Abstract
In this study we developed a multilocus sequence typing (MLST) scheme for bacteria of the Bacillus cereus group. This group, which includes the species B. cereus, B. thuringiensis, B. weihenstephanensis, and B. anthracis, is known to be genetically very diverse. It is also very important because it comprises pathogenic organisms as well as bacteria with industrial applications. The MLST system was established by using 77 strains having various origins, including humans, animals, food, and soil. A total of 67 of these strains had been analyzed previously by multilocus enzyme electrophoresis, and they were selected to represent the genetic diversity of this group of bacteria. Primers were designed for conserved regions of housekeeping genes, and 330- to 504-bp internal fragments of seven such genes, adk, ccpA, ftsA, glpT, pyrE, recF, and sucC, were sequenced for all strains. The number of alleles at individual loci ranged from 25 to 40, and a total of 53 allelic profiles or sequence types (STs) were distinguished. Analysis of the sequence data showed that the population structure of the B. cereus group is weakly clonal. In particular, all five B. anthracis isolates analyzed had the same ST. The MLST scheme which we developed has a high level of resolution and should be an excellent tool for studying the population structure and epidemiology of the B. cereus group.
The Bacillus cereus group comprises the species B. cereus, B. thuringiensis, B. weihenstephanensis, and B. anthracis, which are important for economical and/or medical reasons. B. cereus is a ubiquitous soil bacterium and an opportunistic human pathogen that causes contamination problems in the dairy industry and paper mills. B. thuringiensis is an insect pathogen that is usually harmless to humans, although human infection may occur (4, 16); it is used industrially as a source of insecticidal toxins, often in the form of spore-containing preparations of crystal protein toxins. B. weihenstephanensis, the most recently described member of this group, is a psycrophilic bacterium that was identified as a separate species on the basis of the sequence of its rRNA genes and cold-shock protein genes (22). The pathogenic potential of this bacterium is not known, but the organism carries the same enterotoxin genes that B. cereus carries (38). B. anthracis is the cause of the acute and often lethal disease anthrax, and it has acquired particular notoriety recently because of its potential use in biological warfare and bioterrorism.
It has previously been demonstrated that B. cereus, B. thuringiensis, and B. anthracis should be regarded as one species based on genetic evidence (13). This group of bacteria is known to be genetically highly diverse (11). However, evidence of a clonal population structure, i.e., a population in which recombination rates are low and separate lineages are made up of closely related isolates (clones) that diversify mainly through mutation, has been found both for clinical isolates of B. cereus and for some serotypes of B. thuringiensis (2), as well as for B. anthracis (12, 13), which is one of the most homogeneous species known, representing basically a single clone (10, 18, 32).
The genetic diversity of the B. cereus group has been studied by using various methods, including multilocus enzyme electrophoresis (MEE), pulsed-field gel electrophoresis, and amplified fragment length polymorphism (3, 11-13, 39, 41). However, these methods are difficult to standardize between laboratories, and it is difficult to compare the results for large strain collections. Thus, our understanding of the population structure and evolution of the B. cereus group would be greatly improved by the use of a standardized method for strain analysis.
Multilocus sequence typing (MLST), which is based on sequencing a number of essential or housekeeping genes spread around the bacterial chromosome, is a method that is unambiguous and truly portable among laboratories. Since the initial development of this technique for Neisseria meningitidis in 1998 (23), MLST schemes have been developed for the most important bacterial pathogens, including Streptococcus pneumoniae (7), Streptococcus pyogenes (8), Haemophilus influenzae (25), Staphylococcus aureus (6), Campylobacter jejuni (5), Enterococcus faecium (14), Bordetella pertussis (40), Escherichia coli (1, 29), and Salmonella (20), and schemes are being developed for many other species. These MLST schemes have been used successfully to explore the population structure of bacteria, to study the evolution of their virulence properties, and to identify antibiotic-resistant strains and epidemic clones. MLST is a direct adaptation of the MEE method (37). In MEE, variation in genes that is believed to be selectively neutral is revealed by differential migration during gel electrophoresis of the gene products, the proteins. In MLST genetic variation is observed directly by sequencing DNA from fragments of various housekeeping genes.
In this study, we designed an MLST scheme for the B. cereus group, and to validate the method, we compared the MLST results with the results obtained by MEE for isolates which were analyzed previously (11-13). The sequence data were analyzed in relation to the evolution, phylogeny, and epidemiology of these bacteria.
MATERIALS AND METHODS
Bacterial isolates.
A total of 77 bacterial strains of the B. cereus group were analyzed (Table 1). Sixty-seven of these isolates were chosen to represent the various lineages identified in a diverse collection of isolates previously analyzed by an MEE scheme based on 13 metabolic enzymes (11-13). The B. cereus, B. thuringiensis, and B. weihenstephanensis strains were obtained from the culture collection of the Biotechnology Centre of Oslo, Oslo, Norway. For B. anthracis, DNA was obtained from the Pasteur Institute, Paris, France (strain 7700) and from the Norwegian Institute of Public Health, Oslo, Norway (strain EPI674, a human isolate [33], and strains NVH87 and NVH93, two animal isolates from Norway). DNA sequences of B. anthracis Ames were obtained from The Institute for Genomic Research (TIGR) web site (http://www.tigr.org).
TABLE 1.
