Abstract
Most lymphoid malignancies are initiated by specific chromosomal translocations between immunoglobulin (Ig)/T cell receptor (TCR) gene segments and cellular proto-oncogenes. In many cases, illegitimate V(D)J recombination has been proposed to be involved in the translocation process, but this has never been functionally established. Using extra-chromosomal recombination assays, we determined the ability of several proto-oncogenes to target V(D)J recombination, and assessed the impact of their recombinogenic potential on translocation rates in vivo. Our data support the involvement of 2 distinct mechanisms: translocations involving LMO2, TAL2, and TAL1 in T cell acute lymphoblastic leukemia (T-ALL), are compatible with illegitimate V(D)J recombination between a TCR locus and a proto-oncogene locus bearing a fortuitous but functional recombination site (type 1); in contrast, translocations involving BCL1 and BCL2 in B cell non-Hodgkin's lymphomas (B-NHL), are compatible with a process in which only the IgH locus breaks are mediated by V(D)J recombination (type 2). Most importantly, we show that the t(11;14)(p13;q32) translocation involving LMO2 is present at strikingly high frequency in normal human thymus, and that the recombinogenic potential conferred by the LMO2 cryptic site is directly predictive of the in vivo level of translocation at that locus. These findings provide new insights into the regulation forces acting upon genomic instability in B and T cell tumorigenesis.
Keywords: LMO2, TAL2, TAL1, BCL2 mbr, BCL1 mtc
Introduction
During B cell differentiation in the bone marrow, and T cell differentiation in the thymus, V(D)J recombination generates somatic assembly of the various noncontiguous V, D, and J gene segments, to form a complete V(D)J exon encoding the variable region of the B and T cell receptors (1). The recombination process is directed by the recombination signal sequence (RSS),* which flank each receptor gene segment. The RSSs allow the recruitment, binding and proper positioning of the recombination activating gene (RAG)-1/2 proteins, which will perform the initial cut at the border of the gene segment and the RSS. This process generates two pairs of morphologically distinct intermediate DNA ends: two blunt RSSs or signal ends and two covalently sealed hairpin coding ends. The two signal ends are generally joined together without further modification to form the signal joint (SJ). In contrast, coding end hairpins are subsequently nicked and extensively modified before ligation. Generation of a processed coding joint (CJ) containing nucleotide deletion and additions, and a flush SJ constitute the hallmark of V(D)J recombination.
Normally, joining takes place only between the V, D, and J gene segments of the Ig or TCR loci. Nevertheless, the fact that the RSS is the necessary and sufficient cis-element for site-specific recognition and recombination paradoxically presents a considerable threat to genomic stability (2). Indeed, fortuitous DNA sequences in the genome resembling an authentic RSS can potentially be targeted by mistake, and lead to illegitimate recombination events such as chromosomal deletion, inversion, or translocations. When such RSSs are located in or near cellular proto-oncogenes, pathologic deregulation can ensue (3, 4). Specific chromosomal translocations between Ig/TCR antigen receptor genes and cellular proto-oncogenes are a central feature of lymphoid neoplasms, and in many cases, illegitimate V(D)J recombination has been proposed to be involved in the process. However, the actual mechanism by which cells undergo illegitimate recombination, and the role of the normal V(D)J recombination in the process are still poorly understood. This is due to the fact that to date, knowledge of chromosomal translocation mechanisms is mainly derived from the posttranslocation observation of breakpoints from the derivative chromosomes. In absence of functional systems, the main criteria which have been traditionally used to define a V(D)J-mediated translocation are therefore based on the similarity of the translocation breakpoints to the products of V(D)J recombination: (a) involvement of an Ig/TCR segment as one of the translocation partners; (b) identification of a RSS-like motif near the germline proto-oncogene sequence; (c) recurrent breaks at the border of the RSS motifs on both immune and nonimmune loci; (d) presence of nucleotide additions and deletions at one of the breakpoints; (e) generation of a SJ at the other breakpoint. However, for various reasons, some due to limitations of study in humans, some inherent to the mechanism of the translocation, fulfillment of all criteria has not always been manageable in vivo. In particular, identification of a RSS-like motif based on the primary sequence can be misleading. RSSs normally consist of highly conserved heptamer and nonamer motifs separated by a spacer sequence of 12 or 23 bp (Table I) . Nevertheless, most RSSs flanking V, D, and J segments contain some degree of polymorphism in their sequence, and this has been shown to be a major factor affecting the relative representation of gene segments in the primary repertoire (5). Depending on the position, alterations in the heptamer, nonamer, or spacer sequences (e.g. Tables I–III) can be extremely deleterious for recombination (6–10). Furthermore, multiple variations from consensus can result in synergistic effects (11), and the many combinations of heptamer, nonamer, and spacer variants have obviously not all been analyzed. Thus, despite abundant in vivo and in vitro studies, apart from the mandatory presence of the heptamer’s 5′CAC3′ (2), there is to date no precise rule allowing the prediction of the recombinogenic potential of a RSS variant based solely on its sequence. This is especially true for fortuitous RSSs bearing poor resemblance to consensus sequences, as often the case in proto-oncogene loci (Table II). To assess if such RSSs are truly involved in V(D)J recombination, they have therefore to be tested in functional assays (12).
