Abstract
The purpose of this investigation was to examine the capacity of the biphenyl catabolic enzymes of Comamonas testosteroni B-356 to metabolize dihydroxybiphenyls symmetrically substituted on both rings. Data show that 3,3′-dihydroxybiphenyl is by far the preferred substrate for strain B-356. However, the dihydrodiol metabolite is very unstable and readily tautomerizes to a dead-end metabolite or is dehydroxylated by elimination of water. The tautomerization route is the most prominent. Thus, a very small fraction of the substrate is converted to other hydroxylated and acidic metabolites. Although 2,2′-dihydroxybiphenyl is a poor substrate for strain B-356 biphenyl dioxygenase, metabolites were produced by the biphenyl catabolic enzymes, leading to production of 2-hydroxybenzoic acid. Data show that the major route of metabolism involves, as a first step, a direct dehydroxylation of one of the ortho-substituted carbons to yield 2,3,2′-trihydroxybiphenyl. However, other metabolites resulting from hydroxylation of carbons 5 and 6 of 2,2′-dihydroxybiphenyl were also produced, leading to dead-end metabolites.
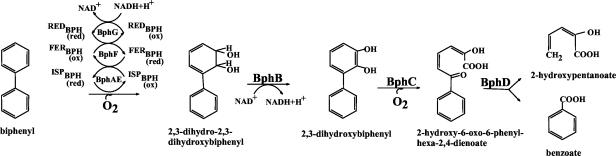
The enzymes of the bacterial biphenyl catabolic pathway are very versatile. They can cometabolically transform several biphenyl analogs, including polychlorinated biphenyls (PCBs) (1, 31), hydroxybiphenyls, and chlorohydroxybiphenyls (33). The four enzymatic steps required to transform biphenyl or its analogs into corresponding benzoic acids are illustrated in Fig. 1. The initial reaction of this pathway is catalyzed by the biphenyl dioxygenase (BPDO) (13, 16). The cis-(2R,3S)-dihydroxy-1-phenylcyclohexa-4,6-diene(cis-biphenyl-2,3-dihydrodiol) (commonly called cis-2,3-dihydro-2,3-dihydroxybiphenyl) generated by the catalytic oxygenation of biphenyl is dehydrogenated by the 2,3-dihydro-2,3-dihydroxybiphenyl 2,3-dehydrogenase (BphB), and the catechol produced is cleaved by the 2,3-dihydroxybiphenyl 1,2-dioxygenase. The 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA) is then hydrolyzed by the HOPDA hydrolase to yield benzoate and hydroxypentanoate. The substrate specificity of BPDO is crucial, because it limits the range of compounds potentially degradable by the catabolic system (14).
FIG. 1.
Biphenyl catabolic pathway of C. testosteroni B-356.
For example, the three symmetrical ortho, meta, or para-substituted dichlorinated biphenyl isomers are not metabolized equally well by various BPDOs. Comamonas testosteroni strain B-356 BPDO oxygenates 3,3′-dichlorobiphenyl much faster and more efficiently than 2,2′-dichlorobiphenyl and 4,4′-dichlorobiphenyl. On the other hand, the catalytic activity of Burkholderia sp. strain LB400 BPDO toward 2,2′-dichlorobiphenyl is much more efficient than that of B-356 BPDO (17), but 3,3′-dichlorobiphenyl and 4,4′-dichlorobiphenyl are poor substrates for LB400 BPDO. Nevertheless, metabolites generated from B-356 and LB400 demonstrating BPDO attack on each of these three substrates have been detected and identified (2, 14, 17). It is noteworthy that, for both strains, BPDO attacks 2,2′-dichlorobiphenyl principally at position 2,3, which results in concomitant dehalogenation of the molecule (2, 14).
Many features of this enzyme have now been well characterized (8, 14, 15, 17, 18, 19, 35, 36). BPDO is a three-component enzymatic system (Fig. 1). The first component, which is an oxygenase, is an iron-sulfur protein (ISPBPH) that catalyzes the addition of molecular oxygen. The second and third components are a flavoprotein reductase (REDBPH) and a ferredoxin (FERBPH), respectively, that are involved in the transfer of electrons from NADH to ISPBPH, which then activates molecular oxygen for insertion into the aromatic substrate. BPDO components are encoded by bphA (α subunit of ISPBPH), bphE (β subunit of ISPBPH), bphF (FERBPH), and bphG (REDBPH) in Burkholderia sp. strain LB400 (4, 11) and in C. testosteroni B-356 (39). Enhancement of BPDO catalytic potency through enzyme engineering toward a range of xenobiotic substrates, including PCBs as well as dibenzodioxins and dibenzofurans and their chlorinated forms, will require a better knowledge of the catalytic capacity of this enzyme toward biphenyl analogs and a better understanding of its mode of attack.
Although it has been shown that metabolism of dibenzofuran by Pseudomonas sp. strain HH69 generates 2,2′,3-trihydroxybiphenyl that is further metabolized to 2-hydroxybenzoic acid (12), there is relatively little information about the metabolism of dihydroxybiphenyls bearing hydroxyl groups on both phenyl rings. The 2-hydroxybiphenyl 3-monooxygenase of Pseudomonas sp. strain HBP1 has been well characterized (37). Beside 2-hydroxybiphenyl this enzyme can also hydroxylate 2,2′-dihydroxybiphenyl to generate 2,3,2′-trihydroxybiphenyl, which is then degraded by a meta-cleavage pathway. In a recent paper, the catalytic oxygenation of 2,2′-dihydroxybiphenyl by Burkholderia strain LB400 BPDO was examined (30). However, very few other investigations have examined the catalytic activity of the biphenyl catabolic pathway on hydroxybiphenyl analogs. In a previous report we have shown that C. testosteroni B-356 is able to metabolize monohydroxybiphenyls and monohydroxychlorobiphenyls through the biphenyl catabolic pathway (33). The transformation involved in the first step is the dioxygenation of vicinal ortho-meta carbons of the unsubstituted ring.
The toxicity of hydroxybiphenyls and substituted hydroxybiphenyls has been exploited for many years in antimicrobial preparations used as biocides for industrial and agricultural purposes (27, 40). However, these compounds are also ubiquitous in the environment. As our understanding of the biological degradation pathways of persistent pollutants progresses, it becomes clear that hydroxybiphenyls are key intermediates produced from multiple sources. For example, they are produced from microbial metabolism of dibenzofuran, fluorene, and carbazole (26). Furthermore, many reports have now shown that plants metabolize major environmental contaminants such as PCBs to generate hydroxylated derivatives (9, 10, 21, 22, 23, 28).
In the context of the potential use of high-performing evolved BPDOs in biological processes for the degradation of persistent pollutants, these enzymes are likely to simultaneously catalyze oxygenation of many analogs of the targeted substrates. Thus, hydroxybiphenyls are likely to be transformed in the course of processes aiming at the destruction of pollutants such as PCBs and polyaromatic hydrocarbons. It is, therefore, essential that the metabolism of hydroxybiphenyls through the biphenyl catabolic pathways be clearly understood.
In this investigation, we have examined the capacity of the biphenyl catabolic pathway of C. testosteroni B-356 to metabolize 2,2′- and 3,3′-dihydroxybiphenyl and provided evidence that metabolism of hydroxybiphenyls can generate dead-end metabolites from rearrangement of the dihydrodiol metabolites. The metabolism of 2,2′-dichlorobiphenyl was found to be similar to that observed when LB400 BPDO was used to catalyze the reaction (14, 30). A preference for the pair of vicinal ortho and meta carbons of 2,2′-dihydroxybiphenyl leads to the formation of 2,3,2′-trihydroxybiphenyl. A small proportion of the substrate was converted to 2,5,2′-trihydroxybiphenyl. The biphenyl catabolic pathway did not further degrade the latter.
On the other hand, catalytic oxygenation of 3,3′-dihydroxybiphenyl by B-356 BPDO generates as a major metabolite 3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one, which is a dead-end metabolite. It was shown, with isopropyl-β-d-thiogalactoside (IPTG)-induced Escherichia coli DH11S expressing B-356 BPDO, that this dead-end metabolite is produced by tautomerization of 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl. Since this compound was a dead-end metabolite resulting from catalytic dioxygenation of 3,3′-dihydroxybiphenyl, we have examined the metabolism of this compound in more detail.
MATERIALS AND METHODS
Bacterial strains, culture media, and chemicals.
The bacterial strains used in this study were C. testosteroni strain B-356 (3) and a variant of strain B-356 described previously (34), which was derived by adaptation through successive transfers on minimal medium containing biphenyl as the sole growth substrate. This strain expresses the biphenyl catabolic enzymes constitutively. Other bacteria used included Pseudomonas putida KT2440 carrying plasmid pDA1 or pDA2, which are described elsewhere (2). Strain KT2440[pDA1] expresses the four biphenyl catabolic enzymes of strain B-356, and KT2440[pDA2] expresses the enzymes of the first three steps of the pathway. E. coli strain DH11S carrying pQE51[LB400-bphFG] plus pDB31[B-356-bphAE] (7) was also used. This strain expresses an active BPDO exhibiting the biochemical features of B-356 BPDO. The media used were minimal medium 30 (MM30) (38), M9 (29), and Luria-Bertani (LB) broth (29) containing the appropriate selective agents for propagation. 2,2′-Dihydroxybiphenyl and 3,3′-dihydroxybiphenyl were from UltraScientific (North Kingstown, R.I.).
Identification of metabolites of hydroxybiphenyls.
Strains B-356 and B-356 biphenyl-adapted were grown at 30°C on MM30 plus biphenyl and/or succinate for the biphenyl-adapted variant. P. putida KT2440 carrying pDA1 or pDA2 was grown on LB broth with appropriate antibiotics (2). Cultures were filtered through packed glass wool to remove particulates, cell aggregates, and crystals of growth substrate. Cells were harvested by centrifugation, washed with phosphate buffer (30 mM, pH 7.2), and resuspended, depending on the experiment, to an optical density at 600 nm (OD600) of 1.0 (5 × 109 cells/ml) or 2.0 (1.0 × 1010 cells/ml) in MM30 plus the appropriate substrate. E. coli DH11S carrying pQE51[LB400-bphFG] plus pDB31[B-356-bphAE] was grown at 37°C to log phase in LB broth. The cultures were induced for 4 h with 0.5 mM IPTG and then washed and suspended to an OD600 of 2.0 in M9 medium containing 0.5 mM IPTG, 0.1% (wt/vol) sodium acetate, and the appropriate hydroxybiphenyl.
The final volume of the cell suspensions was 30 ml in 125-ml Erlenmeyer flasks for experiments involving B-356 and P. putida and 2 ml in 10-ml test tubes for experiments involving E. coli strains. The cultures were incubated at 30°C (37°C for E. coli) with shaking. At intervals, the whole content of each culture was extracted with ethyl acetate at neutral pH and then at pH 3 for gas chromatographic-mass spectrometric analysis according to protocols described previously (33). The metabolites were analyzed as butylboronate or trimethylsilyl (TMS) derivatives according to protocols described previously (17, 33). Occasionally, metabolites were derivatized with 9H2-TMS. 1H nuclear magnetic resonance (NMR) spectra were obtained at the NMR spectrometry center of the Université de Montréal, Montréal, Canada, with a Bruker WH 400-mHz spectrometer. The analysis was carried out in D2O at room temperature.
Ames test.
The Ames test was performed to examine the potential of mutagenicity of (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one, which is a major metabolite of 3,3′-dihydroxybiphenyl. The test was performed in triplicate according to the protocol described by Maron and Ames (24) with the four standard tester strains TA97, TA98, TA100, and TA102 of Salmonella enterica serovar Typhimurium. Tests were done with and without activation by an Aroclor 1254-induced rat liver S9 fraction (Microbiological Associates, Inc., Bethesda, Md.), lot R-469. The S9 fraction was added at concentrations of 20 and 50 μl per plate. (3S,4R)-3,4-Dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one was incorporated directly into the agar overlay at concentrations of 0.2, 2, 20, 50, 100, 250, and 500 μg per plate. The positive controls used to test the strains and the activity of the S9 fraction were sodium azide, 2-aminofluorene, and benzo(a)pyrene. Each strain responded as expected (24) when 1.5, 10, and 1 μg of sodium azide, 2-aminofluorene, and benzo(a)pyrene, respectively, were incorporated directly into the agar overlay of each plate with or without the S9 fraction.
Microtox assay.
The luminescence inhibition of Photobacterium phosphoreum by (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one was examined using the Microtox assay (Microbics Corp., Carlsbad, Calif.) performed on a Microtox model 500 analyzer. The compound was dissolved in dimethyl sulfoxide, and control assays with dimethyl sulfoxide and phenol were run in parallel. The assays were performed according to the exact recommendations of the company (Microtox manual, Microbics Corp., 1992). The 50% effective concentrations (EC50) were calculated using the Microtox version 6.2 data system software.
RESULTS
Oxygen uptake by washed cell suspensions as an initial measure of substrate preference.
B-356 exhibits substrate preferences towards symmetrical meta-substituted dihydroxybiphenyls. This was initially evaluated using the oxygen uptake rates of biphenyl-induced resting cell suspensions of strain B-356 or of its biphenyl-adapted variant. When 4,4′-dihydroxybiphenyl was used as the substrate, oxygen uptake rates were 12.1 and 9.0 μmol of O2/min/ml for B-356 and its biphenyl-adapted variant, respectively. 3,3′-Dihydroxybiphenyl was oxygenated the fastest of the three isomers by resting cell suspensions, 18.1 and 27.1 μmol of O2/min/ml for biphenyl-induced B-356 and the biphenyl-adapted variant, respectively. The oxygen uptake rate was 9.0 μmol of O2/min/ml for both suspensions when 2,2′-dihydroxybiphenyl was the substrate. Thus, similarly to the chlorinated biphenyls, the meta-substituted congeners are the preferred substrates for oxygenases of the biphenyl catabolic pathway of strain B-356 as judged by oxygen uptake in washed cell suspensions.
Metabolism of 2,2′-dihydroxybiphenyl by the biphenyl catabolic enzymes of strain B-356.
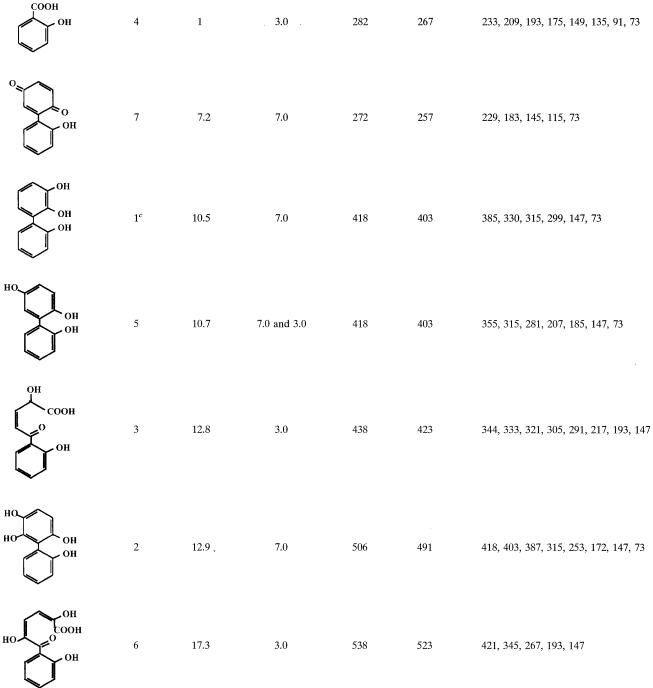
Resting cell suspensions of IPTG-induced E. coli DH11S expressing B-356 BPDO generated two metabolites from 2,2′-dihydroxybiphenyl. From gas chromatography-mass spectrometry analysis (Table 1) they were identified as 2,3,2′-trihydroxybiphenyl (metabolite 1) for the major metabolite and presumably 2,6,2′- or 2,5,2′-trihydroxybiphenyl (metabolite 5) for the minor metabolite. Data showed that the mode of attack on 2,2′-dihydroxybiphenyl by B-356 BPDO is similar to that of LB400 BPDO (30).
TABLE 1.
Mass spectral features of TMS-derived metabolitesa generated from 2,2′-dihydroxybiphenyl
Only major metabolites and those that help to elucidate the metabolic pathway of 2,2′-dihydroxybiphenyl are shown.
Retention time relative to the retention time of 2-hydroxybenzoic acid.
Metabolite found only in cell suspensions of E. coli cells expressing BPDO.