Origins of the 77 B. cereus group isolates examined by MLST
| Strain | ST | Origin | Analyzed by MEE |
|---|---|---|---|
| EPI674 | 1 | B. anthracis, heroin abuser, muscle, Norway | NDa |
| Ames | 1 | B. anthracis Ames | ND |
| NVH87 | 1 | B. anthracis, bovine isolate, Norway, 1987 | ND |
| NVH93 | 1 | B. anthracis, bovine isolate, Norway, 1993 | ND |
| 7700 | 1 | B. anthracis, Pasteur Institute, Paris, France | Yes |
| AH 1293 | 2 | Blood, Norway | ND |
| AH 1294 | 2 | Patient isolate, blood, Norway | ND |
| AH 716 | 2 | Patient isolate, pus, Norway | Yesb |
| AH 889 | 2 | Patient isolate, blood, Norway | Yes |
| AH 75 | 3 | B. cereus ATCC 10987 | Yesb |
| AH 187 | 4 | B. cereus F4810-72 | Yes |
| AH 817 | 4 | Patient isolate, periodontitis, Norway | Yesb |
| AH 226 | 5 | B. cereus ATCC 4342 | Yesb |
| AH 230 | 6 | B. cereus ATCC 21282 | Yes |
| AH 251 | 7 | B. thuringiensis subsp. kenya | Yes |
| AH 267 | 8 | Patient isolate, Norway | Yes |
| AH 403 | 9 | Dairy, Finland | Yesb |
| AH 407 | 10 | Dairy, Finland | Yesb |
| AH 408 | 11 | Dairy, Finland | Yesb |
| AH 572 | 12 | Soil, Moss, Norway | Yesc |
| AH 519 | 12 | Soil, Moss, Norway | Yesc |
| AH 536 | 12 | Soil, Moss, Norway | Yesc |
| AH 542 | 13 | Soil, Moss, Norway | Yesc |
| AH 546 | 14 | Soil, Moss, Norway | Yesc |
| AH 547 | 15 | Soil, Moss, Norway | Yesc |
| AH 553 | 16 | Soil, Moss, Norway | Yesc |
| AH 607 | 17 | Dairy, Norway | Yesb |
| AH 614 | 18 | Dairy, Norway | Yesb |
| AH 627 | 19 | Soil, Sætereng, Norway | Yesc |
| AH 632 | 20 | Soil, Sætereng, Norway | Yesc |
| AH 663 | 20 | Soil, Brekkvasselv, Norway | Yesc |
| AH 641 | 21 | Soil, Sætereng, Norway | Yesc |
| AH 645 | 22 | Soil, Steigen, Norway | Yesc |
| AH 650 | 23 | Soil, Steigen, Norway | Yesc |
| AH 664 | 24 | Soil, Brekkvasselv, Norway | Yesc |
| AH 676 | 24 | Soil, Tromsø, Norway | Yesc |
| AH 675 | 25 | Soil, Tromsø, Norway | Yesc |
| AH 678 | 26 | Soil, Tromsø, Norway | Yesc |
| AH 681 | 27 | Soil, Tromsø, Norway | Yesc |
| AH 685 | 28 | Soil, Tromsø, Norway | Yesc |
| AH 718 | 29 | Patient isolate, nose, Norway | Yesb |
| AH 721 | 30 | Patient isolate, urine, Norway | Yesb |
| AH 726 | 31 | Patient isolate, urine, Norway | Yesb |
| AH 727 | 32 | Patient isolate, pus, Norway | Yesb |
| AH 728 | 33 | Patient isolate, urine, Norway | Yesb |
| AH 810 | 34 | Patient isolate, periodontitis, Norway | Yesb |
| AH 811 | 35 | Patient isolate, periodontitis, Brazil | Yesb |
| AH 812 | 36 | Patient isolate, periodontitis, Norway | Yesb |
| AH 813 | 37 | Patient isolate, periodontitis, Brazil | Yesb |
| AH 816 | 37 | Patient isolate, periodontitis, Norway | Yesb |
| AH 818 | 37 | Patient isolate, periodontitis, Brazil | Yesb |
| AH 820 | 37 | Patient isolate, periodontitis, Norway | Yesb |
| AH 819 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 823 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 824 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 825 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 826 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 827 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 828 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 829 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 831 | 38 | Patient isolate, periodontitis, Norway | Yesb |
| AH 830 | 39 | Patient isolate, periodontitis, Norway | Yesb |
| AH 840 | 40 | B. thuringiensis subsp. thuringiensis 4A3 | Yes |
| AH 1031 | 41 | B. thuringiensis Bt407 | Yes |
| AH 1123 | 42 | Patient isolate, France | Yes |
| AH 1127 | 43 | Patient isolate, France | Yes |
| AH 1129 | 44 | Patient isolate, eye, Oklahoma | Yes |
| AH 1131 | 45 | Patient isolate, eye, Oklahoma | Yes |
| AH 1132 | 46 | Patient isolate, eye, Oklahoma | Yes |
| AH 1135 | 47 | Soil, France | Yes |
| AH 1145 | 48 | B. weihenstephanensis type strain WSBC10204 | Yes |
| AH 1247 | 49 | Soil, 9779, Pasteur Institute, Paris, France | Yes |
| AH 1270 | 50 | Patient isolate, cervix, Iceland | ND |
| AH 1271 | 51 | Secretary lamp, university hospital, Iceland | ND |
| AH 1272 | 52 | Patient isolate, amniotic fluid, Iceland | ND |
| AH 1273 | 52 | Patient isolate, blood, Iceland | ND |
| AH 923 | 53 | B. cereus type strain ATCC 14579 | Yes |
Preparation of DNA.
B. anthracis DNA was obtained from the Pasteur Institute and the Norwegian Institute of Public Health. All B. cereus and B. thuringiensis isolates were grown overnight in Luria-Bertani broth at 30°C. A loopful of cells of each strain was then suspended in lysis buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% Triton X-100) and boiled for 10 to 15 min. After centrifugation at 16,000 × g for 10 min, the supernatant solution was kept at −20°C.
Loci.
A range of loci encoding housekeeping genes were identified from preliminary DNA contigs from B. cereus ATCC 14579 (obtained from Integrated Genomics, Inc., Chicago, Ill.), B. cereus ATCC 10987, and B. anthracis Ames (obtained from TIGR). Primers were designed for conserved areas in these sequences, delimiting partial segments within the genes, by using the Primer3 software (35). A total of 13 loci were tested initially, and 7 of these loci, for which the primer pairs gave PCR products relatively easily for a subset of diverse strains tested, were selected; the primers used are listed in Table 2.
TABLE 2.
Primers used for PCR amplification and sequencing of B. cereus group housekeeping genes
| Gene | Forward primer | Reverse primer | Annealing temp (°C) |
|---|---|---|---|
| adk | 5′CAGCTATGAAGGCTGAAACTG3′ | 5′CTAAGCCTCCGATGAGAACA3′ | 57 |
| ccpA | 5′GTTTAGGATACCGCCCAAATG3′ | 5′TGTAACTTCTTCGCGCTTCC3′ | 59 |
| ftsA | 5′TCTTGACATCGGTACATCCA3′ | 5′GCCTGTAATAAGTGTACCTTCCA3′ | 57 |
| glpT | 5′TGCGGCTGGATGAGTGA3′ | 5′AAGTAAGAGCAAGGAAGA3′ | 52 |
| pyrE | 5′TCGCATCGCATTTATTAGAA3′ | 5′CCTGCTTCAAGCTCGTATG3′ | 57 |
| recF | 5′GCGATGGCGAAATCTCATAG3′ | 5′CAAATCCATTGATTCTGATACATC3′ | 59 |
| sucC | 5′GGCGGAACAGAAATTGAAGA3′ | 5′TCACACTTCATAATGCCACCA3′ | 59 |
The seven genes chosen for the MLST scheme were adk (encoding adenylate kinase), ccpA (catabolite control protein A), ftsA (cell division protein), glpT (glycerol-3-phosphate permease), pyrE (orotate phosphoribosyltransferase), recF (DNA replication and repair protein), and sucC (succinyl coenzyme A synthetase, beta subunit). The only locus used in both our previous MEE studies and the MLST scheme described here is adk. The positional distribution of the seven loci used for MLST on the chromosome of the type strain of B. cereus, ATCC 14579, are (from the origin of replication, oriC) as follows: recF, 3.3 kb; adk, 137.0 kb; glpT, 651.8 kb; sucC, 3,813.2 kb; pyrE, 3,861.6 kb; ftsA, 3,886.8 kb; and ccpA, 4,611.7 kb. Genomic data are available at http://www.integratedgenomics.com. The minimum distance between the loci was 24.5 kb. The relative positions are similar in the B. anthracis Ames genome (31).
Amplification and nucleotide sequencing.
The PCR conditions used for the amplification reactions were as follows. A PCR was performed for 40 cycles with a 50-μl mixture containing each deoxynucleoside triphosphate at a concentration of 0.8 mM, each primer at a concentration of 0.4 μM, 50 ng of genomic DNA, and 1 U of Dynazyme (Finnzymes Oy, Espoo, Finland). An initial denaturation at 94°C for 5 min was followed by 94°C for 1 min. The primer annealing temperature for each primer set is shown in Table 2. The annealing time was 1 min, and this was followed by extension for 1 min at 72°C. After 40 cycles, the reaction was completed by a final extension at 72°C for 7 min.