Table I. Authentic RSSs Flanking V(D)J Gene Segments.
| RSS | Heptamer | Spacer | Nonamer |
|---|---|---|---|
| Consensus | CACAGTG | ACAAAAACC | |
| VκA2b | CACAGAG | 12 | ACAGAAACC |
| VκA27 | CACAGTG | 12 | ACAAAAACC |
| Jκl | CACAGTG a | 23 | ACAAAAACC |
| JH6 b | CACAATG a | 22 | ACAAAAACC |
| Jβ2-7 | CACGGAG a | 12 | ATGCAAACC |
| Jδl | CACAGCA a | 12 | CCAAAAACC |
| DH3.10 | CACAGTG | 12 | TCAAAAACC |
| Dδ2 | CACAGTG | 23 | ACAAAAACT |
| Dβ1 | CACAATG | 23 | ACAAAAACC |
Mismatches from consensus are underlined.
Sequence shown in reverse complement orientation.
Indicates changes from the wt sequence in the constructs (see Materials and Methods).
Table III.
Fortuitous RSS in the Core Plasmid
| RSS | Heptamer | Spacer | Nonamer |
|---|---|---|---|
| Consensus | CACAGTG | ACAAAAACC | |
| ψ150 | CACATTAa | 12 | ATTAATTGT |
| ψ200 | CACAGGA | 12 | GACATTGAG |
| ψ250 | CACAACA | 12 | ATCGGCAGG |
| ψ1200 | CACAGTC | 12 | CCTGGGTCG |
| ψ3490 | CACAGCG | 12 | GTAAGTATC |
| ψ6131 | CACAACAa | 12 | GCATAAAGT |
Only the fortuitous RSSs which underwent recombination are represented.
Sequence shown in reverse complement orientation.
Table II. Fortuitous RSSs at the Vicinity of Proto-oncogene Breakpoints.
| Proto-oncogene | Fortuitous RSS | ||
|---|---|---|---|
| Heptamer | Spacer | Nonamer | |
| Consensus | CACAGTG | ACAAAAACC | |
| TAL2 (9q32) | CACTGTGa | 13 | ATAAAAATA |
| LMO2 (11p13) | CACAGTA a | 12 | GCAATAATT |
| TAL1 (1p34) | CACACCG | 22 | CGAAAAAGG |
| BCL1 mtc (11q13) | CACATCG | 12 | TTGCGTGGA |
| CACTGCA | 12 | CTGTGATTA | |
| CACCTGG | 12 | CGTGAACGA | |
| CACATCC | 12 | CAGGACCTG | |
| CACGCCA a | 23 | AGCTCTTGC | |
| CACGAAGa | 23 | ACGCAGATA | |
| CACGCAGa | 23 | TGTTTCAGA | |
| CACTTTT a | 23 | CGAATATGC | |
| CACAGTC a | 23 | TTATGCTCC | |
| CACGCAA a | 23 | GCAGCCTTA | |
| CACACCGa | 23 | AATGACCAA | |
| BCL mbr (18q21) | CACAGAC | 12 | CTCCTGCCC |
| CACCCAG | 12 | CTCCTTCCG | |
| CACCAAG | 12 | CTGTGGTAT | |
| CACAGGA a | 23 | TTGACAATG | |
| CACTTTGa | 23 | AATATTTTG | |
| CACAGGT a | 23 | AACGACCAC | |
| CACTGCA a | 23 | GCCATGAGA | |
| CACGTAA a | 23 | ACCATAGAT | |
| CACCATA a | 23 | CCATCTGGA | |
Maximum matches of the nonamer are shown with an appropriate 12 or 23-RSS ± 1 bp.
Sequence shown in reverse complement orientation.
In this report, we used an extra-chromosomal V(D)J recombination assay to directly and functionally test which of several proto-oncogene sequences is able to target V(D)J recombination. The breakpoint regions and flanking genomic sequences of five typical translocations presumably resulting from V(D)J recombination mis-targeting, were therefore assayed for recombination: BCL2/IgH fusion in t(14;18)(q32;q21) (13–15), which is the most frequent translocation in B cell non-Hodgkin’s lymphomas (NHL), and the hallmark of follicular lymphomas (FLs); BCL1/IgH fusion in t(11;14)(q13;q32) (16), also present in a substantial fraction of mature B-NHL; TCRβ/TAL2 fusion in t(7;9)(q34;q32) (17), LMO2/ TCRδ fusion in t(11;14)(p13;q11) (18–21), and TAL1/ TCRδ fusion in t(1;14)(p34;q11) (22–25), all recurrently involved in the development of pediatric T cell acute lymphoblastic leukemia (T-ALL; reference 26). Our results clearly show that fortuitous sites in the LMO2 and TAL2 proto-oncogenes can target V(D)J recombination at a frequency comparable to physiological RSSs. Similarly, the region of the TAL1 proto-oncogene involved in most translocation breakpoints also contains a functional recombination site, although triggering V(D)J recombination at a much lower frequency. In contrast, fortuitous sites present in the BCL2 major breakpoint region (mbr) and BCL1 major translocation cluster (mtc), are not able to initiate V(D)J recombination at relative frequencies comparable to the ones corresponding to the breaks observed in vivo. Thus, our results support the presence of at least two distinct mechanisms by which V(D)J recombination leads to translocation in lymphoid neoplasms: in some translocations, illegitimate V(D)J recombination would occur between a legitimate Ig or TCR locus and an illegitimate proto-oncogene locus bearing a fortuitous, but functional RSS; in a second category, however, only the breaks at the immune locus would be mediated by V(D)J recombination. Breaks at the nonimmune locus bearing the proto-oncogene, would be initiated by other (yet unknown) mechanisms, and would subsequently invade the V(D)J synaptic complex during the rearrangement process. Most importantly, our precise quantification of the recombinogenic potentials of the TAL2 and LMO2 proto-oncogene breakpoint regions show that mis-targeting by the V(D)J recombination can constitute a considerable risk of genomic instability. Indeed, we report that the t(11;14)(p13;q11) involving Dδ2/LMO2 recombination can be detected at a surprisingly high frequency in thymus of healthy individuals, and that the recombinogenic potential of LMO2 is directly predictive of the actual in vivo rate of translocation.
Materials and Methods
Recombination Substrates.
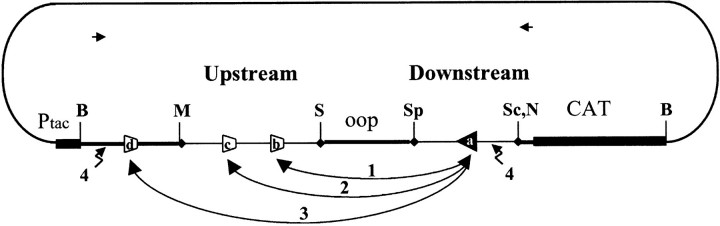
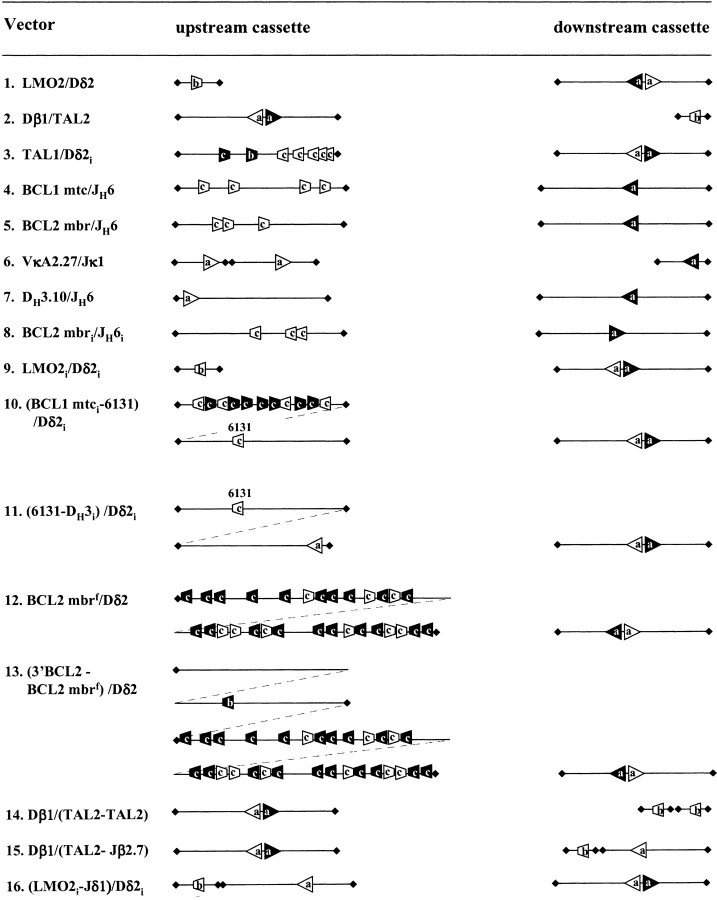
The organization of our recombination plasmids was described previously (27; Fig. 1). Briefly, the two sequences to be tested for VDJ recombination are separated by a termination signal (oop). The Ptac promoter will transcribe the chloramphenicol acetyltransferase (CAT) gene when transformed in E. coli only when the termination signal was previously deleted by recombination in eukaryotic cells. All constructs used in this study were derived from Comp21 (10). Each sequence tested for recombination is flanked by unique restriction sites and forms therefore a cassette which can be easily exchanged (upstream cassette: Mlu I/Sal I, downstream cassette: SpeI/Sac II or Not I; Fig. 1). Cassettes were PCR-amplified from human genomic DNA (primers are listed in Table IV). