Abstract
To assess genetic diversity in Cryptosporidium oocysts from Canada geese, 161 fecal samples from Canada geese in the United States were analyzed. Eleven (6.8%) were positive for Cryptosporidium spp. following nested PCR amplification of the hypervariable region of the 18S rRNA gene. Nine PCR products from geese were cloned and sequenced, and all nine diverged from previously reported Cryptosporidium 18S rRNA gene sequences. Five sequences were very similar or identical to each other but genetically distinct from that of Cryptosporidium baileyi; two were most closely related to, but genetically distinct from, the first five; and two were distinct from any other sequence analyzed. One additional sequence in the hypervariable region of the 18S rRNA gene isolated from a cormorant was identical to that of C. baileyi. Phylogenetic analysis provided evidence for new genotypes of Cryptosporidium species in Canada geese. Results of this study suggest that the taxonomy of Cryptosporidium species in geese is complex and that a more complete understanding of genetic diversity among these parasites will facilitate our understanding of oocyst sources and species in the environment.
Cryptosporidium parvum is a protozoan parasite that causes self-limiting gastrointestinal disease in otherwise healthy adults but severe, prolonged, and potentially life-threatening illness in immunocompromised individuals. This parasite has a broad range of animal hosts and can be transmitted either anthroponotically, zoonotically, or by ingestion of contaminated food or water. Molecular characterization of C. parvum has revealed extensive genetic polymorphism, and the species has been classified into two distinct genotypes: human-adapted genotype 1 and animal-adapted genotype 2 (2, 4, 18, 21, 24). Sufficient variation among members of the animal-adapted C. parvum group has led to further classifications of bovine, dog, pig, mice, deer, ferret, marsupial, and monkey C. parvum genotypes (15, 19, 27).
Traditional taxonomic classification of Cryptosporidium oocysts is based on oocyst morphology, host specificity, and the anatomical site of infection (7). More recently, a polyphasic approach to taxonomy has included molecular-genetic characterization as well as the traditional criteria (8). The small number of Cryptosporidium oocysts recovered from environmental samples often precludes traditional taxonomic analysis, resulting in species identification that is based solely on molecular characterization of one or more genes (13, 19, 28, 30). Genes encoding actin (22), the 70-kDa heat shock protein (HSP70) (23), the Cryptosporidium oocyst wall protein (COWP) (29), and the 18S small-subunit rRNA (26) have all been used for molecular-genetic characterization of Cryptosporidium spp.
Although the molecular-genetic polymorphism of C. parvum has been extensively characterized, diversity in other Cryptosporidium species is not as well studied. Currently, there are over 140 18S rRNA gene sequence entries for C. parvum in the GenBank database (3); by contrast, there are only 34, 12, 6, 5, 3, and 2 entries for C. meleagridis, C. muris, C. serpentis, C. baileyi, C. wrairi, and C. andersoni, respectively. Given the paucity of molecular-genetic information for Cryptosporidium species other than C. parvum, identification of oocyst species from environmental samples by using DNA sequence data alone can be difficult.
We hypothesize that the level of genetic polymorphism seen in C. parvum may also exist in other Cryptosporidium species. To test this hypothesis, the genetic variability of the 18S rRNA gene of Cryptosporidium oocysts in the feces of Canada geese was determined. Geese were chosen as the target animal host because they are ubiquitous and impact surface water quality. In addition, geese are known to be a host for non-C. parvum species: birds are susceptible to infection with only two Cryptosporidium species, C. meleagridis and C. baileyi, which are sufficiently different at the genetic level that they can be unambiguously distinguished by DNA sequence analysis of 18S rRNA gene fragments. Confining this study to geese eliminated the uncertainty of oocyst source and allowed comparison of DNA sequences among particular species of Cryptosporidium from the environment.
Here we report the prevalence of Cryptosporidium oocysts in geese from different parts of the United States and describe 18S rRNA gene polymorphisms in oocysts recovered from goose feces. These findings will improve our understanding of the role of geese in the transmission of waterborne cryptosporidiosis and of the genetic variability of Cryptosporidium spp. in this host. A more complete understanding of the phylogeny of Cryptosporidium spp. will facilitate the identification of likely sources of oocysts detected in the environment by molecular-genetic methods.
MATERIALS AND METHODS
Fecal collection.
Fresh fecal pellets from geese were collected from August 2001 to October 2002 at various geographic locations in the United States (Table 1). All fecal pellets were handled with disposable gloves to prevent cross-contamination of samples. Individual fecal pellets were stored on ice in sterile 50-ml polypropylene conical tubes and shipped to the D. B. Shauer laboratory at the Massachusetts Institute of Technology (MIT) within 24 h of collection. One additional avian fecal sample, collected in August 2000 from a cormorant in Massachusetts, was included in the study as well.
