodegradation of a few mono- and dichlorinated chlorinated anisyl metabolites (CAMs) produced by fungi (30, 40, 46), and bromophenols produced by marine organisms (21, 35), very little is known about the anaerobic microbial metabolism of natural organohalides. This is especially so for the extensively chlorinated fungal CHMs. Since many CHMs are heavily chlorinated, they have the potential to serve as more thermodynamically effective electron acceptors than less chlorinated aromatic compounds. Herein we describe the microbial pathways for anaerobic biodegradation of fungal CHMs with a focus on TCMP. The results establish fundamental pathways in the global halogen cycle associated with the biodegradation of natural organohalides.
MATERIALS AND METHODS
Chemicals and synthesis of CHMs.
The following compounds were purchased from Sigma-Aldrich: TCHQ, 2,5-dichloro-1,4-dihydroquinone (2,5-DCHQ), 2-chloro-1,4-dihydroquinone (2-CHQ), 1-4-dihydroquinone (hydroquinone [HQ]), 2-chloro-4-methoxyphenol (2-CMP), 4-methoxyphenol, 2,3,5,6-tetrachloro-1,4-benzoquinone, 2,5-dichloro-1,4-benzoquinone, 2,6-dichloro-1,4-benzoquinone, and 2-chloro-1,4-benzoquinone.
2,3,5-Trichloro-1,4-dihydroquinone (TriCHQ) and 2,3,5-trichloro-1,4-benzoquinone were prepared from 2,6-dichloro-1,4-benzoquinone as described by Renner and Hopfer (29). 2,3-Dichloro-1,4-hydroquinone (2,3-DCHQ) and 2,3-dichloro-1,4-benzoquinone were prepared from hydroquinone as described by Yu and Mattern (51). 2,6-Dichloro-1,4-dihydroquinone (2,6-DCHQ) was prepared from 2,6-dichloro-1,4-benzoquinone by the method described by Yu and Mattern (51). TCDB, TCMP, and 2,5-dichloro-4-methoxyphenol (2,5-DCMP) were prepared as described by Ramirez et al. (28). Reaction of 2,3-dichloro-1,4-benzoquinone with trimethyl phosphite under Ramirez's conditions (benzene and alkaline hydrolysis) yielded 2,3-dichloro-4-methoxyphenol (2,3-DCMP). Under similar reaction conditions, 2,3,5-trichloro-1,4-benzoquinone yielded a mixture of 2,3,6-trichloro-4-methoxyphenol (2,3,6-TriCMP) and 2,3,5-trichloro-4-methoxyphenol (2,3,5-TriCMP) (1:4). Reaction of 2,6-dichloro-1,4-benzoquinone with trimethyl phosphite resulted in the isomeric 2,6-dichloro-4-methoxyphenol (2,6-DCMP) and 3,5-dichloro-4-methoxyphenol (3,5-DCMP) in a 6:1 ratio. Preparative thin-layer chromatography and column chromatography (with SiO2, ethyl acetate, and hexanes) were used to purify compounds to ≥90% purity (based on 1H-nuclear magnetic resonance spectroscopic and gas chromatographic-mass spectrometry analysis). Spectroscopic data for the chlorinated methoxyphenols were identical to those reported by Knuutinen et al. (22). All compounds were stored in the dark.
Sample preparation for CHM analysis.
Whole cultures (10 ml) were extracted twice with 2 ml of high-pressure liquid chromatography-grade ethyl acetate (Fisher Scientific) by shaking for 1 h. The organic layers were pooled in a gas chromatography vial and evaporated under a stream of N2. The residue was derivatized in 300 μl of ethyl acetate with 100 μl of bis(trimethylsilyl)trifluoroacetamide (BSTFA) (Sigma-Aldrich) and analyzed immediately on a gas chromatograph-mass spectrometer (GC-MS).
Recovery of CHMs and their degradation products was approximately 60 to 70% at time zero from both axenic and active sediment-free cultures. Recovery of CHM substrates from abiotic (sterile) controls remained unchanged after 60 days, and no degradation products were ever observed.
CHM analysis.
CHMs were identified by matching retention times, molecular weights, relative natural isotopic abundance of 35Cl/37Cl (49), and fragmentation patterns with those of authentic standards. Relative retention times and molecular masses of each compound and its derivative with BSTFA are presented in Table 1. Analysis was conducted with a Hewlett-Packard 6890-5972A GC-MS (Agilent) equipped with a HP-5MS 5% phenylmethyl Siloxane capillary column (30 m by 0.25 mm by 0.25 μm [film thickness]) and the following oven program: initial temperature gradient at 20°C/min from 80 to 180°C, temperature hold at 180°C for 16 min, final temperature gradient at 50°C/min from 180 to 280°C, final temperature hold at 280°C for 2 min. When 3,5-DCMP and 2-CHQ required baseline separation, the oven program was as described above except that the initial temperature gradient was run at 2.5°C/min.
TABLE 1.
Relative retention times of trimethylsilyl derivatized CHMs
| Compound | Relative RTa |
|---|---|
| TCHQ | 1.000 |
| TriCHQ | 0.699 |
| 2,6-DCHQ | 0.539 |
| 2,5-DCHQ | 0.524 |
| 2,3-DCHQ | 0.575 |
| 2-CHQb | 0.453 |
| HQ | 0.392 |
| TCMP | 0.762 |
| 2,3,6-TriCMP | 0.645 |
| 2,3,5-TriCMP | 0.563 |
| 2,5-DCMP | 0.483 |
| 3,5-DCMPb | 0.455 |
| 2,3-DCMP | 0.526 |
| 2,6-DCMP | 0.481 |
| 2-CMP | 0.412 |
| 4-MP | 0.352 |
| TCDB | 0.600 |
Retention time (RT) relative to TCHQ, defined as 1.000.