PCR products were rinsed with an exonuclease-alkaline phosphatase kit (ExoSAP-IT; USB Corporation, Cleveland, Ohio) as described by the manufacturer. PCR products were then sequenced with an ABI Prism BigDye terminator cycle sequencing Ready Reaction kit (v2.0) used as described by the manufacturer, except that the volume was only one-quarter of the recommended volume. The primers used to generate the PCR products were used to sequence the DNA on both strands. DNA sequencing was done with an ABI Prism 377 DNA sequencer by using 5% Long Rangers (Cambrex, East Rutherford, N.J.).
Data analysis.
After electrophoresis, the complementary strands were aligned, and the sequences were edited by using the AutoAssembler software (PE Applied Biosystems, Foster City, Calif.). For each gene, the sequence fragments of the 77 strains were compared, and different sequences were assigned arbitrary allele numbers (identical sequences were assigned the same allele number). Each strain was then characterized on the basis of its combination of alleles at the seven loci, which defined an allelic profile, and distinctive (unique) profiles were designated multilocus sequence types (STs). The rationale for this approach is that in the absence of information concerning whether sequence differences are due to mutation or recombination, the total amount of divergence may not be suitable for differentiating strains.
Standard MLST statistics (e.g., number of polymorphic sites, allele frequencies) were computed by using the START v1.0.5 software (17). Pairwise ratios of nonsynonymous substitutions to synonymous substitutions (dN/dS) in sequences were calculated by using the method of Nei and Gojobori (28) and the START software.
The index of association (IA) (24), which was an estimate of the degree of linkage disequilibrium among loci, was computed at the MLST web site (http://www.mlst.net). The statistical significance of IA was assessed by comparing the observed variance of IA with the maximum variance obtained from 1,000 randomized data sets.
To assess the genetic relatedness among the strains, a dendrogram based on the allelic profiles was constructed by using the unweighted pair group method with arithmetic means (UPGMA) applied to a matrix of pairwise distances, defined as the percentages of allelic differences between profiles, by means of the MEGA 2.1 analysis package (21). To improve the discrimination between divergent lineages, similar analyses were conducted after each gene was split into three segments whose lengths were nearly the same; when dividing the gene lengths produced decimal values, the smallest integer was used as the segment length. Allele numbers were assigned independently for each gene region, and the allelic profiles were then combinations of 21 numbers. Allele assignment and computation of distance matrices were carried out by using computer scripts written by one of us (N.J.T.).
Phylogenetic clustering of the strains based on the total numbers of differences among gene sequences was also performed by using either the UPGMA or the neighbor-joining (NJ) method (36) and the MEGA 2.1 package. This was done mainly to study congruence among gene trees and to detect possible recombination. For NJ trees, pairwise distances between sequences were computed by using Kimura's (19) two-parameter model, which takes into account multiple substitutions at a given site and transition-transversion rate bias. The bootstrap technique (9) was employed to assess support for the various groups by resampling the sequence alignment 1,000 times.
Nucleotide sequence accession numbers.
Nucleotide sequences of the internal fragment genes used in this analysis have been deposited in the GenBank data bank under accession numbers AY387859 to AY388397.
RESULTS
Allelic profiles and STs.
The lengths of the fragments analyzed ranged from 330 bp (glpT) to 504 bp (sucC) (Table 3). The average G+C contents of the different gene fragments ranged from 34 to 45%. Examination of genomic data for B. anthracis Ames (from TIGR database [http://www.tigr.org]) (31), B. cereus ATCC 14579 (from the Integrated Genomics, Inc., database [http://www.integratedgenomics.com]) (15), and B. cereus ATCC 10987 (D. A. Rasko, J. Ravel, O. A. Økstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A.-B. Kolstø, C. M. Fraser, and T. D. Read, unpublished data) revealed that this range was greater than the range observed for the complete gene sequences (35 to 40%). However, the values were still within the range of G+C contents computed for the whole gene sets in these genomes. For each gene, the nucleotide frequencies were nearly identical among the strains.
TABLE 3.
Genetic diversity at the seven loci examined in this study
| Locus | Fragment length (bp)a | No. of polymorphic sitesb | % of variable nucleotide sites | No. of alleles | Avg G + C content (%) | Avg dN/dS ratio | Avg distance between allelesc |
|---|---|---|---|---|---|---|---|
| adk | 450 (648) | 55 (36) | 12.2 | 27 | 38.5 | 0.0809 | 0.031 (0.012) |
| ccpA | 418 (996) | 67 (48) | 16.0 | 38 | 36.1 | 0.0454 | 0.034 (0.014) |
| ftsA | 401 (1,299) | 37 (36) | 9.2 | 25 | 41.4 | 0.0024 | 0.023 (0.014) |
| glpT | 330 (1,347) | 86 (62) | 26.1 | 40 | 44.8 | 0.0494 | 0.080 (0.040) |
| pyrE | 404 (630) | 60 (45) | 14.9 | 40 | 40.3 | 0.0328 | 0.023 (0.009) |
| recF | 470 (1,125) | 110 (77) | 23.4 | 33 | 34.0 | 0.0420 | 0.072 (0.043) |
| sucC | 504 (1,158) | 85 (67) | 16.9 | 34 | 36.7 | 0.0378 | 0.040 (0.017) |
The numbers in parentheses are total lengths of the genes.
The numbers in parentheses are numbers of synonymous polymorphic sites.
Percentage of nucleotide differences. The numbers in parentheses are standard deviations.
The sequences had from 9% (ftsA) to 26% (glpT) variable sites, which resulted in 25 to 40 distinct alleles for the different loci (Table 3). The average number of alleles per locus was 33.9. No insertions or deletions were observed in any of the sequences. All of the average dN/dS values shown in Table 3 are much less than 1, indicating that most of the sequence variability identified is selectively neutral. Synonymous substitutions were at least 12 (1/0.0809) times more frequent than amino acid changes at any locus. Remarkably, 36 of the 37 polymorphic sites in ftsA were synonymous, and only a single amino acid difference was observed in a single strain (AH 650) for the entire ftsA-encoded protein alignment. This means that virtually all of the variation in our ftsA data set is neutral.
Altogether, 53 unique STs were observed in the 77 isolates (see Table 4), and 44 of the STs (83%) were identified only once. The most frequent ST, ST 38, was found in nine (12%) of the strains examined, and it was found exclusively in the periodontitis strains (AH 819, AH 823 to AH 829, and AH 831) (Table 1). The five B. anthracis strains had identical alleles at all seven loci (ST 1). The seven additional STs identified more than once were ST 37 (four periodontitis patient isolates from Norway and Brazil), ST 2 (four patient isolates from Norway), ST 4 (B. cereus F4810-72 and a periodontal strain), ST 52 (two patient isolates from Iceland), and ST 12, ST 20, and ST 24 (three, two, and two soil isolates from Norway, respectively).
TABLE 4.
Allele numbers at the seven loci assigned for entire gene fragments
| ST | Allele no. at the following loci:
|
||||||
|---|---|---|---|---|---|---|---|
| adk | ccpA | ftsA | glpT | pyrE | recF | sucC | |
| 1 | 25 | 36 | 2 | 18 | 19 | 9 | 12 |
| 2 | 3 | 13 | 4 | 13 | 38 | 30 | 3 |
| 3 | 12 | 16 | 16 | 36 | 16 | 7 | 9 |
| 4 | 10 | 14 | 5 | 14 | 7 | 4 | 4 |
| 5 | 11 | 15 | 6 | 15 | 8 | 5 | 5 |
| 6 | 12 | 16 | 16 | 16 | 16 | 7 | 9 |
| 7 | 13 | 17 | 7 | 17 | 27 | 6 | 6 |
| 8 | 14 | 18 | 8 | 18 | 16 | 11 | 21 |
| 9 | 15 | 19 | 23 | 19 | 28 | 19 | 22 |
| 10 | 5 | 20 | 9 | 20 | 9 | 20 | 7 |
| 11 | 16 | 21 | 10 | 21 | 29 | 21 | 23 |
| 12 | 7 | 22 | 7 | 22 | 10 | 22 | 24 |
| 13 | 7 | 22 | 7 | 22 | 11 | 22 | 24 |
| 14 | 17 | 23 | 11 | 23 | 30 | 23 | 25 |
| 15 | 5 | 20 | 9 | 24 | 31 | 20 | 26 |
| 16 | 7 | 24 | 24 | 25 | 32 | 24 | 24 |
| 17 | 18 | 25 | 7 | 26 | 10 | 25 | 27 |
| 18 | 16 | 26 | 12 | 27 | 33 | 26 | 28 |
| 19 | 19 | 21 | 10 | 28 | 12 | 21 | 23 |
| 20 | 5 | 20 | 9 | 29 | 9 | 20 | 7 |
| 21 | 20 | 20 | 9 | 19 | 13 | 27 | 29 |
| 22 | 21 | 27 | 9 | 30 | 34 | 20 | 29 |
| 23 | 22 | 28 | 25 | 31 | 35 | 27 | 30 |
| 24 | 23 | 29 | 9 | 19 | 14 | 20 | 31 |
| 25 | 24 | 20 | 9 | 29 | 9 | 20 | 7 |
| 26 | 23 | 30 | 13 | 32 | 36 | 28 | 32 |
| 27 | 22 | 31 | 9 | 8 | 13 | 29 | 16 |
| 28 | 23 | 20 | 9 | 8 | 37 | 14 | 33 |
| 29 | 3 | 32 | 4 | 13 | 38 | 30 | 3 |
| 30 | 3 | 33 | 7 | 33 | 39 | 31 | 6 |
| 31 | 3 | 13 | 1 | 13 | 38 | 30 | 3 |
| 32 | 7 | 34 | 14 | 34 | 40 | 32 | 34 |
| 33 | 10 | 35 | 15 | 35 | 15 | 4 | 8 |
| 34 | 26 | 37 | 17 | 37 | 15 | 8 | 10 |
| 35 | 3 | 38 | 1 | 38 | 38 | 33 | 3 |
| 36 | 10 | 35 | 18 | 14 | 17 | 4 | 11 |
| 37 | 2 | 35 | 2 | 39 | 18 | 9 | 1 |
| 38 | 10 | 35 | 18 | 14 | 7 | 4 | 11 |
| 39 | 27 | 35 | 16 | 40 | 16 | 7 | 9 |
| 40 | 10 | 14 | 5 | 36 | 7 | 4 | 4 |
| 41 | 1 | 1 | 1 | 1 | 20 | 10 | 13 |
| 42 | 2 | 2 | 2 | 2 | 1 | 11 | 14 |
| 43 | 2 | 3 | 2 | 3 | 2 | 1 | 1 |
| 44 | 3 | 4 | 1 | 4 | 3 | 12 | 15 |
| 45 | 4 | 5 | 3 | 5 | 4 | 2 | 2 |
| 46 | 3 | 6 | 1 | 6 | 21 | 13 | 3 |
| 47 | 2 | 7 | 2 | 7 | 5 | 1 | 1 |
| 48 | 5 | 8 | 9 | 8 | 22 | 14 | 16 |
| 49 | 6 | 9 | 19 | 9 | 23 | 15 | 17 |
| 50 | 7 | 10 | 20 | 10 | 24 | 16 | 18 |
| 51 | 8 | 11 | 21 | 11 | 25 | 17 | 19 |
| 52 | 9 | 12 | 22 | 12 | 26 | 18 | 20 |
| 53 | 3 | 13 | 4 | 13 | 6 | 3 | 3 |
Interestingly, even though B. anthracis strains did not have the same ST as any other B. cereus group isolate, some B. cereus strains (in particular, all isolates from patients except AH 1135) shared one or more alleles with B. anthracis. Strains AH 813, AH 816, AH 818, AH 820, AH 1123, AH 1127, and AH 1135 had the same ftsA allele as B. anthracis; strain AH 267 had the same glpT allele; and strains AH 813, AH 816, AH 818, and AH 820 had the same recF allele.
The allele frequencies for STs showed that for some loci (adk, ccpA, ftsA, recF, and sucC), one or two alleles were dominant in the population (with a frequency of 11 to 19%), while for other loci (glpT and pyrE), the allelic distribution was more even (data not shown). On average, the 53 STs differed from each other at more than six of the seven loci studied (average number of allelic mismatches, 6.8).
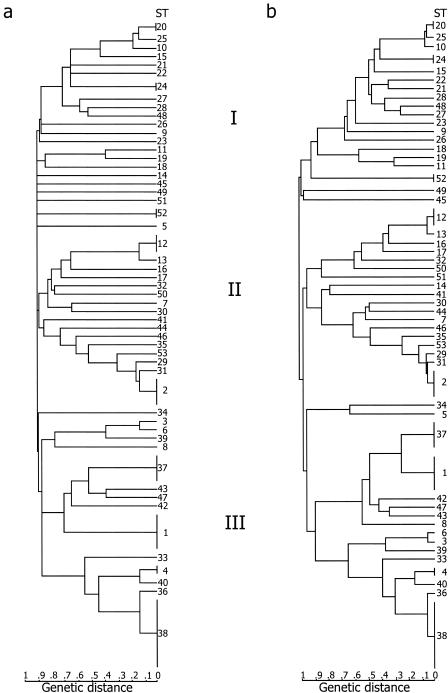
Clustering of the STs revealed three major groups (Fig. 1b). In group I all isolates except AH 1131, AH 1272, and AH 1273 were from soil or dairy sources and joined the type strain of B. weihenstephanensis. Group II contained a mixture of isolates from patients, soil, and dairy sources, as well as the B. cereus type strain, and group III contained almost exclusively B. anthracis and patient isolates, except for AH 75, AH 230, AH 226, and AH 1135.
FIG. 1.
Genetic relationships among 77 isolates of the B. cereus group. Strains were clustered by UPGMA applied to a distance matrix of pairwise differences between allelic profiles. Alleles were identified either for entire gene fragments (Table 4) (a) or after fragments were split into three equal-length parts (Table 5) (a). The scale bars indicate the percentages of mismatches between allelic profiles.
Congruence and recombination analysis.
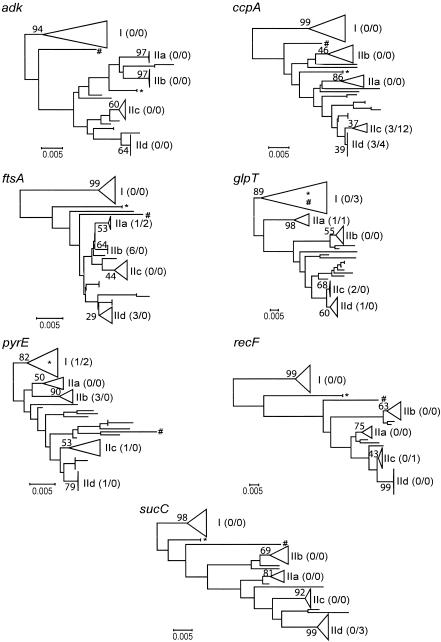
To analyze whether recombination events are frequent in the B. cereus group, trees based on the total amount of nucleotide differences were constructed for each gene separately by using the NJ method and Kimura's two-parameter model (UPGMA trees based on observed numbers of nucleotide differences had similar shapes [data not shown]). Incongruence among gene trees would indicate that there had been frequent recombination. For all genes, lineages could be divided into two major clusters, clusters I and II, corresponding to the groups identified by MEE (11-13). Strains belonging to cluster I differed from strains belonging to cluster II at all seven loci. The mean sequence divergence between clusters I and II was 0.061 substitution per site (for concatenated sequences), whereas the within-group average sequence divergence values were 0.025 and 0.013 substitution per site for clusters I and II, respectively. Both major clusters included nearly the same isolates in all dendrograms; the few exceptions were related most noticeably to strains AH 1272 and AH 1273 (ST 52) and strain AH 1247 (ST 49), which clustered either within cluster I or within cluster II or between the two groups depending on the gene examined (Fig. 2). AH 1247 (ST 49) joined cluster I once (for glpT) and cluster II once (for pyrE) and five times was outside the two clusters. AH 1272 and AH 1273 (ST 52) grouped within cluster I for two loci (glpT and pyrE), within cluster II for two loci (adk and ccpA), and between the two clusters for three loci. AH 627 (ST 19) grouped within cluster II for pyrE only. Furthermore, as Fig. 2 shows, cluster II was divided into four subclusters, clusters IIa, IIb, IIc, and IId, for each locus. Basically, a given subcluster for the most part contained the same isolates, and there were a few additional or missing strains depending on the locus analyzed (detailed trees and/or compositions of the clusters are available from us). Branching within and among the groups was more variable.
FIG. 2.
Phylogenetic relationships among 77 B. cereus group isolates inferred from individual genes. Trees were constructed by using the NJ method applied to pairwise distances among strains, computed by using Kimura's (19) nucleotide substitution model. Five groups of strains that were recovered for each tree were designated I, IIa, IIb, IIc, and IId. For the sake of simplicity, strain designations and detailed relationships among isolates within the groups are not shown (detailed trees and compositions of the clusters are available from us). For each of the five groups, the numbers in parentheses indicate the number of missing isolates/number of additional isolates compared to the number of isolates in the adk tree, and the numbers above the branches indicate bootstrap values, expressed as percentages (based on 1,000 replicates). In the adk tree, clusters I, IIa, IIb, IIc, and IId contain 18, 8, 12, 13, and 14 strains, respectively. B. anthracis belongs to cluster IIc. Isolates AH 1272 and AH 1273 (ST 52) and AH 1247 (ST 49), which cluster within cluster I, within cluster II, or between the two groups depending on the gene analyzed, are indicated by an asterisk and a number sign, respectively. The scale bars indicate 0.005 nucleotide substitution per site.
The recovery of five similar clusters for all seven genes suggests that the observed groups represent true genetic relatedness among the isolates and that the level of recombinational exchange is low.
The IA was calculated to estimate the degree of association and recombination between alleles at different loci based on the allelic profile data. The IA computed by using all 77 isolates was 3.54 (P < 0.001), indicating that there was significant linkage disequilibrium, which suggests that there were limited recombinational events and a clonal population structure in the B. cereus group. The IA was still significantly different from zero when only one representative of each ST was included in the computation, which removed bias due to taxonomic sampling, although the IA was smaller (1.90; P < 0.001).
MLST versus MEE.
The MLST and MEE results were compared for the 67 strains that were previously analyzed by MEE (11-13; Helgason, unpublished data).
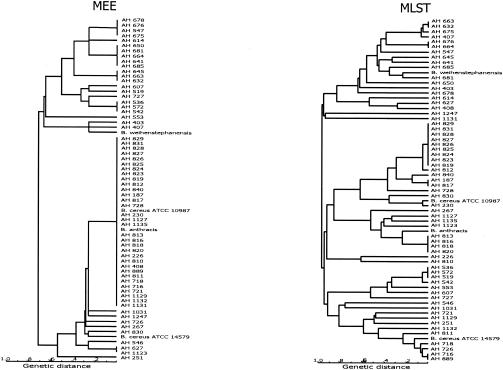
Although only seven loci were examined by MLST, compared to the 13 loci examined by MEE, the number of STs identified by MLST (50 STs) was more than twice the number of electrophoretic types (22 electrophoretic types) identified by MEE for the 67 isolates. A cluster of 34 isolates that were not distinguishable by MEE and thus had the same electrophoretic type were resolved into 20 STs by MLST. Most of these isolates, however, were part of the same MLST cluster. Dendrograms for MEE and MLST data were somewhat congruent in that major clusters I and II (as well as a few other closely related groups) were recovered by both methods. One difference is that the MLST dendrograms contained three main clusters and that strains AH 536, AH 572, AH519, AH542, AH 553, AH 607, and AH 722, which were grouped in cluster I by MEE, were grouped in cluster II by MLST (Fig. 3). The deep branches for the clusters were not in agreement when the MEE and MLST dendrograms were compared. The individual gene trees based on nucleotide sequence comparisons were in agreement with the main MEE clusters, clusters I and II.
FIG. 3.
Comparison of MEE and MLST data for a subset of 67 B. cereus group isolates (out of the 77 strains used in this study). Both dendrograms were generated by the UPGMA method from a matrix of coefficients of genetic distances. The scale bars indicate the percentages of allelic differences between electrophoretic types for MEE and between STs for MLST. The MEE dendrogram was based on 13 enzyme loci as described previously (11-13).
Gene splitting.
Since the results described above suggested that recombination has only a weak overall impact on the generation of diversity among strains, we split genes into three regions that were the same length and assigned alleles separately for each segment, thus taking into account differences in the distribution of mutations and variable sites. (The divergence did not increase; it decreased, and splitting grouped together strains that were completely different before. Rather, splitting took into account where the differences were in a gene [, i.e., in which part of the gene].) Note that splitting did not change the STs. Examination of the split allelic profiles revealed some relationships among isolates that were not seen when whole gene fragments were used to assign alleles. For example, AH 226 (ST 5) and AH 810 (ST 34) had no allele in common (Table 4); however, after gene splitting their genetic distance was 38% (Fig. 1b and Table 5), which is more similar to the relatedness observed in the previous analysis in which MEE was used (12). Phylogenetic clustering of split profiles provided dendrograms that were much more highly resolved than the standard MLST tree (compare Fig. 1a and b). Major groups were conserved, and the trees were similar to those obtained by concatenating all gene sequences together and clustering strains based on total numbers of nucleotide differences (data not shown). Furthermore, MLST trees obtained by using genes split into three parts had a resolution similar to or higher than the resolution of trees constructed from MEE data, while the standard MLST tree had more unresolved branches.