TABLE 1.
Summary of fecal sample collections.
| Date | Location | No. of samples | No. positivea | No. cloned | Sequence identifier(s) |
|---|---|---|---|---|---|
| August 2001 | New York | 12 | 4 | 1 | Goose no. 7 |
| August 2001 | Illinois | 25 | 4 | 4 | Geese no. 1, 2, 3 (a and b), 5 |
| March 2002 | Massachusetts | 10 | 0 | ||
| June 2002 | Colorado | 15 | 0 | ||
| June 2002 | Virginia | 7 | 2 | 2 | Geese no. 8, 9 |
| June 2002 | Washington | 5 | 0 | ||
| July 2002 | Massachusetts | 25 | 0 | ||
| July 2002 | Colorado | 1 | 0 | ||
| August 2002 | Pennsylvania | 15 | 0 | ||
| August 2002 | Oklahoma | 13 | 0 | ||
| September 2002 | Massachusetts | 19 | 1 | 1 | Goose no. 6 |
| October 2002 | Colorado | 14 | 0 | ||
| Total | 161 | 11 | 8 |
Positive for Cryptosporidium spp. oocysts by nested PCR targeting the hypervariable region of the 18S rRNA gene.
Oocyst isolation.
Upon arrival of the specimens at the Shauer laboratory, a 1- to 3-g sample of each fecal pellet was suspended in 20 ml of laboratory-grade water (Milli-Q System; Millipore Corp., Bedford, Mass.). A second 1- to 3-g sample of one fecal pellet for each batch of samples was included as a positive control for the detection assay. This positive-control pellet was resuspended in 19.5 ml of laboratory-grade water and spiked with 500 μl of a 104-oocyst-ml−1 suspension. Each fecal suspension was vortexed for 30 s to homogenize the fecal slurry and was then allowed to settle for 3 min to remove large fecal particles (mostly grass). Next, 10 ml of supernatant was transferred to a glass Leighton tube (Bellco Glass, Inc., Vineland, N.J.); oocysts were purified from each fecal sample by immunomagnetic separation (IMS) using a Crypto-Scan IMS kit (ImmuCell, Portland, Maine) according to the manufacturer's recommendations. After dissociation from the magnetic beads, oocysts were transferred to a microcentrifuge tube and treated with 5 μl of 1 N NaOH to neutralize the pH. The oocysts were pelleted for 2 to 3 min at 16,000 × g, resuspended in 50 μl of laboratory-grade water, and stored at 4°C.
Positive and negative IMS controls were processed with each set of fecal samples. Positive IMS controls consisted of 9.5 ml of laboratory-grade water spiked with 500 μl of a 104-oocyst-ml−1 suspension; negative IMS controls consisted of 10 ml of laboratory-grade water. IMS controls were processed as described above.
Genomic DNA extraction.
Oocysts were lysed by adding 25 μl of IMS product to 475 μl of Tris-EDTA (TE) buffer containing 0.2 g of proteinase K liter−1 and 0.4% sodium dodecyl sulfate and incubating the mixture overnight at 45°C. Positive and negative DNA extraction controls were included for each set of fecal samples. Positive DNA extraction controls consisted of 25 μl of a 104-oocyst-ml−1 suspension in 475 μl of TE buffer; negative DNA extraction controls consisted of 25 μl of laboratory-grade water in 475 μl of TE buffer. DNA was extracted with phenol-chloroform, precipitated with 0.2 M NaCl and 2 volumes of absolute ethanol, and resuspended in 30 μl of TE buffer.
Nested PCR assay.
Nested-PCR amplification of the hypervariable region of the 18S rRNA gene was performed as previously described (13) with minor modifications. The concentration of each deoxynucleoside triphosphate (Perkin-Elmer, Wellesley, Mass.) was 0.15 mM. The initial amplification reaction was performed with 15 μl of DNA template, and 1 μl of the initial amplification product was used as the template in the secondary PCR. Positive and negative PCR controls were included with each set of fecal samples. For the initial amplification reaction, positive PCR controls contained 14 μl of laboratory-grade water and 1 μl of genomic C. parvum DNA (at a concentration equivalent to 104 oocysts μl−1) while negative PCR controls contained 15 μl of laboratory-grade water. For the secondary amplification reaction, positive PCR controls contained 1 μl of genomic C. parvum DNA (at a concentration equivalent to 104 oocysts μl−1) while negative PCR controls contained 1 μl of laboratory-grade water.
Both amplification reactions used forward and reverse oligonucleotide primers that are complementary to all Cryptosporidium 18S rRNA gene sequences. For the primary PCR, an approximately 1,056-bp product (dependent on Cryptosporidium species) was obtained using forward and reverse primers KLJ1 and KLJ2, respectively (13); for the secondary PCR, an approximately 434-bp product was obtained using forward and reverse primers CPB-DIAGF and CPB-DIAGR, respectively (14). Cycling conditions for both the primary and secondary PCRs consisted of an initial denaturation (5 min at 80°C, followed by 30 s at 98°C), 25 cycles of amplification (denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 1 min at 72°C), and a final extension (10 min at 72°C). Secondary-PCR products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide.
Cloning.
Secondary PCR products were cloned into the pGEM-T Easy Vector System (Promega Corporation, Madison, Wis.) and used to transform XL-1 Blue Escherichia coli cells (Stratagene, La Jolla, Calif.). Clones were selected on Luria-Bertani (LB) agar supplemented with 100 μg of ampicillin ml−1 and cultured overnight in LB broth supplemented with 100 μg of ampicillin ml−1. Plasmid DNA was isolated from clones by using a QIAPrep Spin Miniprep Kit (Qiagen, Inc., Valencia, Calif.) and digested with NotI (New England Biolabs, Beverly, Mass.) to verify the presence of the secondary PCR amplicon insert and with NdeI (New England Biolabs) to identify any heterogeneity among the clones (13). Restriction digestion was carried out in a 20-μl volume containing 4 μl of plasmid DNA, 20 U of NotI, 10 U of NdeI, 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, and 100 μg of bovine serum albumin ml−1, followed by incubation at 37°C for 1 h. Digestion products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide.
Sequencing of secondary PCR products.
Representative clones of the secondary PCR products were sequenced on an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems, Foster City, Calif.), using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase, FS (PE Applied Biosystems). If multiple NdeI digestion patterns existed among clones from a given sample, at least one clone for each digestion pattern was sequenced. With the exception of goose no. 7, at least three clones for each positive sample were sequenced, and their identities were confirmed by sequencing both strands. For goose no. 7, one clone was successfully sequenced, and its identity was confirmed by sequencing both strands. The consensus sequences for the clones recovered from each bird were used in the phylogenetic analysis.
Phylogenetic analysis.
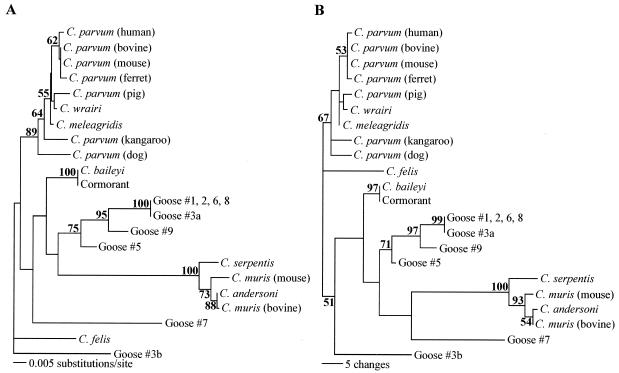
Sequences were aligned manually, based on the secondary structure of the 18S rRNA, using the GCG sequence editor (Genetics Computer Group, Madison, Wis.). Variable-length regions were masked and excluded from the phylogenetic analysis. Phylogenetic Analysis Using Parsimony (PAUP), beta version 4.0 (25), was used to create both neighbor-joining and parsimony trees from the GCG alignments. Construction of neighbor-joining trees was based on the evolutionary distances between different isolates, calculated by the Kimura two-parameter analysis, and the designation of C. felis as an outgroup. Statistical support for the resulting trees was tested using 1,000 pseudoreplicates of the bootstrap test; only values above 50% were reported, and bootstrap values greater than 70% were considered significant (12). GenBank accession numbers used in the phylogenetic analyses are noted in the figure legends (see Fig. 2 and 3).
FIG.2.
Neighbor-joining (A) and parsimony (B) trees based on the hypervariable region of the 18S rRNA gene (created with PAUP 4.0 software). C. felis was designated an outgroup. Evolutionary distances were determined by the Kimura two-parameter method. GenBank accession numbers of sequences included in the trees are AB089285 (C. andersoni), L19068 (C. baileyi), AF112575 (C. felis), AF112574 (C. meleagridis), L19069 (C. muris bovine genotype), AB089284 (C. muris murine genotype), AF093489 (C. parvum human genotype), AF093493 (C. parvum bovine genotype), AF112571 (C. parvum mouse genotype), AF112572 (C. parvum ferret genotype), AF115377 (C. parvum pig genotype), AF112576 (C. parvum dog genotype), AF112570 (C. parvum kangaroo genotype), AF093499 (C. serpentis), U11440 (C. wrairi), AY324634 (cormorant), AY324635 (goose no. 1), AY324636 (goose no. 2), AY324637 (goose no. 3 [sequence a]), AY324638 (goose no. 3 [sequence b]), AY324639 (goose no. 5), AY324640 (goose no. 6), AY324641 (goose no. 7), AY324642 (goose no. 8), and AY324643 (goose no. 9). Bootstrap values greater than 50% are indicated in bold at each respective node.
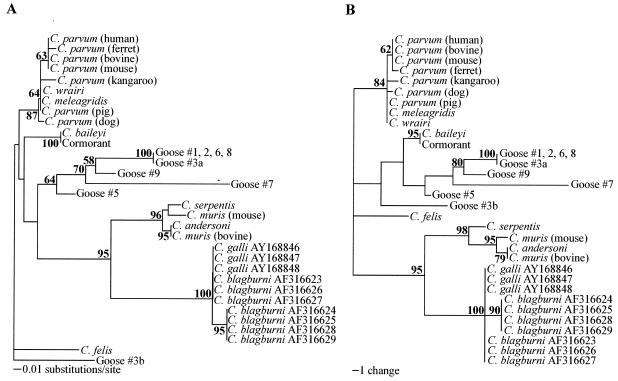
FIG. 3.
Phylogenetic analysis of the partial hypervariable region of the 18S rRNA gene to assess the relationships between the goose-derived sequences in the present study and C. galli and C. blagburni in finches. Shown are neighbor-joining (A) and most parsimonious (B) trees created with PAUP 4.0. C. felis was designated an outgroup. Evolutionary distances were determined by the Kimura two-parameter method. GenBank accession numbers of sequences included in the trees are AB089285 (C. andersoni), L19068 (C. baileyi), AF112575 (C. felis), AF112574 (C. meleagridis), L19069 (C. muris bovine genotype), AB089284 (C. muris murine genotype), AF093489 (C. parvum human genotype), AF093493 (C. parvum bovine genotype), AF112571 (C. parvum mouse genotype), AF112572 (C. parvum ferret genotype), AF115377 (C. parvum pig genotype), AF112576 (C. parvum dog genotype), AF112570 (C. parvum kangaroo genotype), AF093499 (C. serpentis), U11440 (C. wrairi), AY168846 through AY168848 (C. galli), AF316623 through AF316629 (C. blagburni), AY324634 (cormorant), AY324635 (goose no. 1), AY324636 (goose no. 2), AY324637 (goose no. 3 [sequence a]), AY324638 (goose no. 3 [sequence b]), AY324639 (goose no. 5), AY324640 (goose no. 6), AY324641 (goose no. 7), AY324642 (goose no. 8), and AY324643 (goose no. 9). Bootstrap values greater than 50% are indicated in bold at each respective node.
Oocyst detection limit.
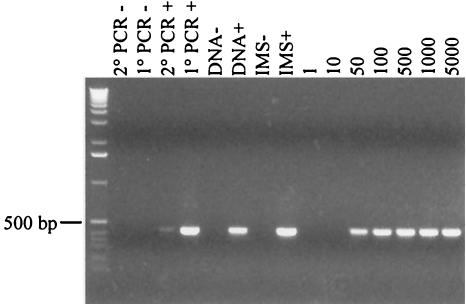
Pooled goose fecal samples negative for Cryptosporidium oocysts were divided into seven 2-g aliquots and seeded with 1, 10, 50, 100, 500, 1,000, or 5,000 C. parvum oocysts. Laboratory-grade water was added to bring each spiked fecal sample to a final volume of 20 ml. Fecal suspensions were vortexed for 30 s to homogenize the fecal slurry and then allowed to settle for 3 min to remove large fecal particles. After particle settling, 10 ml of each sample supernatant was transferred to a glass Leighton tube and processed as described above (IMS, DNA extraction, and nested PCR). Secondary PCR products were visualized after electrophoresis on a 1.2% agarose gel stained with ethidium bromide to identify the sensitivity (oocysts per gram of feces) of the detection assay.
RESULTS
Oocyst detection limit.
Using a nested PCR amplification method, the lower limit of detection for C. parvum was found to be 25 oocysts per gram of feces (i.e., 50 oocysts spiked into 2 g of feces) (Fig. 1). The detection limit assay was performed twice with identical results.
FIG.1.
The oocyst detection limit (oocysts per gram of feces) was determined by spiking fecal samples with decreasing numbers of oocysts. Secondary PCR products are shown after electrophoresis on a 1.2% agarose gel stained with ethidium bromide. From left to right, the lanes are as follows: molecular size standards; negative controls (−) for secondary (2°) and initial (1°) PCRs, respectively; positive (+) controls for 2° and 1° PCRs, respectively; negative and positive controls for DNA extraction, respectively; negative and positive controls for IMS, respectively; and fecal samples spiked with 1, 10, 50, 100, 500, 1,000, and 5,000 oocysts, respectively.
Prevalence of Cryptosporidium oocysts in geese.
In total, 161 fecal samples from geese were collected and examined for the presence of Cryptosporidium oocysts, and 11 (6.8%) were positive for the parasite (Table 1). Of the 11 positive fecal specimens, 8 were successfully cloned and sequenced. For one sample, goose no. 3 from Illinois, two different 18S rRNA gene sequences (designated a and b) were identified.
Phylogenetic analysis.
Both neighbor-joining and parsimony trees were created to determine the phylogenetic relationship of the parasites obtained from geese (Fig. 2). Several distinct taxa of Cryptosporidium spp. are evident from the phylogenetic trees: C. parvum, C. meleagridis, and C. wrairi form one clade; C. andersoni, C. muris, and C. serpentis form another clade; and C. baileyi and C. felis are each on their own distinct branch. Evolutionary distances (Table 2) between clades are relatively large, ranging from 0.087 to 0.103 substitutions per site between the C. andersoni and C. parvum clades, 0.087 between C. baileyi and the C. andersoni clade, 0.035 to 0.042 between C. baileyi and the C. parvum clade, and 0.106 to 0.115 between C. felis and the C. andersoni clade. Within a clade, evolutionary distances are much smaller, with a range of 0.010 to 0.017 substitutions per site within the C. andersoni clade and 0.002 to 0.007 within the C. parvum clade.
TABLE 2.
Kimura two-parameter distance matrix (substitutions per site)a
| Sequence source | Evolutionary distance for:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. felis | C. andersoni | C. muris (murine) | C. serpentis | C. baileyi | Goose no. 1 | Goose no. 3a | Goose no. 3b | Goose no. 5 | Goose no. 7 | Goose no. 9 | C. meleagridis | C. parvum (human) | C. parvum (bovine) | C. wrairi | |
| C. felis | — | ||||||||||||||
| C. andersoni | 0.106 | — | |||||||||||||
| C. muris (murine) | 0.115 | 0.010 | — | ||||||||||||
| C. serpentis | 0.112 | 0.017 | 0.017 | — | |||||||||||
| C. baileyi | 0.050 | 0.087 | 0.087 | 0.087 | — | ||||||||||
| Goose no. 1 (Ill.) | 0.082 | 0.107 | 0.112 | 0.098 | 0.050 | — | |||||||||
| Goose no. 3a (Ill.) | 0.083 | 0.107 | 0.113 | 0.099 | 0.050 | 0.000 | — | ||||||||
| Goose no. 3b (Ill.) | 0.063 | 0.098 | 0.103 | 0.098 | 0.063 | 0.096 | 0.096 | — | |||||||
| Goose no. 5 (Ill.) | 0.069 | 0.079 | 0.084 | 0.071 | 0.027 | 0.032 | 0.032 | 0.068 | — | ||||||
| Goose no. 7 (N.Y.) | 0.085 | 0.124 | 0.129 | 0.127 | 0.074 | 0.077 | 0.077 | 0.099 | 0.074 | — | |||||
| Goose no. 9 (Va.) | 0.077 | 0.104 | 0.104 | 0.095 | 0.042 | 0.024 | 0.025 | 0.088 | 0.027 | 0.074 | — | ||||
| C. meleagridis | 0.040 | 0.087 | 0.095 | 0.092 | 0.035 | 0.066 | 0.066 | 0.052 | 0.042 | 0.069 | 0.055 | — | |||
| C. parvum (human) | 0.042 | 0.095 | 0.103 | 0.100 | 0.042 | 0.069 | 0.069 | 0.060 | 0.050 | 0.077 | 0.058 | 0.007 | — | ||
| C. parvum (bovine) | 0.045 | 0.092 | 0.100 | 0.097 | 0.040 | 0.071 | 0.072 | 0.058 | 0.047 | 0.074 | 0.060 | 0.005 | 0.002 | — | |
| C. wrairi | 0.045 | 0.089 | 0.098 | 0.095 | 0.037 | 0.069 | 0.069 | 0.058 | 0.045 | 0.069 | 0.058 | 0.005 | 0.007 | 0.005 | — |
GenBank accession numbers are AF112575 (C. felis), AB089285 (C. andersoni), AB089284 (C. muris murine genotype), AF093499 (C. serpentis), L19068 (C. baileyi), AF112574 (C. meleagridis), AF093489 (C. parvum human genotype), AF093493 (C. parvum bovine genotype), U11440 (C. wrairi), AY324635 (goose no. 1), AY324637 (goose no. 3a), AY324638 (goose no. 3b), AY324639 (goose no. 5), AY324641 (goose no. 7), and AY324643 (goose no. 9).
The sequence recovered from the cormorant was 100% identical to C. baileyi L19068. Sequences of oocysts from geese no. 1, 2, 6, and 8 were 100% identical to each other and had an evolutionary distance (Table 2) to C. baileyi L19068 (0.050) identical to the evolutionary distance between C. baileyi L19068 and C. felis AF112574. One of the sequences from goose no. 3 (sequence a) was also very closely related to the sequences of geese no. 1, 2, 6, and 8, and these five sequences formed a clade with bootstrap values of 100 and 99%, respectively, by the neighbor-joining and parsimony methods (Fig. 2). The sequence from goose no. 9 clustered with the those of the clade of geese no. 1, 2, 3 (sequence a), 6, and 8, with significant bootstrap values of 95 and 97% for neighbor-joining and parsimony analyses, respectively. The sequence from goose no. 5 also clustered with the sequence from goose no. 9 and those of the clade of geese no. 1, 2, 3 (sequence a), 6, and 8 by both the neighbor-joining and parsimony analyses. The evolutionary distances of the sequences from geese no. 5 and 9 to those of the group including geese no. 1, 2, 6, and 8 were 0.032 and 0.024, respectively, greater than the distances between C. parvum AF093489 and C. wrairi U11440 (0.007) or between C. serpentis AF093499 and C. muris AB089284 (0.017). Similarly, the evolutionary distance between the goose no. 5 and goose no. 9 sequences (0.027) was greater than the distance between distinct Cryptosporidium species.
Two additional sequences, from goose no. 7 and goose no. 3 (sequence b), were very different from the sequences recovered from the other geese and from GenBank sequences. The evolutionary distances between the sequences from goose no. 7 and goose no. 1 (0.077), the sequence from goose no. 7 and C. baileyi (0.074), and the sequence from goose no. 7 and C. meleagridis AF112574 (0.069) were greater than the evolutionary distance between C. parvum AF093489 and C. baileyi L19068 (0.042). In addition, the evolutionary distance between sequence b from goose no. 3 and all other sequences in the phylogenetic analysis ranged from 0.052 to 0.103 (Table 2), similar to the range of evolutionary distances between C. felis and the other sequences in the trees (0.040 to 0.115).
DISCUSSION
A previous study by Graczyk et al. (11) reported the average concentration of C. parvum oocysts in goose feces to be 370 ± 197 oocysts g−1. Since birds are refractory hosts of C. parvum (9), even larger numbers of C. baileyi or C. meleagridis oocysts would be expected in goose feces. Thus, the detection limit of the current assay (25 per gram of feces) was acceptable for the concentrations of Cryptosporidium oocysts expected in geese.
At the time of this study, only five C. baileyi 18S rRNA gene sequences had been deposited in GenBank; these five sequences are from United States, Australian, and Hungarian strains and are identical. Furthermore, the HSP70 and COWP gene sequences of these isolates are identical, suggesting that the genome of C. baileyi may be conserved. The genetic distinctness of C. baileyi was further supported by the recovery of an identical 18S rRNA gene sequence from a cormorant in Massachusetts during this study.
Unique 18S rRNA gene sequences, suggestive of new Cryptosporidium genotypes, were identified in goose feces as well. Identical sequences from geese no. 1, 2, 6, and 8 (from Illinois, Illinois, Massachusetts, and Virginia, respectively) were recovered, showing conservation of a new 18S rRNA gene sequence across broad geographic areas. The evolutionary distance of these sequences (and sequence a from goose no. 3) to C. baileyi was similar to the evolutionary distance of C. felis to C. baileyi, suggesting that this clade of sequences (Fig. 2) represents a new genotype or perhaps even a distinct species of Cryptosporidium in geese. The sequences from geese no. 5 and 9 were most closely related to this clade, yet the evolutionary distances between goose no. 5 and 9 sequences and those of this clade were greater than the distance between C. serpentis and C. muris. Thus, the oocysts recovered from geese no. 5 and 9 may represent two new genotypes, or two distinct but closely related species, of the taxonomic group represented by the clade of goose no. 1, 2, 3 (sequence a), 6, and 8 sequences. A definitive taxonomic classification of these oocysts requires morphological and biological characterizations that are not feasible given the limited oocyst quantities in environmental samples.