2-CHQ and 3,5-DCMP achieve baseline separation (not shown) with an alternate GC-MS method (see Materials and Methods).
Cultures and media.
Pure cultures of dehalogenating bacteria were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) or American Type Culture Collection (ATCC): Desulfitobacterium chlororespirans Co23 (DSM 11544), Desulfitobacterium dehalogenans JW/DU1 (ATCC 51507), Desulfitobacterium hafniense DCB2 (DSM 10664), D. hafniense DB7 (DSM 13498), D. hafniense PCP1 (ATCC 700357), D. hafniense TCE1 (DSM 12704), Desulfitobacterium sp. strain PCE1 (DSM 10344), and Dehalospirillum multivorans (DSM 12446). Each strain was initially cultured in its recommended DSMZ or ATCC medium. For comparative purposes, all organisms were grown in DSMZ 720. DSMZ 720 contains the following salts and solutions (per liter): 1 g of NH4Cl, 0.1 g of NaCl, 0.1 g of MgCl2 · 6H2O, 0.05 g of CaCl2 · 2H2O, 0.3 g of K2HPO4, 1 g of yeast extract, 2.6 g of NaHCO3, 0.3 g of Na2S · 9H2O, and 1 ml of trace element solution SL-10 (DSMZ 320) with 0.5 g of Na2-EDTA per liter, 10 ml of resazurin at 0.1 g/liter, 10 ml of vitamin solution (DSMZ 141) (50), 10 ml of 25% (wt/vol) CH3COCOONa, 5 ml of 25% (wt/vol) Na2S2O3 · 5H2O. The solutions were prepared anaerobically under 80% N2 plus 20% CO2 as described previously (3), and the pH was adjusted to 7.0 with HCl. Ten-milliliter cultures were inoculated with 5 × 105 exponentially growing cells. Following inoculation, the CHM was delivered in filter-sterilized 60% (vol/vol) ethanol (5 mM final concentration in medium) via a digital microdispenser (Drummond, Broomall, Pa.) for a final concentration of 163 μM TCDB, 173 μM TCMP, or 183 μM TCHQ. The cultures were maintained and transferred under anaerobic conditions in anaerobic culture tubes (Belco, Vineland, N.J.) sealed with Teflon-coated butyl stoppers (West Co., Lionville, Pa.) secured by an aluminum crimp seal. All cultures were incubated in the dark at 30°C.
Enrichment cultures.
Initial inocula were prepared by suspending estuarine (Baltimore Harbor, 39°16.8′N, 76°36.1′W) and freshwater (pond in the Cumberland Mountains of Tennessee, 36°21.72′N, 84°42.07′W) sediment (20% [vol/vol]) into anaerobic estuarine (3) or freshwater (34) media. Following shaking and settling for clarification (approximately 10 min), 1% (vol/vol) of the supernatant from these anaerobic suspensions was transferred to sediment-free media. This constituted the first in a series of three or four sequential transfer cultures. Each subsequent transfer culture was made as a 1% (vol/vol) transfer after 30 days of incubation. TCDB, TCMP, or TCHQ was delivered in ethanol to each culture as described above. All cultures were transferred under anaerobic conditions as described above and incubated in the dark at 30°C.
RESULTS
Biodegradation of CHMs by enriched microbial communities.
Several series of sediment-free enrichment cultures were established with microorganisms from freshwater or estuarine sediment. Each enrichment culture was sequentially transferred three or four times (single cultures) in series with TCDB, TCMP, or TCHQ. TCHQ is not reported to be synthesized by fungi, but it was included in the study because of its structural similarity to the CHMs and because it was detected in preliminary examinations of sediment microcosms incubated with TCDB or TCMP (data not shown).
Table 2 summarizes the results of CHM biodegradation obtained with each enrichment culture series. In general, the microbial communities enriched from both environments exhibited similar degradation patterns for CHMs. At no time were less chlorinated methoxyphenols detected in any of the enrichment cultures incubated with TCDB or TCMP, indicating that dechlorination was not the first CHM transformation process performed by the mixed microbial communities. Instead, demethylation to TCHQ was observed with all but one of the enrichment cultures maintained with TCDB or TCMP. Following demethylation to TCHQ, TriCHQ was produced by all four sequential transfer cultures inoculated with estuarine microorganisms and grown with TCMP. Further dechlorination to 2,5-DCHQ was observed in the fourth sequential transfer culture of this series. The freshwater microorganisms dechlorinated TCHQ but not TCMP. Since demethylation and dechlorination occurred with both freshwater and estuarine enrichment cultures maintained with either TCMP or TCHQ, it is apparent that microorganisms from both environments sequentially demethylate and dechlorinate CHMs.
TABLE 2.
CHM degradation products observed with enrichment cultures at 18 days
| Enrichment Culture | Concn (μM) ofa:
|
|||
|---|---|---|---|---|
| TCMP | TCHQ | TriCHQ | 2,5-DCHQ | |
| Estuarine | ||||
| With TCDB | <5-20 (2/3) | <5-50 (2/3) | <5 | <5 |
| With TCMP | 10-100 (4/4) | 10-30 (4/4) | <5-5 (1/4) | |
| With TCHQ | <5-20 (2/4) | <5-20 (2/4) | ||
| Freshwater | ||||
| With TCDB | 10-20 (3/3) | 10 (3/3) | <5 | <5 |
| With TCMP | 10-50 (3/3) | <5 | <5 | |
| With TCHQ | <5-40 (3/4) | 10-150 (3/4) | ||
Enrichment culture data are summarized for three or four enrichment cultures transferred sequentially and grown with one of the three CHMs. The number of sequential transfer enrichment cultures in which the compound was detected is in parentheses. Similar results were observed in replicate experiments. The detection limit was less than 5 μM.