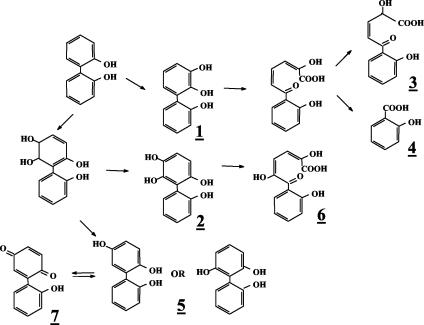
Evidence that strain B-356 metabolizes 2,2′-dihydroxybiphenyl through a pathway involving BPDO as a first step was provided from analysis of 2,2′-dihydroxybiphenyl metabolites generated by washed cell suspensions. The spectral features of the metabolites found in these washed cell suspensions are presented in Table 1. Trace amounts of metabolite 2, 2,5,6,2′-tetrahydroxybiphenyl, and its meta-cleavage derivative (metabolite 6) confirmed that catalytic oxygenation on carbons 5 and 6 had occurred. Metabolite 5, 2,5,2′-trihydroxybiphenyl, which is a dead-end metabolite, was likely to result from 5,6-dihydro-2,5,6,2′-tetrahydroxybiphenyl by elimination of water, and its identity was confirmed from the presence of 2-(2-hydroxyphenyl)-1,4-benzoquinone (metabolite 7). No 2,3,2′-dihydroxybiphenyl was found in the cell suspensions. However, the presence of 2-hydroxybenzoic acid (metabolite 4) and 2-hydroxy-5-oxo-5-(2-hydroxyphenyl)-3-pentanoic acid (metabolite 3) confirmed dioxygenation of the ortho and meta carbons on the substituted side of the ortho-hydroxylated ring. The fact that strain B-356 can degrade 2-hydroxybenzoate (33) makes it likely that part of the substrate is mineralized by the strain. Based on these data, the proposed pathway for the metabolism of 2,2′-dihydroxybiphenyl is shown in Fig. 2.
FIG. 2.
Proposed pathway for production of metabolites generated from 2,2′-dihydroxybiphenyl.
Metabolism of 3,3′-dihydroxybiphenyl by the biphenyl catabolic pathway of strain B-356. (i) Metabolites from IPTG-induced E. coli DH11S expressing B-356 BPDO.
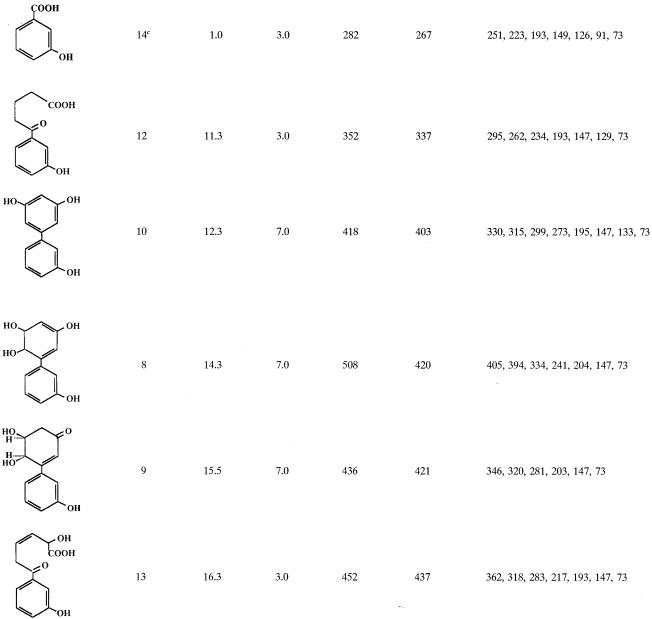
IPTG-induced E. coli cells expressing B-356 BPDO transformed 3,3′-dihydroxybiphenyl essentially into two metabolites. One metabolite, which was produced only in trace amounts, was identified as a trihydroxybiphenyl from the mass spectral features of its TMS derivative (M+ at m/z 418 plus characteristic ions at 403, 330, and 315). The major metabolite, (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one (metabolite 9, Table 2), accounted for more than 99% of the total metabolites generated from 3,3′-dihydroxybiphenyl.
TABLE 2.
Mass spectral features of TMS-derived metabolitesa generated from 3,3′-dihydroxybipheny
Only major metabolites and those that help to elucidate the metabolic pathway of 3,3′-dihydroxybiphenyl are shown.
Retention time relative to the retention time of 3-hydroxybenzoic acid.
Metabolite found only in cell suspensions of P. putida KT2440[pDA1].
(ii) Production of (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one.
Metabolite 9 was also the major metabolite obtained when 3,3′-dihydroxybiphenyl was transformed by resting cell suspensions of biphenyl-induced B-356 or its biphenyl-adapted variant. The latter strain is derepressed for the biphenyl catabolic enzymes; it does not require biphenyl for induction, preventing any cross-contamination of hydroxybiphenyl metabolites resulting from growth on biphenyl. Under optimal conditions, 30 ml of a resting cell suspension adjusted to an OD600 of 1.0 (5 × 109 cells/ml) was able to convert 2.5 mg of 3,3′-dihydroxybiphenyl completely into metabolite 9 within 24 h (rate of 120 μg/h). The optimal conditions for production of metabolite 9 were established as follows. Cells of strain B-356 were grown in MM30 containing biphenyl as the sole growth substrate. Log-phase cells (18-h cultures) were washed and suspended in MM30 without carbon source, to 5 × 109 cells/ml. Thirty milliliters of the cell suspension was transferred into a 125-ml Erlenmeyer flask to which 2.5 mg of 3,3′-dihydroxybiphenyl was added. The flask was incubated at 30°C with rotatory shaking at 250 rpm for 24 h. Under these conditions, metabolite 9 was the only compound extracted with ethyl acetate at neutral pH from the cell suspension. Gas chromatography-mass spectrometry analysis of these extracts showed no trace of 3,3′-dihydroxybiphenyl and no trace of any other metabolite. When metabolite 9 (0.1 to 3 mg in 30 ml) was added to washed-cell suspensions of B-356, adapted variant, and/or P. putida KT2440[pDA1] or KT2440[pDA2], expressing the enzymes of the biphenyl catabolic pathway, no further metabolite was detected in cultures (data not shown). Therefore, this is a dead-end biotransformation product of BPDO from 3,3′-dihydroxybiphenyl.
Identification of (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one.
From X-ray analysis of crystals the structure of metabolite 9 was determined to be (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one, C12H12O4 (32). The mass spectral features of the TMS-derived compound are shown in Table 2. The spectra of its TMS derivative showed prominent peaks at m/z 436 (M+), 421 (M-15), and 320 (M-116, OTMS-CH2-CH). The mass shift by 27 to 463 for the (2H9)TMS derivative is indicative of a trihydroxylated metabolite (data not shown). The spectra of the butylboronate derivative exhibited prominent ions at m/z 358 (M+), which is compatible with the presence of two vicinal hydroxyl groups (data not shown).
The proton H+ NMR spectrum (data not shown) corroborates the structure determined by X-ray crystal analysis. When D2O was the solvent, the 1H NMR at 400 MHz yielded the following data: 7.33 ppm [dd, J = 8 Hz, 8 Hz, 1H, HC(5′)]; 7.18 ppm [d, J = 8 Hz, 1H, HC(4′ or 6′)]; 7.07 ppm [dd, J = 2 Hz, 2 Hz, 1H, HC(2′)]; 6.95 ppm [ddd, J = 0.5 Hz, 2 Hz, 5 Hz, 1H, HC(4′ or 6′)]; 6.31 ppm [s, deshielded, 1H, HC(6)]; 4.87 ppm [d, J = 3.3 Hz, H, HC(4)]; 4.21 ppm [ddd, J = 3.4 Hz, 1.2 Hz, 3.4 Hz, 1H, HC(3)]; 2.75 ppm [dd, J = −16.8 Hz, 10 Hz, 1H, HC(2)]; 2.68 ppm [dd, J = −16.8 Hz, 4.7 Hz, 1H, HC(2)]. There were two coupling systems showing four adjacent protons each, one of which was an aromatic system as indicated by its chemical shift between 7 and 8 ppm.
Other metabolites generated from 3,3′-dihydroxybiphenyl by the biphenyl catabolic pathway of strain B-356.
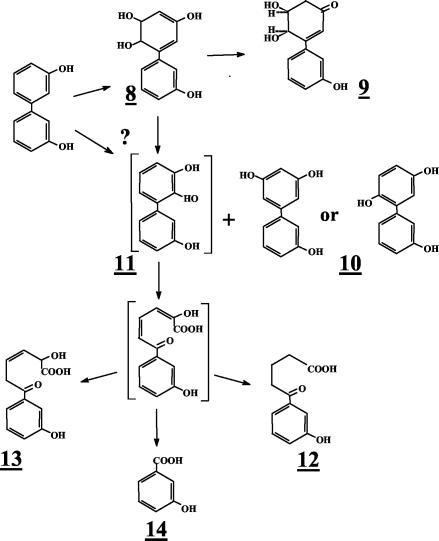
When the concentration of 3,3′-dihydroxybiphenyl was raised over 100 μg/ml of cell suspension of strain B-356 or of its adapted variant, the rate of transformation of substrate into metabolite 9 was lower (ca. 75 μg/h for suspension adjusted to 1010 cells/ml). Furthermore, several other metabolites were generated in the growth medium. The metabolites that were found in 48-h-old resting cell suspensions of B-356 are shown in Table 2 along with the mass spectral features of their TMS derivatives. Typically, approximately 70 to 80% of the substrate was transformed into metabolite 9. Metabolite 10 was the second most abundant in the culture extracts, representing approximately 10 to 15% of the substrate converted. From the mass spectral features of its TMS derivative, metabolite 10 was identified as trihydroxybiphenyl. Butylboronate derivatization of metabolite 10 revealed that none of its hydroxyl groups were adjacent. By deduction, it must be the 3,5,3′-trihydroxybiphenyl or the 3,6,3′-trihydroxybiphenyl that would be generated by elimination of water from 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl. On the other hand, the total absence of 2-(3-hydroxyphenyl)-1,4-benzoquinone strongly suggests that it is 3,5,3′-trihydroxybiphenyl. All other metabolites were present in small amounts (less than 1% of total mass).
A small amount of a dihydrotetrahydroxybiphenyl which is presumed to be 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl (metabolite 8) was found in the extracts of suspensions prepared with the biphenyl-adapted variant of strain B-356. 2,3,3′-Trihydroxybiphenyl (metabolite 11, Fig. 3) was not detected in the extracts; however, the presence of metabolites 12 and 13 in pH 3.0 extracts provides the evidence that the elimination of water from 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl or catalytic oxygenation on carbons 2 and 3 of 3,3′-dihydroxybiphenyl can generate 2,3,3′-trihydroxybiphenyl as well. Metabolite 12 was found in suspensions of strain B-356 whereas metabolite 13 was detected in suspensions of P. putida KT2440[pDA2]. Metabolites 12 and 13 must have been generated by further nonspecific transformation of 2-hydroxy-6-oxo-6-(3-hydroxyphenyl)-hexa-2,4-dienoic acid through pathways described previously for the corresponding chlorinated biphenyls and hydroxybiphenyls (25, 33). The same metabolites were also present in extracts from resting cell suspensions of B-356 and P. putida KT2440[pDA1] and KT2440[pDA2] when 3-hydroxybiphenyl was used as the substrate (data not shown). The absence of 3-hydroxybenzoate (metabolite 14, Table 2) in resting cell suspensions of strain B-356 with 3,3′-dihydroxybiphenyl can be explained by the ability of this strain to use 3-hydroxybenzoate for growth (33). P. putida KT2440[pDA1] is unable to metabolize 3-hydroxybenzoate (33). It expresses the four enzymes required to transform biphenyl into benzoate, but the expression is poor (2). The conversion rate of 3,3′-dihydroxybiphenyl by a cell suspension of P. putida KT2440[pDA1] is 10 times lower than that of strain B-356. In spite of the fact that less than 10% of the total 3,3-dihydroxybiphenyl added was converted into metabolite 9, a small amount of 3-hydroxybenzoate was found in pH 3.0 extracts of 48-h-old resting cell suspensions of P. putida KT2440[pDA1] (data not shown). This observation provides further evidence that, although minor, an alternative metabolic route for the degradation of 3,3′-dihdyroxybiphenyl involves oxygenation on carbons 2 and 3 to ultimately generate 3-hydroxybenzoate that is mineralized by strain B-356.
FIG. 3.
Proposed pathway for production of metabolites generated from 3,3′-dihydroxybiphenyl. Metabolites in brackets were not detected in extracts of resting cell suspensions prepared as described in Materials and Methods. The question mark indicates the uncertainty about the pathway for production of metabolite 11, which could also be generated by oxygenation of the ring on carbon 2 and 3.
Cytotoxicity and genotoxicity of (3S,4R)-3,4-dihydroxy-5-(3′-hydroxyphenyl)-5-cyclohexen-1-one.
Because metabolite 9 is a dead-end metabolite produced in large amounts from 3,3′-dihydroxybiphenyl, we have examined some of its toxic properties. Metabolite 9 did not exhibit any mutagenic properties in the Ames test (24). The assay was done in the presence and absence of Aroclor-induced fraction S-9, with strains TA97A, TA98, TA100, and TA102 of S. enterica serovar Typhimurium. All strains exhibited the expected responses when benzo(a)pyrene, sodium azide, and 2-aminofluorene were used as controls. However, no change in the number of prototrophs recovered per plate was found for any of the plates in which 0.2 to 500 μg of metabolite 9 was incorporated into the top agar layer.
Metabolite 9 was also tested for its toxicity toward the fluorescent bacterium P. phosphoreum in the Microtox assay. Under the conditions described in Materials and Methods, its EC50 was determined as 20 μg/ml compared to an EC50 of 15 μg/ml for phenol. The toxicity of metabolite 9 for cells is thus of the same order as that for phenol.
DISCUSSION
Strain B-356 BPDO was shown to oxygenate 3,3′-dichlorobiphenyl at higher rates than those for 2,2′-dichlorobiphenyl and 4,4′-dichlorobiphenyl (17). The fact that B-356 cells also metabolize 3,3′-dichlorobiphenyl more efficiently than 2,2′-dichlorobiphenyl or 4,4′-dichlorobiphenyl reflects this feature (6). It was thus of interest to verify if strain B-356 exhibits similar substrate preferences towards symmetrical ortho-, meta-, or para-substituted dihydroxybiphenyls.
In this study we have shown that the catabolic pathway of C. testosteroni B-356 can metabolize dihydroxybiphenyls that are symmetrically substituted on both rings. Similar to the reaction involving the symmetrically substituted dichlorobiphenyls (17), strain B-356 BPDO shows a preference for the meta-substituted substrate, 3,3′-dihydroxybiphenyl. However, the enzyme can also oxygenate 2,2′- and 4,4′-dihydroxybiphenyls.
In the case of 2,2′-dihydroxybiphenyl, the major products of oxidation are the same as those generated by Burkholderia sp. strain LB400. BPDO catalytic oxygenation, with IPTG-induced E. coli DH11S expressing B-356 BPDO, generates principally 2,3,2′-trihydroxybiphenyl. It has been proposed that production of 2,3,2′-trihydroxybiphenyl is the result of a catalytic oxygenation on the substituted side of the ring (30). However, production of 2,3,2′-trihydroxybiphenyl from 5,6-dihydro-2,5,6,2′-tetrahydroxybiphenyl by spontaneous elimination of water resulting in loss of a hydroxyl group in position 2 is an alternative pathway that cannot be excluded.
Production of trace amounts of tetrahydroxybiphenyl (metabolite 2) and 2,5-dihydroxy-6-oxo-6-(2-hydroxyphenyl)-hexa-2,4-dienoic acid (metabolite 6) also provides evidence for oxygenation of at least a portion of the substrate on carbons 5 and 6 of the phenyl ring. The presence of metabolite 5 (2,5,2′-trihydroxybiphenyl) shows that under our experimental conditions some of the 5,6-dihydro-2,5,6,2′-tetrahydroxybiphenyl spontaneously eliminates water, reconstituting the aromatic ring, to produce this minor metabolite. In our experiments metabolite 5 was extracted at pH 7. The instability of dihydrodiols under acidic conditions or at high temperatures has been well documented (5, 20, 41). Furthermore, some carboxycyclic cis-dihydrodiol metabolites of heterocyclic arene such as those generated from dioxygenation of benzofuran [5,6-(OH)2], benzothiophene [4,5-(OH)2], and isoquinoline [5,6-(OH)2] have been reported to aromatize spontaneously at ambient temperature at neutral pH (5). However, traditionally it is believed that cis-dihydrodiols derived from dioxygenation of aromatic compounds are stable under physiological conditions. Therefore, data suggest that the type and position of substitutes present on the biphenyl ring influence the stability of the dihydrodiol metabolites produced from dioxygenation of biphenyl analogs. This observation is clearly demonstrated from analysis of dihydrodiol metabolites produced from 3,3′-dihydroxybiphenyl.
In the case of 3,3′-dihydroxybiphenyl, data showed that, among the three dihydroxybiphenyls symmetrically substituted on both rings, it is by far the most preferred substrate for B-356 BPDO. Although 3,3′-dihydroxybiphenyl is rapidly oxygenated by B-356 BPDO, this compound cannot be efficiently mineralized by the biphenyl catabolic pathway of strain B-356. The dihydrodiol produced from 3,3′-dihydroxybiphenyl is readily tautomerized to a more stable form (metabolite 9) which is not metabolized further by the biphenyl catabolic pathway. However, a fraction of the 3,3′-dihydroxybiphenyl substrate is converted to the trihydroxylated derivative by spontaneous elimination of water to generate metabolite 10 (Fig. 3). At this time, it is not clear whether the water elimination reaction is also responsible for the production of 2,3,3′-trihydroxybiphenyl or whether it is produced directly through an oxygenation reaction on carbons 2 and 3 of 3,3′-dihydroxybiphenyl. The fact is that resting cell suspensions of strain B-356 or of P. putida expressing the biphenyl catabolic enzymes of strain B-356 are producing metabolites 12, 13, and 14 (Fig. 3), which could occur only if 2,3,3′-trihydroxybiphenyl was produced.
The tautomerization reaction observed for 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl and the reactions involving water elimination under physiological conditions for 5,6-dihydro-2,5,6,2′-tetrahydroxybiphenyl and 5,6-dihydro-3,5,6,3′-tetrahydroxybiphenyl have significant consequences, as they lead to rapid accumulation of dead-end recalcitrant metabolites such as metabolite 9 with little information on its biological activities. Data show that the position and type of substitute on the biphenyl ring can strongly influence the stability of the cis-dihydrodiol metabolites produced by dioxygenation of biphenyl analogs. The dihydrodiol metabolite resulting from catalytic dioxygenation of 2,2′-dichlorobiphenyl has been detected in the reaction medium (14). The same is true for the dihydrodiol metabolite derived from 3,3′-dichlorobiphenyl (data not shown). However, when the rings are symmetrically substituted on the meta carbon by hydroxyl groups, the dihydrodiol metabolite is unstable. Thus, in spite of the fact that strain B-356 grows on 3-hydroxybenzoate and that B-356 BPDO catalyzes the oxygenation of 3,3′-dihydroxybiphenyl rather efficiently, this strain is unable to grow on it and to mineralize this compound. This observation could have significant consequences for biological processes involved in the degradation of substituted biphenyl analogs and deserves further examination.
Acknowledgments
This work was supported in part by NSERC grant OGP0039579.
REFERENCES
- 1.Abramowicz, D. A. 1994. Aerobic PCB biodegradation and anaerobic PCB dechlorination in the environment. Res. Microbiol. 145:42-46. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, D., M. Sylvestre, and M. Sondossi. 1991. Subcloning of bph genes from Pseudomonas testosteroni B-356 in Pseudomonas putida and Escherichia coli: evidence for dehalogenation during initial attack on chlorobiphenyls. Appl. Environ. Microbiol. 57:2880-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad, D., R. Massé, and M. Sylvestre. 1990. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene 86:53-61. [DOI] [PubMed] [Google Scholar]
- 4.Asturias, J. A., and K. N. Timmis. 1993. Three different 2,3-dihydroxybiphenyl-1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J. Bacteriol. 175:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, S. A., N. Bowers, D. R. Boyd, N. D. Sharma, L. Hamilton, R. Austin, S. McMordie, and H. Dalton. 1998. The potential role of cis-dihydrodiol intermediates in bacterial aromatic hydroxylation and the NIH Shift. J. Chem. Soc. Perkin Trans. 1:3443-3451. [Google Scholar]
- 6.Barriault, D., C. Pelletier, Y. Hurtubise, and M. Sylvestre. 1997. Substrate selectivity pattern of Comamonas testosteroni strain B-356 towards dichlorobiphenyls. Int. Biodeterior. Biodegrad. 39:311-316. [Google Scholar]
- 7.Barriault, D., M. M. Plante, and M. Sylvestre. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broadus, R. M., and J. D. Haddock. 1998. Purification and characterization of the NADH:ferredoxin BPH oxidoreductase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. Arch. Microbiol. 170:106-112. [DOI] [PubMed] [Google Scholar]
- 9.Chroma, L., M. Moeder, P. Kucerova, T. Macek, and M. Mackova. 2003. Plant enzymes in metabolism of polychlorinated biphenyls. Fresenius Environ. Bull. 12:291-295. [Google Scholar]
- 10.Chroma, L., M. Mackova, P. Kucerova, C. in der Wiesche, J. Burkhard, and T. Macek. 2002. Enzymes in plant metabolism of PCBs and PAHs. Acta Biotechnol. 22:34-41. [Google Scholar]
- 11.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortnagel, P., H. Harms, R.-M. Wittich, S. Krohn, H. Meyer, V. Sinwell, H. Wikess, and W. Francke. 1990. Metabolism of dibenzofuran by Pseudomonas sp. strain HH69 and mixed culture HH27. Appl. Environ. Microbiol. 56:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddock, J. D., and D. T. Gibson. 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddock, J. D., J. R. Horton, and D. T. Gibson. 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddock, J. D., D. A. Pelletier, and D. T. Gibson. 1997. Purification and properties of ferredoxin(BPH), a component of biphenyl 2,3-dioxygenase of Pseudomonas sp. strain LB400. J. Ind. Microbiol. Biotechnol. 19:355-359. [DOI] [PubMed] [Google Scholar]
- 16.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1996. Characterization of active recombinant his-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J. Biol. Chem. 271:8152-8156. [DOI] [PubMed] [Google Scholar]
- 17.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1998. Involvement of the terminal oxygenase β subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J. Bacteriol. 180:5828-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtubise, Y., D. Barriault, J. Powlowski, and M. Sylvestre. 1995. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J. Bacteriol. 177:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imbeault, N. Y., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 20.Joern, J. M., T. Sakamoto, A. A. Arisawa, and F. H. Arnold. 2001. A versatile high throughput screen for dioxygenase activity using solid-phase digital imaging. J. Biomol. Screen. 6:219-223. [DOI] [PubMed] [Google Scholar]
- 21.Köller, G., M. Moeder, and K. Czihal. 2000. Peroxidative degradation of selected PCB: a mechanistic study. Chemosphere 41:1827-1834. [DOI] [PubMed] [Google Scholar]
- 22.Kucerova, P., C. in der Wiesche, M. Wolter, T. Macek, F. Zadrazil, and M. Mackova. 2001. The ability of different plant species to remove polycyclic aromatic hydrocarbons and polychlorinated biphenyls from incubation media. Biotechnol. Lett. 23:1355-1359. [Google Scholar]
- 23.Lee, I., and J. S. Fletcher. 1992. Involvement of mixed function oxidase systems in polychlorinated biphenyl metabolism by plant cells. Plant Cell Rep. 11:97-100. [DOI] [PubMed] [Google Scholar]
- 24.Maron, D. M., and B. N. Ames. 1983. Revised methods for the salmonella mutagenicity test. Mutat. Res. 113:173-215. [DOI] [PubMed] [Google Scholar]
- 25.Massé, R., F. Messier, C. Ayotte, M.-F. Lévesque, and M. Sylvestre. 1989. A comprehensive gas chromatographic/mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed. Environ. Mass Spectrom. 18:27-47. [Google Scholar]
- 26.Nojiri, H., and T. Omori. 2002. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci. Biotechnol. Biochem. 66:2001-2016. [DOI] [PubMed] [Google Scholar]
- 27.Prindle, R. F. 1983. Phenolic compounds, p. 197-224. In S. S. Block (ed.), Disinfection, sterilization and preservation. Lea and Febiger, Philadelphia, Pa.
- 28.Puri, R. K., Q. P. Ye, S. Kapila, W. R. Lower, and V. Puri. 1997. Plant uptake and metabolism of polychlorinated biphenyls (PCBs), p. 481-513. In W. Wang, J. W. Gorsuch, and J. S. Hughes (ed.), Plants for environmental studies. CRC Press, Inc., Boca Raton, Fla.
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Seeger, M., B. Camara, and B. Hofer. 2001. Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase. J. Bacteriol. 183:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Degradation of chlorobiphenyls catalysed by the bph-encoded biphenyl-2,3-dioxygenase and biphenyl-2,3-dihydrodiol-2,3-dehydrogenase of Pseudomonas sp. LB400. FEMS Microbiol. Lett. 133:259-264. [DOI] [PubMed] [Google Scholar]
- 32.Sondossi, M., B. A. Lloyd, D. Barriault, M. Sylvestre, and M. Simard. 1995. Microbial transformation of a dihydroxybiphenyl. Acta Crystallogr. Sect. C 51:491-494. [Google Scholar]
- 33.Sondossi, M., M. Sylvestre, D. Ahmad, and D. Barriault. 1993. Importance of stringent control of biphenyl-induced biphenyl and chlorobenzoate catabolic pathways for chlorobiphenyl degradation in enhanced strain of Pseudomonas testosteroni B-356, p. 456-461. In A. Bousher, M. Chandra, and R. Edivean (ed.). Biodeterioration and biodegradation 9. Proceedings of the 9th International Symposium of the International Biodeterioration Association. The Institution of Chemical Engineers, Rugby, Warwickshire, United Kingdom.
- 34.Sondossi, M., M. Sylvestre, D. Ahmad, and R. Massé. 1991. Metabolism of hydroxybiphenyl and chloro-hydroxybiphenyl by biphenyl/chlorobiphenyl degrading Pseudomonas testosteroni, strain B-356. J. Ind. Microbiol. 7:77-88. [Google Scholar]
- 35.Suenaga, H., M. Goto, and K. Furukawa. 2001. Emergence of multifunctional oxygenase activities by random priming recombination. J. Biol. Chem. 276:22500-22506. [DOI] [PubMed] [Google Scholar]
- 36.Suenaga, H., T. Watanabe, M. Sato, Ngadiman, and K. Furukawa. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suske, W. A., W. J. H. vanBerkel, and H. P. E. Kohler. 1999. Catalytic mechanism of 2-hydroxybiphenyl 3-monooxygenase, a flavoprotein from Pseudomonas azelaica HBP1. J. Biol. Chem. 274:33355-33365. [DOI] [PubMed] [Google Scholar]
- 38.Sylvestre, M., and J. Fauteux. 1982. A new facultative anaerobe capable of growth on chlorobiphenyls. J. Gen. Appl. Microbiol. 28:61-72. [Google Scholar]
- 39.Sylvestre, M., M. Sirois, Y. Hurtubise, J. Bergeron, D. Ahmad, F. Shareck, D. Barriault, I. Guillemette, and J. M. Juteau. 1996. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene 174:195-202. [DOI] [PubMed] [Google Scholar]
- 40.Trotz, S. I., and J. J. Pitts. 1984. Industrial antimicrobial agents, p. 223-253. In D. Eckroth (ed.), Kirk-Othmer encyclopedia of chemical technology, 3rd ed., vol. 13. John Wiley & Sons, Inc., New York, N.Y.
- 41.Wackett, L. P., and D. T. Gibson. 1983. Rapid method for the detection and quantification of hydroxylated aromatic intermediates produced by microorganisms. Appl. Environ. Microbiol. 45:1144-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]