Further evidence for new Cryptosporidium genotypes in geese was found in the unique 18S rRNA gene sequences recovered from geese no. 7 and 3 (sequence b). The integrity of the 18S rRNA secondary structure, given the nucleotide changes observed in the sequences from geese no. 3 and 7, was verified, and the possibility of Taq polymerase error during PCR was eliminated as an explanation for the observed sequence differences. The sequences recovered from geese no. 7 and 3 (sequence b) are valid and most likely represent two previously uncharacterized species of Cryptosporidium. The genetic heterogeneity observed among Cryptosporidium oocysts from geese in this study supports the increasing level of diversity often reported for this genus (5, 17, 19, 28).
Two new species of Cryptosporidium in birds have been recently proposed (16, 20). Oocysts isolated from finches have been named C. blagburni on the basis of the unique localization of the oocysts in the proventriculus of these birds and phylogenetic analyses of both the 18S rRNA and HSP70 genes (16). In a separate study, partial sequences for the 18S rRNA gene of oocysts isolated from finches have been submitted to GenBank under the name C. galli. Phylogenetic analysis of C. blagburni, C. galli, and the goose-derived sequences from the present study, at the 18S rRNA locus (Fig. 3), shows that the sequences from the present study are genetically distinct from those of C. blagburni and C. galli and also suggests that C. blagburni and C. galli may represent the same taxonomic group.
Although we set out to characterize the level of genetic heterogeneity in the 18S rRNA genes of C. baileyi and C. meleagridis, we ultimately revealed an increasing level of genetic heterogeneity within the genus. Because the majority of 18S rRNA gene sequences recovered in this study were distinct from existing Cryptosporidium sequences, little has been discovered about the level of genetic variation among C. baileyi and C. meleagridis from geese. A more exhaustive sampling of birds will be required to ascertain the level of genetic heterogeneity of C. baileyi and C. meleagridis in the environment, since only 11 of 161 geese in this study were positive for Cryptosporidium oocysts. It is important to note that some of the negative fecal sample results reported in this study may have been due to poor PCR amplification caused by the high level of genetic variability observed in the amplified portion of the 18S rRNA.
Although some conservation of Cryptosporidium 18S rRNA gene sequences at different geographic locations was observed in the present study, the data suggest that geographic location is not predictive of 18S rRNA gene sequence, since distinct 18S rRNA gene sequences were recovered from geese in the same geographic area. Different 18S rRNA gene sequences were recovered from closely related oocysts in geese from Illinois (geese no. 1, 3 [sequence a], and 5) and Virginia (geese no. 8 and 9), and one goose (no. 3) shed oocysts with two distinct 18S rRNA gene sequences. The data suggest that geese can be carriers of more than one species of oocyst simultaneously.
The heterogeneity observed among Cryptosporidium 18S rRNA gene sequences from geese highlights the need for additional studies and offers insight into the use of 18S rRNA gene sequence data for species and source identification of oocysts in the environment. To date, the most-well-characterized member of the genus is C. parvum, the species of primary concern for human health. Because non-C. parvum species have recently been associated with human disease and are frequently encountered in the environment, additional information about other Cryptosporidium species is needed to interpret the results of environmental studies. As the present study showed, it is likely that many novel Cryptosporidium genotypes will be found in the environment. Only with a broader knowledge of the genetic heterogeneity of each species, and the genus as a whole, will it be possible to utilize the results of environmental studies for the development of appropriate watershed management strategies to protect surface waters from oocyst contamination.
Goose feces have been clearly identified as potential sources of microbiological contamination of surface waters (1, 6). Graczyk et al. (9, 10) showed that C. parvum oocysts retained infectivity for neonatal BALB/c mice after intestinal passage through Pekin ducks and Canada geese. A later field study performed near Chesapeake Bay (11) identified infectious zoonotic C. parvum oocysts in goose feces, indicating that waterfowl can serve as mechanical vectors of C. parvum and disseminate infectious oocysts to the environment. Although we did not see evidence of C. parvum oocysts in goose feces in this study, we did isolate novel gene sequences of uncharacterized Cryptosporidium spp. oocysts with unknown potential to cause disease in humans. These Cryptosporidium oocysts are not necessarily goose parasites, as disease in the geese was not evident from the appearance of the animals or the quality of the fecal samples. At the very least, however, geese may serve as mechanical vectors of the novel Cryptosporidium isolates recovered in this study. Further studies to rigorously characterize the extent of diversity of Cryptosporidium spp. in goose feces, the ability of those species to cause infection in humans, and the role of geese in the epidemiology of waterborne cryptosporidiosis are warranted.
Acknowledgments
We thank Giovanni Widmer (Tufts University School of Veterinary Medicine, North Grafton, Mass.) for materials and technical assistance. We gratefully acknowledge the efforts of the following individuals in the collection of fecal samples: Barbara Kostroff (Diagnostic Specialty Lab, New City, N.Y.), Monica Tischler (Benedictine University, Lisle, Ill.), Thomas Lustig (National Wildlife Foundation, Boulder, Colo.), Jo Ann Jellison, Richard Ting, Ralph Capriola, Jacqueline Fletcher (Oklahoma State University, Stillwater), and Thomas Roderick.
This work was supported by a U.S. EPA “Science to Achieve Results” (STAR) fellowship.
REFERENCES
- 1.Alderisio, K. A., and N. DeLuca. 1999. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada geese (Branta canadensis). Appl. Environ. Microbiol. 65:5628-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad-El-Kariem, F. M., H. A. Robinson, D. A. Dyson, D. Evans, S. Wright, M. T. Fox, and V. McDonald. 1995. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology 110:129-132. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., M. S. Boguski, D. J. Lipman, J. Ostell, and B. F. F. Ouellette. 1998. GenBank. Nucleic Acids Res. 26:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carraway, M., S. Tzipori, and G. Widmer. 1996. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl. Environ. Microbiol. 62:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalmers, R. M., K. Elwin, W. J. Reilly, H. Irvine, A. L. Thomas, and P. R. Hunter. 2002. Cryptosporidium in farmed animals: the detection of a novel isolate in sheep. Int. J. Parasitol. 32:21-26. [DOI] [PubMed] [Google Scholar]
- 6.Dieter, R. A., Jr., R. S. Dieter, R. A. Dieter III, and G. Gulliver. 2001. Zoonotic diseases: health aspects of Canadian geese. Int. J. Circumpolar Health 60:676-684. [PubMed] [Google Scholar]
- 7.Dubey, J. P., C. A. Speer, and R. Fayer. 1990. Cryptosporidiosis of man and animals. CRC Press, Boca Raton, Fla.
- 8.Egyed, Z., T. Sreter, Z. Szell, B. Beszteri, M. Dobos-Kovacs, K. Marialigeti, A. W. C. A. Cornelissen, and I. Varga. 2002. Polyphasic typing of Cryptosporidium baileyi: a suggested model for characterization of cryptosporidia. J. Parasitol. 88:237-243. [DOI] [PubMed] [Google Scholar]
- 9.Graczyk, T. K., M. R. Cranfield, R. Fayer, and M. S. Anderson. 1996. Viability and infectivity of Cryptosporidium parvum oocysts are retained upon intestinal passage through a refractory avian host. Appl. Environ. Microbiol. 62:3234-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graczyk, T. K., M. R. Cranfield, R. Fayer, J. Trout, and H. J. Goodale. 1997. Infectivity of Cryptosporidium parvum oocysts is retained upon intestinal passage through a migratory water-fowl species (Canada goose, Branta canadensis). Trop. Med. Int. Health 2:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graczyk, T. K., R. Fayer, J. M. Trout, E. J. Lewis, C. A. Farley, I. Sulaiman, and A. A. Lal. 1998. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl. Environ. Microbiol. 64:2736-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 13.Jellison, K. L., H. F. Hemond, and D. B. Schauer. 2002. Sources and species of Cryptosporidium oocysts in the Wachusett Reservoir watershed. Appl. Environ. Microbiol. 68:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan, U. M., P. T. Monis, R. Fayer, P. Deplazes, and R. C. Thompson. 1999. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J. Parasitol. 85:1126-1133. [PubMed] [Google Scholar]
- 16.Morgan, U. M., P. T. Monis, L. Xiao, J. Limor, I. Sulaiman, S. Raidal, P. O'Donoghue, R. Gasser, A. Murray, R. Fayer, B. L. Blagburn, A. A. Lal, and R. C. A. Thompson. 2001. Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int. J. Parasitol. 31:289-296. [DOI] [PubMed] [Google Scholar]
- 17.Ong, C. S. L., D. L. Eisler, A. Alikhani, V. W. K. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a corvine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. L. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perz, J. F., and S. M. LeBlancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York state. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan, U. M., L. Xiao, C. Read, I. M. Sulaiman, P. Monis, A. A. Lal, R. Fayer, and I. Pavlasek. 2003. A redescription of Cryptosporidium galli Pavlasek 1999. (Apicomplexa: Cryptosporidiidae) from birds. J. Parasitol. 89:809-813. [DOI] [PubMed] [Google Scholar]
- 21.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. LeBlancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographic origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulaiman, I. M., A. A. Lal, and L. Xiao. 2002. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88:388-394. [DOI] [PubMed] [Google Scholar]
- 23.Sulaiman, I. M., U. M. Morgan, R. C. A. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4. Sinauer Associates, Sunderland, Mass.
- 26.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, L., J. Limor, U. M. Morgan, I. M. Sulaiman, R. C. A. Thompson, and A. A. Lal. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol. 66:5499-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]