Dechlorination products beyond 2,5-DCHQ were never observed in any of the enrichment cultures. After 60 days the CHMs and degradation products were undetectable. Degradation products were never observed in abiotic (sterile) controls, and the starting CHMs were always recoverable. The experiments were qualitative and not designed to ensure the acquisition of a final mass balance. Therefore, it is quite possible that further degradation, including the transformation to HQ or even CO2, may have occurred with live cultures. Similar sets of experiments with additional enrichment cultures (data not shown) yielded the same results, i.e., demethylation of the CHMs followed by dechlorination to 2,5-DCHQ.
Biodegradation of TCMP by axenic cultures.
Eight strains of anaerobic bacteria were tested for their ability to dechlorinate TCMP or TCHQ. These included four strains of Desulfitobacterium capable of dechlorinating chlorophenols (4, 5, 10, 23, 30, 40, 41), a Desulfitobacterium strain that is not capable of dechlorinating organohalides (strain DP7) (43), and three bacterial species that dechlorinate chlorinated ethenes (11, 12, 32), including one that dechlorinates tetrachloroethene and some chlorophenols (Desulfitobacterium strain PCE1). These experiments served two purposes: (i) to identify bacterial species capable of carrying out the reactions identified in the enrichment cultures, and (ii) to identify alternative pathways of anaerobic CHM biodegradation. The dechlorinating capabilities of all eight strains are summarized in Table 3. D. hafniense strains PCP1 and DCB2, D. dehalogenans strain JW/DU1, and D. chlororespirans strain Co23 readily dechlorinated TCMP and TCHQ within 3 days. D. hafniense strain DP7, which has a high phylogenetic similarity to strains PCP1 and DCB2 (43), was incapable of degrading the fungal CHMs within 30 days in this study. This is consistent with the organism's inability to dechlorinate chlorophenols or tetrachloroethene (43). Furthermore, TCMP completely inhibited the growth of strain DP7. None of the other strains dechlorinated the fungal CHMs.
TABLE 3.
Dechlorinating capability of species tested with fungal CHMsa
| Bacterium | Result with dehalogenation substrate
|
||||||
|---|---|---|---|---|---|---|---|
| This study
|
Previous studies
|
||||||
| TCMP | TCHQ | 3-Cl-4-OHPA | PCP | 3,4,5-TriCP | Other CPs | PCE/TCE | |
| Dehalospirillum multivorans | − | − | ND | ND | ND | ND | +/+ |
| Desulfitobacterium chlororespirans Co23 | + | + | + | − | ND | + | ND |
| Desulfitobacterium dehalogenans JW/DU1 | + | + | + | + | − | + | −/− |
| Desulfitobacterium hafniense DCB2 | + | + | + | + | − | + | −/− |
| Desulfitobacterium hafniense DP7 | − | − | − | − | ND | − | −/− |
| Desulfitobacterium hafniense PCP1 | + | + | − | + | + | + | −/− |
| Desulfitobacterium hafniense TCE1 | − | − | − | ND | ND | ND | +/+ |
| Desulfitobacterium sp. strain PCE1 | − | − | + | − | ND | + | +/− |
Sources for the data are as follows: TCMP and TCHQ data (all strains), this study; Desulfitobacterium sp. strain PCE1, references 11 and 12; D. hafniense PCP1, reference 10; D. dehalogenans, references 40 and 41; D. hafniense TCE1, reference 11; D. hafniense DP7, reference 43; D. chlororespirans, reference 30; D. multivorans, reference 32; D. hafniense DCB2, reference 5. 3-Cl-4-OHP, 3-chloro-4-hydroxyphenyl acetate; PCE and TCE, tetra- and trichloroethene; 3,4,5-TriCP, 3,4,5-trichlorophenol. Other chlorophenols (CPs) exclude PCP and 3,4,5-TriCP. ND, not determined or reported in the literature.
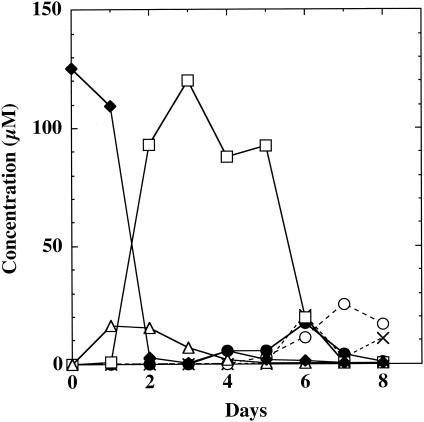
Each CHM-dechlorinating Desulfitobacterium strain degraded TCMP to HQ within 9 days, but the HQ recovered accounted for only a small percentage of the TCMP added to the cultures. After 14 days none of the TCMP or any of its degradation products, including HQ, were detected. To more fully characterize the degradation of TCMP, D. hafniense strain DCB2 was examined in greater detail. Figure 1 shows the time dependent transformation of TCMP by replicate cultures of strain DCB2. The degradation products were identified by matching retention times and mass spectra with those of authentic standards (data not shown). Degradation products were not observed in sterile controls. Within 1 day of incubation, TriCMP was detected and its formation preceded that of 3,5-DCMP. The dichloro- isomer accumulated to levels accounting for >68% of the TCMP added to the cultures, and it remained at these levels for several days. Demethylation, which preceded dechlorination in the mixed microbial communities of the enrichment cultures, occurred after the formation and accumulation of 3,5-DCMP in the pure culture of strain DCB2. GC-MS analysis of the dechlorination homologs of TCMP produced by the other CHM-dechlorinating bacteria demonstrated that all of these organisms accumulated 3,5-DCMP (data not shown). Both 2-CHQ and HQ were produced shortly after the demethylation of 3,5-DCMP to 2,6-DCHQ, but these hydroquinones did not accumulate to the levels of 3,5-DCMP. The other isomers of the mono- or dichloro-methoxyphenols and Tri-CHQ or TCHQ were not detected.
FIG. 1.
Time course of TCMP dechlorination and demethylation by D. hafniense strain DCB2. ♦, TCMP; ▵, 2,3,5-TriCMP; □, 3,5-DCMP; •, 2,6-DCHQ; ○, 2-CHQ; ×, HQ.
Biodegradation of TCHQ by axenic cultures.
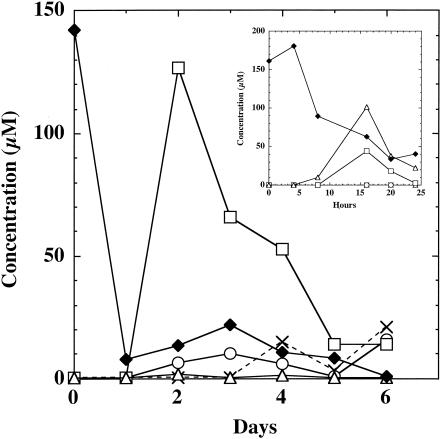
Since TCHQ is structurally similar to TCMP and is produced by the enrichment cultures, its dechlorination was also examined. Strain DCB2 was again used to illustrate the dechlorination time course of TCHQ (Fig. 2). Recovery of TCHQ and its dechlorination products was low after 24 h of incubation in replicate cultures. The experiment was repeated with additional sampling over the first 24 h (Fig. 2, inset). The results were similar, i.e., low recovery of substrate and products at 24 h, but the inset data demonstrate that TriCHQ and 2,5-DCHQ formed within 16 h. The data prove that the TriCHQ was an intermediate and accumulated to a modest but detectable level. The decrease in recovery of chlorinated hydroquinones at 24 h followed by an increase in recovery was observed with CHM-dechlorinating and nondechlorinating strains, e.g., strain DP7 (data not shown). Since the recovery procedure was equally effective for all purified intermediates, one possible explanation for the low recovery of chlorinated hydroquinones in the culture at 24 h is that the compounds are bound to an unknown (e.g., the vessel walls or a cellular component) that impedes their effective extraction from the cultures or subsequent derivatization. Within the next 24 h most of the TCHQ added to dechlorinating cultures was recovered as 2,5-DCHQ (Fig. 2) or as TCHQ in nondechlorinating cultures (data not shown). These results suggest that the dechlorinating and nondechlorinating cultures are able to increase the accessibility of the chlorinated hydroquinones as they grow, thus rendering the compound available for metabolism and extraction. Data from 48 h and later show that 2-CHQ and eventually HQ were produced, but similar to the results with TCMP, recovery of these dechlorination products was low. Recovery of 2-CHQ and HQ when added to cultures was not problematic. Therefore, the decreased detection of these metabolites may be due to degradation to products not detectable by the methods used. Within 10 days, all four strains of CHM-dechlorinating desulfitobacteria produced HQ via 2,5-DCHQ, but not 2,6-DCHQ as observed with the degradation of TCMP.
FIG. 2.
Time course of TCHQ dechlorination by D. hafniense strain DCB2. ♦, TCHQ; ▵, TriCHQ; □, 2,5-DCHQ; ○, 2-CHQ; ×, HQ.
DISCUSSION
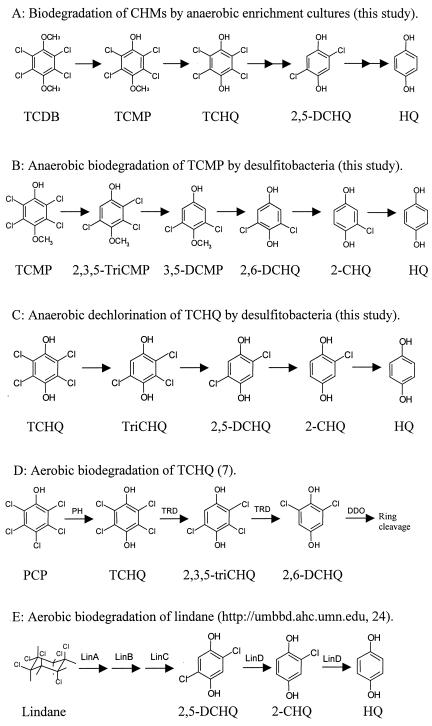
The evidence presented here shows that pure cultures of Desulfitobacterium spp. are capable of the complete demethylation and dechlorination of fungal CHMs. Degradation also occurred with mixed microbial communities of anaerobic enrichment cultures examined in this study. However, the dechlorination and demethylation substrates and products differed from those of the axenic cultures, and it is likely that more than one species of bacterium contribute to the total degradation pathway in the enrichment cultures. The eventual elimination of all of the hydroquinone metabolites by anaerobic bacteria suggests that these natural compounds are more readily degraded than some anthropogenic halogenated organohalides (e.g., PCP and polychlorinated biphenyls) and may be biomineralized or converted into biomass. Pathways of anaerobic biodegradation of the extensively chlorinated hydroquinones to HQ by enrichment cultures and Desulfitobacterium spp. are presented in Fig. 3.