TABLE 5.
Allele numbers at the seven loci assigned after gene fragments were split into three equal-length parts
| ST | Allele no. at the following loci:
|
||||||
|---|---|---|---|---|---|---|---|
| adk | ccpA | ftsA | glpT | pyrE | recF | sucC | |
| 1 | 16, 2, 2 | 2, 2, 14 | 2, 2, 2 | 3, 3, 16 | 12, 8, 13 | 8, 1, 7 | 9, 1, 1 |
| 2 | 3, 3, 3 | 5, 13, 4 | 1, 4, 1 | 11, 4, 12 | 4, 3, 15 | 23, 3, 3 | 3, 3, 3 |
| 3 | 11, 9, 11 | 12, 16, 7 | 1, 14, 7 | 14, 13, 28 | 11, 1, 10 | 4, 5, 4 | 4, 9, 8 |
| 4 | 9, 8, 9 | 2, 14, 2 | 1, 5, 2 | 12, 3, 13 | 5, 1, 6 | 4, 4, 4 | 4, 4, 4 |
| 5 | 10, 2, 10 | 11, 15, 2 | 1, 6, 4 | 13, 12, 14 | 1, 4, 7 | 5, 1, 5 | 5, 5, 5 |
| 6 | 11, 9, 11 | 12, 16, 7 | 1, 14, 7 | 14, 13, 15 | 11, 1, 10 | 4, 5, 4 | 4, 9, 8 |
| 7 | 12, 3, 3 | 13, 13, 8 | 1, 1, 5 | 15, 4, 4 | 11, 1, 20 | 6, 3, 3 | 6, 6, 3 |
| 8 | 13, 2, 2 | 2, 17, 9 | 2, 7, 1 | 3, 3, 16 | 11, 1, 10 | 8, 1, 1 | 1, 17, 1 |
| 9 | 14, 10, 12 | 14, 18, 10 | 5, 8, 6 | 16, 14, 7 | 15, 15, 6 | 16, 8, 10 | 12, 18, 14 |
| 10 | 5, 5, 5 | 14, 8, 5 | 4, 8, 6 | 16, 15, 17 | 6, 1, 6 | 17, 8, 10 | 7, 7, 6 |
| 11 | 14, 10, 13 | 15, 19, 5 | 5, 9, 6 | 17, 16, 18 | 16, 15, 6 | 18, 13, 15 | 17, 19, 14 |
| 12 | 7, 7, 7 | 16, 20, 2 | 1, 1, 5 | 18, 17, 19 | 7, 5, 8 | 19, 14, 16 | 18, 15, 12 |
| 13 | 7, 7, 7 | 16, 20, 2 | 1, 1, 5 | 18, 17, 19 | 7, 5, 9 | 19, 14, 16 | 18, 15, 12 |
| 14 | 1, 3, 14 | 17, 21, 11 | 1, 10, 1 | 19, 18, 20 | 4, 16, 21 | 9, 15, 17 | 19, 20, 10 |
| 15 | 5, 5, 5 | 14, 8, 5 | 4, 8, 6 | 20, 14, 7 | 17, 17, 22 | 17, 8, 10 | 20, 18, 15 |
| 16 | 7, 7, 7 | 18, 20, 2 | 1, 18, 5 | 21, 9, 21 | 7, 5, 23 | 19, 14, 18 | 18, 15, 12 |
| 17 | 7, 7, 15 | 19, 22, 2 | 1, 1, 5 | 22, 17, 10 | 7, 5, 8 | 19, 14, 19 | 21, 15, 12 |
| 18 | 14, 10, 13 | 15, 23, 5 | 5, 11, 6 | 23, 19, 22 | 18, 1, 6 | 20, 8, 15 | 22, 21, 14 |
| 19 | 14, 10, 16 | 15, 19, 5 | 5, 9, 6 | 17, 20, 23 | 8, 1, 10 | 18, 13, 15 | 17, 19, 14 |
| 20 | 5, 5, 5 | 14, 8, 5 | 4, 8, 6 | 16, 15, 24 | 6, 1, 6 | 17, 8, 10 | 7, 7, 6 |
| 21 | 5, 11, 17 | 14, 8, 5 | 4, 8, 6 | 16, 14, 7 | 9, 6, 6 | 11, 8, 20 | 12, 18, 6 |
| 22 | 5, 12, 18 | 14, 24, 5 | 4, 8, 6 | 16, 14, 25 | 9, 18, 6 | 17, 8, 10 | 12, 18, 6 |
| 23 | 5, 11, 5 | 14, 8, 12 | 4, 8, 11 | 6, 21, 7 | 9, 19, 6 | 11, 8, 20 | 23, 18, 14 |
| 24 | 5, 13, 5 | 20, 8, 5 | 4, 8, 6 | 16, 14, 7 | 10, 7, 6 | 17, 8, 10 | 24, 22, 6 |
| 25 | 15, 5, 5 | 14, 8, 5 | 4, 8, 6 | 16, 15, 24 | 6, 1, 6 | 17, 8, 10 | 7, 7, 6 |
| 26 | 5, 13, 5 | 21, 8, 5 | 6, 8, 6 | 16, 14, 26 | 9, 6, 24 | 21, 16, 21 | 24, 22, 16 |
| 27 | 5, 11, 5 | 22, 8, 5 | 4, 8, 6 | 6, 7, 7 | 9, 6, 6 | 22, 8, 10 | 12, 13, 6 |
| 28 | 5, 13, 5 | 14, 8, 5 | 4, 8, 6 | 6, 7, 7 | 19, 20, 25 | 11, 8, 10 | 12, 18, 17 |
| 29 | 3, 3, 3 | 23, 13, 4 | 1, 4, 1 | 11, 4, 12 | 4, 3, 15 | 23, 3, 3 | 3, 3, 3 |
| 30 | 3, 3, 3 | 24, 4, 1 | 1, 1, 5 | 24, 1, 4 | 20, 21, 11 | 24, 3, 3 | 6, 6, 3 |
| 31 | 3, 3, 3 | 5, 13, 4 | 1, 1, 1 | 11, 4, 12 | 4, 3, 15 | 23, 3, 3 | 3, 3, 3 |
| 32 | 7, 7, 7 | 18, 25, 2 | 7, 12, 5 | 25, 22, 27 | 7, 1, 8 | 25, 14, 18 | 25, 15, 12 |
| 33 | 9, 8, 9 | 2, 2, 13 | 1, 13, 7 | 26, 3, 13 | 1, 4, 11 | 4, 4, 4 | 4, 8, 7 |
| 34 | 17, 2, 2 | 25, 15, 2 | 8, 6, 4 | 27, 13, 14 | 1, 4, 11 | 7, 6, 6 | 8, 10, 9 |
| 35 | 3, 3, 3 | 23, 6, 4 | 1, 1, 1 | 1, 4, 12 | 4, 3, 15 | 26, 3, 3 | 3, 3, 3 |
| 36 | 9, 8, 9 | 2, 2, 13 | 1, 15, 7 | 12, 3, 13 | 5, 1, 12 | 4, 4, 4 | 4, 9, 4 |
| 37 | 2, 2, 2 | 2, 2, 13 | 2, 2, 2 | 3, 3, 29 | 2, 8, 13 | 8, 1, 7 | 1, 1, 1 |
| 38 | 9, 8, 9 | 2, 2, 13 | 1, 15, 7 | 12, 3, 13 | 5, 1, 6 | 4, 4, 4 | 4, 9, 4 |
| 39 | 18, 14, 9 | 2, 2, 13 | 1, 14, 7 | 14, 23, 28 | 11, 1, 10 | 4, 5, 4 | 4, 9, 8 |
| 40 | 9, 8, 9 | 2, 14, 2 | 1, 5, 2 | 14, 13, 28 | 5, 1, 6 | 4, 4, 4 | 4, 4, 4 |
| 41 | 1, 1, 1 | 1, 1, 1 | 1, 1, 1 | 1, 1, 1 | 8, 9, 14 | 9, 3, 8 | 10, 11, 10 |
| 42 | 2, 2, 2 | 2, 2, 2 | 2, 2, 2 | 2, 2, 2 | 1, 1, 1 | 8, 1, 1 | 1, 12, 1 |
| 43 | 2, 2, 2 | 2, 3, 3 | 2, 2, 2 | 3, 3, 3 | 2, 2, 1 | 1, 1, 1 | 1, 1, 1 |
| 44 | 3, 3, 3 | 3, 4, 1 | 1, 1, 1 | 1, 4, 4 | 3, 1, 2 | 10, 3, 3 | 11, 3, 3 |
| 45 | 4, 4, 4 | 4, 5, 2 | 3, 3, 3 | 4, 5, 5 | 4, 1, 3 | 2, 2, 2 | 2, 2, 2 |
| 46 | 3, 3, 3 | 5, 6, 4 | 1, 1, 1 | 1, 1, 4 | 13, 10, 15 | 3, 7, 9 | 3, 3, 3 |
| 47 | 2, 2, 2 | 2, 7, 3 | 2, 2, 2 | 5, 6, 6 | 1, 1, 4 | 1, 1, 1 | 1, 1, 1 |
| 48 | 5, 5, 5 | 6, 8, 5 | 4, 8, 6 | 6, 7, 7 | 9, 11, 16 | 11, 8, 10 | 12, 13, 6 |
| 49 | 6, 6, 6 | 7, 9, 5 | 9, 16, 8 | 7, 8, 8 | 4, 12, 17 | 12, 9, 11 | 13, 14, 11 |
| 50 | 7, 7, 7 | 8, 10, 2 | 1, 1, 9 | 8, 9, 9 | 7, 1, 18 | 13, 10, 12 | 14, 15, 12 |
| 51 | 7, 2, 4 | 9, 11, 6 | 1, 1, 2 | 9, 10, 10 | 14, 13, 19 | 14, 11, 13 | 15, 15, 13 |
| 52 | 8, 3, 8 | 10, 12, 5 | 10, 17, 10 | 10, 11, 11 | 6, 14, 6 | 15, 12, 14 | 16, 16, 14 |
| 53 | 3, 3, 3 | 5, 13, 4 | 1, 4, 1 | 11, 4, 12 | 4, 3, 5 | 3, 3, 3 | 3, 3, 3 |
DISCUSSION
In this paper, we describe development of an MLST scheme based on seven housekeeping genes (adk, ccpA, ftsA, glpT, pyrE, recF, and sucC) for molecular typing and population genetic analyses of bacterial strains of the B. cereus group, which includes B. anthracis, B. cereus, B. thuringiensis, and B. weihenstephanensis. In an attempt to build a solid MLST tool we focused on choosing a set of isolates covering the known diversity of this bacterial group. The system which we developed is based on one primer pair for each gene to obtain PCR products and sequence DNA from this phylogenetically diverse group of bacteria. Genomic data for B. anthracis Ames (31), B. cereus ATCC 14579 (15), and B. cereus ATCC 10987 (D. A. Rasko et al., unpublished data) showed that these genetic loci are scattered in different areas of the main chromosome. Although three genes, sucC, pyrE, and ftsA, are close to each other and have minimum intergenic distances of 47.2 and 24.5 kb, no annotated insertion sequence elements or transposons were found in the area including these loci in the B. cereus type strain and B. anthracis Ames genomes; thus, they are unlikely to be involved in joint horizontal transfer of two or more loci.
For all seven loci, the amount of synonymous substitutions was at least 12 times (1/0.0809) larger than the amount of nonsynonymous substitutions, as shown by the low dN/dS ratios (Table 3). This means that most of the observed variability is selectively neutral at the protein level, which makes the loci chosen suitable for MLST analysis (23). Remarkably, a single nonsynonymous site was identified in the ftsA alignment. This gene, which encodes a cell division protein, seems to be subject to very high levels of purifying selection limiting changes in the amino acid sequence. The average number of alleles per locus, 33.9, relative to the number of strains analyzed was much higher than the numbers observed in several other bacterial species studied by MLST, including N. meningitidis (23), S. pneumoniae (7), C. jejuni (5), and E. faecium (14). For example, 33.3 alleles per locus were identified in 194 isolates (155 STs) of C. jejuni, and only 25.1 alleles per locus were found in 295 S. pneumoniae strains (143 STs). The differences reflect the great diversity of the isolates examined here and the extensive diversity of this group of bacteria. The genetic variability within the B. cereus group is high, not only in the DNA sequences but also when the patterns of presence and absence of genes are examined. The gene content varies between strains and is related to the phylogenetic structure of the group, as shown by analyses of S-layer proteins (26), suppression subtractive hybridization (30), and comparative genome hybridization (31).
In contrast, B. anthracis is monomorphic (10, 18, 32). Accordingly, not a single nucleotide polymorphism was detected at any locus in the five B. anthracis isolates examined (Tables 4 and 5). The B. anthracis strains share one or two alleles with a few other B. cereus strains at three loci, ftsA, glpT, and recF. Interestingly, all but one (AH 1135) of the B. cereus strains that share alleles with B. anthracis were isolated from patients (AH 813, AH 816, AH 818, AH 820, AH 1123, AH 1127, and AH 267), and four of these strains were isolated from patients affected by periodontitis (Table 1). Besides the B. anthracis lineage, a few B. cereus and B. thuringiensis isolates that have the same ST (Fig. 1 and Table 1) were identified. Most of these strains were isolated from patients and might be recently emerged virulent types. Our study confirmed the overall genetic relationships within this group of bacteria and that particular B. cereus and B. thuringiensis strains are more closely related to B. anthracis than to any other B. cereus or B. thuringiensis strain. This fact has been used to support the contention that B. anthracis, B. cereus, and B. thuringiensis should be considered members of the same species (13). The close relationship of B. cereus and B. anthracis has recently been confirmed on a genomic scale (15, 31).
Congruence analyses of sequence trees (Fig. 2), in which the same five clusters were obtained in dendrograms for individual genes, and examination of the association among loci in allelic profiles (IA) suggested that genetic exchanges through recombination may not have played a major role in generating the diversity among the strains studied. The fact that the overall structure of the MLST tree was conserved when genes were each split into three equal-length segments further strengthens this assumption (Fig. 1). These results indicate that the population structure of the B. cereus group as a whole is clonal in spite of extensive genetic diversity, in agreement with the conclusions drawn from MEE studies (12). The main clonal groups identified are the B. anthracis and B. cereus periodontal lineages.
For a weakly recombining population, splitting the genes clearly increased the resolution of the dendrograms by clustering strains that were impossible to classify based on their single-sequence profiles, and this might be a simple alternative to increasing the number of genetic loci sequenced. Some groups obtained by splitting genes may better reflect real genetic relationships among strains, because conservation of different parts of a sequenced fragment is taken into account. For example, B. cereus strains AH 226 and AH 810, which clustered at a genetic distance of approximately 0.2 in our MEE analysis (12), were unrelated in the single-sequence tree (Fig. 1a). However, they had two common regions in ccpA, ftsA, and pyrE and a common region in adk and glpT (Table 5). All major clusters in the triple-sequence tree (Fig. 1b) were supported by bootstrap values greater than 90% when trees were based on the total amounts of nucleotide differences between concatenated sequences.
In this study we focused on finding DNA sequences that were relatively easy to amplify by PCR. In the chromosomal area from approximately 90° to 250°, it was difficult to amplify DNA of all isolates with the same primers. This might be because the sequence variability in this area is even higher than that found near oriC (31) and might imply that the level of genetic diversity of the B. cereus group is even higher than that estimated by this study. This phenomenon is also known in other bacterial species. In Pseudomonas aeruginosa there are indications that there is an increasing gradient in the genomic diversity from the origin of replication (34), and in E. coli and Salmonella genes close to the replication terminus exhibit 50% higher divergence at synonymous sites than genes near the replication origin exhibit (27).
MLST provided greater discrimination than MEE. For example, only one MLST allele (ccpA allele 35) (Table 4) was found in two periodontal groups, ST 37 (AH 813, AH 816, AH 818, AH 820) and ST 38 (AH 819,AH 823, AH 824, AH 825, AH 826, AH 827, AH 828, AH829, AH 831 38), whereas MEE analysis showed that all periodontal isolates were identical (12). However, the MLST data were still consistent with a clonal population structure with geographically widespread and potentially highly virulent clones, such as the B. anthracis clone and a number of B. cereus clones associated with disease in humans (12, 13).
Strains AH 1272 and AH 1273 provide an interesting example. These two isolates possess the same ST (ST 52), which is completely different from all other STs observed in our data set (Fig. 1 and Table 4). AH 1272 and AH 1273 were isolated from two different patients 1.5 years apart in two different buildings of the same hospital in Iceland (Table 1). AH 1293 and AH 1294 were isolated from contaminated blood and from a patient who received the contaminated blood, respectively. Both isolates were ST 2 isolates, as were two other patient isolates from Norway. Furthermore, these strains are closely related to two other patient isolates, AH 718 (ST 29) and AH 726 (ST 31), as well as to the B. cereus type strain. Thus, MLST should be a useful tool for confirming the source of an infection.
MLST also could be helpful for selecting isolates of B. thuringiensis with high biopesticide potential but low risk as human pathogens. It could also be used to detect isolates that may be potential human pathogens. The discrimination level is convincing when we look at isolates AH 817 and AH 831. These two isolates were not distinguishable by MEE but had different pulsed-field gel electrophoresis profiles when they were digested with different enzymes (12). When MLST was used, strains AH 817 (ST4) and AH 831 (ST38) were distinguished because they had two different, but still closely related, STs (Fig. 1).
We believe that we have developed a robust MLST scheme that is suitable for the highly diverse B. cereus group; it has high-resolution power, its results are in agreement with results obtained previously by MEE, and it detects clonal lineages represented by isolates from different sources that might be virulent. This scheme should therefore significantly contribute to a better understanding of the population structure of the B. cereus group.
Acknowledgments
This work was supported by grants to E.H. and A.-B.K. from The Norwegian Research Council.
Johan Biørnstad and Pétur Jónsson are thanked for skillful technical assistance. We thank Sigfús Karlsson (Landspítali, University Hospital, Reykjavik, Iceland) for providing strains.
REFERENCES
- 1.Adiri, R. S., U. Gophna, and E. Z. Ron. 2003. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 222:199-203. [DOI] [PubMed] [Google Scholar]
- 2.Ankarloo, J., D. A. Caugant, B. M. Hansen, A. Berg, A. B. Kolstø, and A. Lövgren. 2000. Genome stability of Bacillus thuringiensis subsp. israelensis isolates. Curr. Microbiol. 40:51-56. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, C. R., D. Caugant, and A.-B. Kolstø. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damgaard, P. H., P. E. Granum, J. Bresciani, M. V. Torregrossa, J. Eilenberg, and L. Valentino. 1997. Characterization of Bacillus thuringiensis isolated from infections in burn wounds. FEMS Immunol. Med. Microbiol. 18:47-53. [DOI] [PubMed] [Google Scholar]
- 5.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 10.Harrell, L. J., G. L. Andersen, and K. H. Wilson. 1995. Genetic variability of Bacillus anthracis and related species. J. Clin. Microbiol. 33:1847-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgason, E., D. A. Caugant, M. M. Lecadet, Y. Chen, J. Mahillon, A. Lövgren, I. Hegna, K. Kvaløy, and A. B. Kolstø. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 12.Helgason, E., D. A. Caugant, I. Olsen, and A. B. Kolstø. 2000. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. Dusko Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, S. G., R. B. Goodbrand, R. Ahmed, and S. Kasatiya. 1995. Bacillus cereus and Bacillus thuringiensis isolated in a gastroenteritis outbreak investigation. Lett. Appl. Microbiol. 21:103-105. [DOI] [PubMed] [Google Scholar]
- 17.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 18.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Lechner, S., R. Mayr, K. P. Francis, B. M. Pruss, T. Kaplan, E. Wiessner-Gunkel, G. S. Stewart, and S. Scherer. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48:1373-1382. [DOI] [PubMed] [Google Scholar]
- 23.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maynard-Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mignot, T., B. Denis, E. Couture-Tosi, A. B. Kolstø, M. Mock, and A. Fouet. 2001. Distribution of S-layers on the surface of Bacillus cereus strains: phylogenetic origin and ecological pressure. Environ. Microbiol. 3:493-501. [DOI] [PubMed] [Google Scholar]
- 27.Mira, A., and H. Ochman. 2002. Gene location and bacterial sequence divergence. Mol. Biol. Evol. 19:1350-1358. [DOI] [PubMed] [Google Scholar]
- 28.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 29.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 32.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 33.Ringertz, S. H., E. A. Hoiby, M. Jensenius, J. Maehlen, D. A. Caugant, A. Myklebust, and K. Fossum. 2000. Injectional anthrax in a heroin skin-popper. Lancet 356:1574-1575. [DOI] [PubMed] [Google Scholar]
- 34.Römling, U., J. Greipel, and B. Tummler. 1995. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol. Microbiol. 17:323-332. [DOI] [PubMed] [Google Scholar]
- 35.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenfors, L. P., and P. E. Granum. 2001. Psychrotolerant species from the Bacillus cereus group are not necessarily Bacillus weihenstephanensis. FEMS Microbiol. Lett. 197:223-228. [DOI] [PubMed] [Google Scholar]
- 39.Ticknor, L. O., A. B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Loo, I. H., K. J. Heuvelman, A. J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilas-Boas, G., V. Sanchis, D. Lereclus, M. V. Lemos, and D. Bourguet. 2002. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]