FIG. 3.
Degradation pathways of CHMs by anaerobic enrichment cultures and axenic cultures of desulfitobacteria in relation to aerobic degradation of TCHQ and lindane. PCP is aerobically degraded by a series of enzymes including pentachlorophenol hydroxylase (PH), TRD, and dichlorohydroquinone dioxygenase. Lindane is degraded aerobically by a series of proteins (LinA, -B, -C, and -D), including the 2,5-DCHQ reductive dehalogenase (LinD). HQ, 1,4-dihydroquinone; 2,6-DCHQ, 2,6-dichlorohydroquinone; 2-CHQ, 2-chlorohydroquinone.
Demethylation.
Demethylation of CHMs was a common reaction for the enrichment cultures initiated with freshwater or estuarine sediment and always preceded dechlorination (Fig. 3A). Anaerobic O-demethylation of methoxylated aromatic compounds (2, 6, 8), including chlorinated aromatics (16), is well established, and the fungal CAM 3,5-dichloro-p-anisyl alcohol is demethylated in methanogenic sludge (46). Demethylation of methoxy groups is observed with homoacetogens that use the reaction for energy and a carbon source. Therefore, the demethylation of TCDB or TCMP by enrichment cultures is consistent with the anaerobic metabolism of fungal CAMs and other chlorinated aromatic compounds. This suggests that TCHQ, and not TCDB or TCMP, would serve as a substrate for reductive dechlorination by anaerobic microorganisms from freshwater and estuarine sediments.
In contrast, axenic cultures of D. chlororespirans strain Co23, D. dehalogenans strain JW/DUI, and D. hafniense strains PCP1 and DCB2 do not demethylate TCMP. Instead, they reductively demethylate the less chlorinated methoxyphenol 3,5-DCMP, which accumulates in the pathway of TCMP biodegradation carried out by several of the desulfitobacteria tested in this study (Fig. 1 and 3B). This suggests that the catalytic specificity of demethylation exhibited by the desulfitobacteria is different from that transforming TCDB or TCMP in the mixed microbial communities of the enrichment cultures. Furthermore, the accumulation of the 3,5-DCMP followed by its relatively rapid transformation suggests that a nonconstitutive demethylase is induced by the dichloromethoxyphenol and not by the tetra- or trichlorinated methoxyphenols.
Dechlorination.
The specificity that dehalogenating bacteria have for organohalide substrates varies substantially even among phylogenetically similar organisms. A stark example of this is the inability of D. hafniense strain DP7 to dechlorinate chlorinated aromatic or aliphatic compounds (43), including the fungal CHMs (this study), even though the organism's 16S rRNA gene sequence is highly similar to that of other dechlorinating D. hafniense strains. Even strains of D. hafniense that are very effective dechlorinators often exhibit a different specificity with chlorinated substrates (Table 3). However, this study showed that if a dehalogenating organism dechlorinated the fungal metabolite TCMP, it would dechlorinate TCHQ. Since D. chlororespirans strain Co23, D. dehalogenans strain JW/DUI, and D. hafniense strains PCP1 and DCB2 exhibit identical degradation pathways for the extensively chlorinated and naturally produced CHMs (Fig. 3B and C), it is likely that the strains use similar catalysts (enzymes and cofactors) for the completion of each pathway. These observations are the basis for the postulate that these pathways are fundamental for the reductive dehalogenation of chlorinated phenolic compounds by anaerobic dehalogenating bacteria.
In general, if a microorganism had been reported in the literature to be capable of dechlorinating a chlorophenol compound, it would dechlorinate TCMP and TCHQ (Table 3). In contrast, if a bacterium had been reported to dechlorinate chlorinated ethenes, it would not dechlorinate TCMP or TCHQ (e.g., Dehalospirillum multivorans). The reverse of the latter statement also held, i.e., species that could dechlorinate TCMP were not reported to dechlorinate ethenes. However, these generalizations were not always supported. For example, 3-chloro-4-hydroxyphenyl acetate, a substrate of the o-chlorophenol dehalogenase (CprA) of D. dehalogenans strain JW/DU1 (44), is not reported to be dechlorinated by the CHM-dechlorinating D. hafniense strain PCP1 (10). In addition, Desulfitobacterium strain PCE1 dechlorinates 3-chloro-4-hydroxyphenyl acetate and tetrachloroethene (12) but does not dechlorinate the CHMs. Therefore, even though the dechlorination of CHMs was common among the desulfitobacteria, not all such species are capable of CHM dehalogenation even when they can dechlorinate aromatic compounds. This prevents the full prediction of which dehalogenases are responsible for CHM dechlorination. Such breadth and variability of specificity among dehalogenating bacteria reaffirms the existence of multiple dehalogenases and suggests that not-yet-defined systems may be required for the anaerobic biodegradation of TCMP and TCHQ.
During the dechlorination of TCHQ, the CHM-dechlorinating desulfitobacteria and the microorganisms in the enrichment cultures selectively remove a chlorine from TriCHQ para to that selected for the dechlorination of TCHQ, thereby producing 2,5-DCHQ (Fig. 3C). This para selection of a chlorine was not observed with the methoxylated TCMP; hence, the dichloro- intermediates of TCHQ and TCMP anaerobic degradation represent a distinct dechlorination pattern. The regiochemistry of the dechlorination of TCHQ by the desulfitobacteria also differs from that carried out by some aerobic bacteria during the degradation of PCP (reviewed in reference 7). In contrast to the anaerobic TCHQ pathway, which proceeds by four reductive dechlorination steps in succession, TCHQ degradation in aerobic bacteria begins with two reductive dechlorination reactions and ends with oxidative ring cleavage of the dichloro- intermediate (Fig. 3D). Therefore, the anaerobic end product HQ is not a component of the aerobic pathway. The other distinct difference between the aerobic and anaerobic dechlorination of TCHQ is that 2,6-DCHQ is the reductive dechlorination product and midpoint intermediate in the aerobic pathway. Therefore, the specificity of the TCHQ reductive dehalogenase (TRD) of aerobic bacteria does not match that of the anaerobic desulfitobacteria nor that of the microorganisms in the anaerobic enrichment cultures described here. Stepwise, the dechlorination of 2,5-DCHQ to HQ during the anaerobic dechlorination of TCHQ is identical to two other reductive dechlorination reactions also found in aerobic bacteria. These reactions are carried out by a 2,5-DCHQ reductive dehalogenase (LinD) as part of the aerobic degradation of gamma-hexachlorocyclohexane, the pesticide known as lindane (http://umbbd.ahc.umn.edu) (24). The pathway of aerobic lindane degradation is presented in Fig. 3E. Although the TRD and LinD of aerobes perform similar reactions to those found in the anaerobic bacteria, it is unlikely that the reductive dehalogenation genes and enzymes in the aerobic bacteria are similar to those in the anaerobic bacteria. The different products formed from TriCHQ within the aerobic and anaerobic pathways support this conclusion. More importantly, a survey of the genome of D. hafniense strain DCB2 (http://www.jgi.doe.gov/JGI_microbial/html/) reveals that this organism does not possess genes with high sequence similarity to those coding for TRD or LinD. The annotated genomic data for strain DCB2 does show that the organism possesses the cprA gene for the o-chlorophenol dehalogenase along with four other uncharacterized cprA-like genes. Since all anaerobic dechlorination of the CHMs require a substrate with a chlorine atom adjacent to a hydroxyl group, it is possible that CprA or a CprA-like protein could perform each reaction. However, further research is required in order to determine which of these enzymes is responsible for each of the dechlorination reactions presented in Fig. 3B and C and how the expression and activity of these dehalogenases are coordinated with demethylation.
Induction of pathways.
All of the desulfitobacteria that can completely dechlorinate and demethylate the CHMs are known to dechlorinate at least some if not several chlorophenols (10, 30, 41). The dechlorination of the chlorophenols is considered inducible, and in some cases specific chlorophenols are required to induce the dechlorination of other chlorophenols (10, 40). During TCHQ dechlorination by the axenic and enrichment cultures tested here, 2,5-DCHQ accumulated and persisted for several days, just as 3,5-DCMP did during the dechlorination of TCMP (Fig. 1 and 2). The accumulation of the dichloro- isomers of methoxyphenol and hydroquinone in these two pathways (Fig. 3B and C) suggests that more than one dehalogenating system may be activated for the pathways to proceed to completion. Although the specificity for anthropogenic or natural substrates of the cprA-like gene products of DCB2 along with their regulatory control has not been established, these gene products along with cprA could act in concert to complete the pathways shown in Fig. 3B and C.
Some reductive dehalogenases are inducible, and putative reductive dehalogenases are suspected of requiring induction (33, 47). Dennie et al. postulated that there are at least two inducible dehalogenase systems in D. hafniense strain PCP1 (formerly Desulfitobacterium frappieri strain PCP1) (10). Due to the phylogenetic similarities of the CHM-degrading Desulfitobacterium spp. and their identical pathways for the degradation of CHMs, it is conceivable that these bacteria share inducible dehalogenases. Dennie et al. did not examine the degradation of fungal CHMs such as TCMP or TCHQ, but they did test strain PCP1 with several extensively chlorinated compounds similar to the CHMs. The compounds they tested are not symmetrical in relation to the positioning of the hydroxy or methoxy substitutions, as are TCHQ and TCMP, but instead possess four adjacent chlorines. When strain PCP1 was supplied with inducing molecules (3,5-dichlorophenol and 2,3,5-trichlorophenol), complete dechlorination of tetrachlorocatechol to catechol was observed. A striking difference between the degradation of the anthropogenic compounds studied with PCP1 by Dennie et al. and that of the CHMs here is that none of the organisms that could degrade TCMP required any inducing chlorophenol for the complete degradation of the compound to HQ. If any induction was required, it was accomplished with the natural compounds alone. This result is consistent with the postulate that the anaerobic degradation of TCMP and TCHQ are fundamental degradation pathways for these desulfitobacteria. Since the CHM-degrading bacteria do not require the addition of chlorophenols to initiate the anaerobic degradation of CHMs, then it is conceivable that these fungal compounds could serve as natural and fully biodegradable inducers of pathways for the degradation of anthropogenic compounds of environmental concern.
Summary.
The present description of anaerobic dechlorination of fungal CHMs by desulfitobacteria is the first of its kind. Until now, examinations of how anaerobic bacteria contribute to a natural halogen cycle have been sparse and primarily limited to the degradation of a small number of less chlorinated anisyl metabolites produced by fungi (44, 52). The data obtained here with enrichment and axenic cultures demonstrate that complete dechlorination in combination with demethylation of the fungal CHMs to readily degradable HQ (degradation of HQ by anaerobes has been reported previously [26, 31, 38]) can be carried out by several species of desulfitobacteria. However, the production of TCHQ from TCMP followed by the formation of less chlorinated HQs by the mixed microbial communities suggests that TCHQ may more often be the natural substrate for dechlorinating organisms in the environment. The CHM-degrading bacteria described here (D. chlororespirans strain Co23, D. dehalogenans strain JW/DUI, and D. hafniense strains PCP1 and DCB2) exhibit identical degradation pathways for the extensively chlorinated and naturally produced CHMs. These microorganisms are not reported to be capable of dechlorinating an aromatic compound unless it is additionally substituted with a nonhalogen (5, 10, 30, 40, 41). Our testing of these strains with polychlorinated biphenyls and benzenes confirmed that the organisms do not dechlorinate such compounds. The data indicate that the complete and rapid degradation of TCMP and TCHQ by the Desulfitobacterium spp. constitute distinct metabolic pathways. These results support the hypothesis that anthropogenic organohalides are degraded as part of the global halogen cycle that has evolved in response to naturally produced organohalides.
Acknowledgments
This study was supported by National Science Foundation grant 0078133 to H.D.M., G.P.M., and K.R.S.
REFERENCES
- 1.Anchel, M. 1952. Identification of drosophilin A as p-methoxytetrachlorophenol. J. Am. Chem. Soc. 74:2493. [Google Scholar]
- 2.Bache, R., and N. Pfennig. 1981. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch. Microbiol. 130:255-261. [Google Scholar]
- 3.Berkaw, M., K. R. Sowers, and H. D. May. 1996. Anaerobic ortho dechlorination of polychlorinated biphenyls by estuarine sediments from Baltimore Harbor. Appl. Environ. Microbiol. 62:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, B., R. Beaudet, R. Villemur, G. McSween, F. Lepine, and J.-G. Bisaillon. 1996. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int. J. Syst. Bacteriol. 46:1010-1015. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen, N., and B. K. Ahring. 1996. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46:442-448. [Google Scholar]
- 6.Cocaign, M., E. Wilberg, and N. Lindley. 1991. Sequential demethoxylation reactions during methylotrophic growth of methoxylated aromatic substrates with Eubacterium limosum. Arch. Microbiol. 155:496-499. [Google Scholar]
- 7.Copley, S. D. 2000. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 6:261-265. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, S. L., Z. Wu, and H. L. Drake. 1988. Growth of thermophilic acetogenic bacteria on methoxylated aromatic acids. FEMS Microbiol. Lett. 52:25-28. [Google Scholar]
- 9.de Jong, E., and J. A. Field. 1997. Sulfur tuft and turkey tail: biosynthesis and biodegradation of organohalogens by basidiomycetes. Annu. Rev. Microbiol. 51:375-414. [DOI] [PubMed] [Google Scholar]
- 10.Dennie, D., I. Gladu, F. Lépine, R. Villemur, J.-G. Bisaillon, and R. Beaudet. 1998. Spectrum of the reductive dehalogenation activity of Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 64:4603-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerritse, J., O. Drzyzga, G. Kloetstra, M. Keijmel, L. P. Wiersum, R. Hutson, M. D. Collins, and J. C. Gottschal. 1999. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 65:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerritse, J., V. Renard, T. M. Pedro Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 13.Gribble, G. W. 1998. Naturally occurring organohalogen compounds. Accounts Chem. Res. 31:141-152. [Google Scholar]
- 14.Gribble, G. W. 2000. The natural production of organobromine compounds. Environ. Sci. Pollut. Res. 7:37-49. [DOI] [PubMed] [Google Scholar]
- 15.Häggblom, M. M. 1992. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol. Rev. 103:29-72. [DOI] [PubMed] [Google Scholar]
- 16.Häggblom, M. M., M. H. Berman, A. C. Frazer, and L. Y. Young. 1993. Anaerobic O-demethylation of chlorinated guaiacols by Acetobacterium woodii and Eubacterium limosum. Biodegradation 4:107-114. [Google Scholar]
- 17.Häggblom, M. M., and I. D. Bossert (ed.). 2003. Dehalogenation: microbial processes and environmental implications. Kluwer Academic Publishers, Boston, Mass.
- 18.Häggblom, M. M., D. Janke, and M. S. Salkinoja-Salonen. 1989. Transformation of chlorinated phenolic compounds in the genus Rhodococcus. Microb. Ecol. 18:147-159. [DOI] [PubMed] [Google Scholar]
- 19.Harvey, P. J., H. E. Schoemaker, and J. M. Palmer. 1986. Veratryl alcohol as a mediator and the role of radical cations in lignin biodegradation by Phanerochaete chrysosporium. FEBS Lett. 195:242-246. [Google Scholar]
- 20.Hileman, B. 1993. Concerns broaden over chlorine and chlorinated hydrocarbons. Chem. Eng. News 1993(April 19):11-20. [Google Scholar]
- 21.King, G. M. 1988. Dehalogenation in marine sediments containing natural sources of halophenols. Appl. Environ. Microbiol. 54:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knuutinen, J., P. Autio, P. Klein, S. Livela, L. Virkki, and M. Lahtipera. 1988. Synthesis and structure verification of chlorinated 4-methoxyphenols, models of metabolites of chlorophenolic compounds. Chemosphere 17:1821-1829. [Google Scholar]
- 23.Madsen, T., and D. Licht. 1992. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl. Environ. Microbiol. 58:2874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyauchi, K., S.-K. Suh, Y. Nagata, and M. Takagi. 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Öberg, G. 2002. The natural chlorine cycle—fitting the scattered pieces. Appl. Microbiol. Biotechnol. 58:565-581. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor, O. A., and L. Y. Young. 1996. Effects of six different functional groups and their position on the bacterial metabolism of monosubstituted phenols under anaerobic conditions. Environ. Sci. Technol. 30:1419-1428. [Google Scholar]
- 27.Pfefferle, W., H. Anke, M. Bross, and W. Steglich. 1990. Inhibition of solubilized chitin synthase by chlorinated aromatic compounds isolated from mushroom cultures. Agric. Biol. Chem. 54:1381-1384. [Google Scholar]
- 28.Ramirez, F., E. H. Chen, and S. Dershowitz. 1959. Reaction of trialkyl phophites with p-benzoquinone and other symmetrically substituted p-quinones. A new synthesis of hydroquinone monoalkyl ethers. J. Am. Chem. Soc. 81:4338-4342. [Google Scholar]
- 29.Renner, G., and C. Hopfer. 1990. Preparations and properties of transformation products of pentachlorophenol (PCP). Toxicol. Environ. Chem. 25:105-108. [Google Scholar]
- 30.Sanford, R. A., J. R. Cole, F. E. Löffler, and J. M. Tiedje. 1996. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling the oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl. Environ. Microbiol. 62:3800-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schink, B., B. Philipp, and J. Müller. 2000. Anaerobic degradation of phenolic compounds. Naturwissenschaften 87:12-23. [DOI] [PubMed] [Google Scholar]
- 32.Scholz-Muramatsu, H., A. Neumann, M. Meβmer, E. Moore, and G. Diekert. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48-56. [Google Scholar]
- 33.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sowers, K. R., J. E. Boone, and R. P. Gunsalus. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steward, C. C., T. C. Dixon, Y. P. Chen, and C. R. Lovell. 1995. Enrichment and isolation of a reductively debrominating bacterium from the burrow of a bromometabolite-producing marine hemichordate. Can. J. Microbiol. 41:637-642. [Google Scholar]
- 36.Suzuki, T. 1983. Methylation and hydroxylation of pentachlorophenol by Mycobacterium sp. isolated from soil. J. Pesticide Sci. 8:419-428. [Google Scholar]
- 37.Swarts, H. J., F. J. M. Verhagen, J. A. Field, and J. B. P. A. Wijnberg. Trichlorinated phenols from Hypholoma elongatum. Phytochemistry 49:203-206.
- 38.Szewzyk, U., R. Szewzyk, and B. Schink. 1985. Methanogenic degradation of hydroquinone and catechol via reductive dehydroxylation to phenol. FEMS Microbiol. Ecol. 31:79-87. [Google Scholar]
- 39.Teunissen, P. J. M., H. J. Swarts, and J. A. Field. 1997. The de novo production of drosophilin A (tetrachloro-4-methoxyphenol) and drosophilin A methyl ether (tetrachloro-1,4-dimethoxybenzene) by ligninolytic basidiomycetes. Appl. Microbiol. Biotechnol. 47:695-700. [DOI] [PubMed] [Google Scholar]
- 40.Utkin, I., D. D. Dalton, and J. Wiegel. 1995. Specificity of reductive dehalogenation of substituted ortho-chlorophenols by Desulfitobacterium dehalogenans JW/IU-DCI. Appl. Environ. Microbiol. 61:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utkin, I., C. Woese, and J. Wiegel. 1994. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int. J. Syst. Bacteriol. 44:612-619. [DOI] [PubMed] [Google Scholar]
- 42.Valli, K., and M. H. Gold. 1991. Degradation of 2,4-dichlorophenol by the lignin-degrading fungus Phanerochaete chrysosporium. J. Bacteriol. 173:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Pas, B. A., H. J. M. Harmsen, G. C. Raangs, W. M. de Vos, G. Schrass, and A. J. M. Stams. 2001. A Desulfitobacterium strain isolated from human feces that does not dechlorinate chloroethenes or chlorophenols. Arch. Microbiol. 175:389-394. [DOI] [PubMed] [Google Scholar]
- 44.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 45.Verhagen, G. J. M., H. J. Swarts, T. W. Kuyper, J. B. P. A. Wijnberg, J. A. Field. 1996. The ubiquity of natural adsorbable organic halogen production among basidiomycetes. Appl. Microbiol. Biotechnol. 45:710-718. [Google Scholar]
- 46.Verhagen, F. J. M., H. J. Swarts, J. B. P. A. Wijnberg, and J. A. Field. 1998. Biotransformation of the major fungal metabolite 3,5-dichloro-p-anisyl alcohol under anaerobic conditions and its role in formation of bis(3,5-dichloro-4-hydroxyphenyl)methane. Appl. Environ. Microbiol. 64:3225-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]
- 48.Wacket, L. P., and C. D. Hershberger. 2001. Biocatalysis and biodegradation: microbial transformation of organic compounds. ASM Press, Washington, D.C.
- 49.Watson, J. T. 1985. Introduction to mass spectrometry, p. 153-172. Raven Press, New York, N.Y.
- 50.Wolin, E. A., M. J. Wolin, and R. S. Wolfe. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882-2886. [PubMed] [Google Scholar]
- 51.Yu, D., and D. L. Mattern. 1999. Convenient preparations of the three 2,3-dihalo-1,4-benzoquinones. Synthetic Commun. 29:821-825. [Google Scholar]
- 52.Zhang, X., and J. Wiegel. 1992. The anaerobic degradation of 3-chloro-4-hydroxybenzoate in freshwater sediment proceeds via either chlorophenol of hydroxybenzoate to phenol and subsequently to benzoate. Appl. Environ. Microbiol. 58:3580-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]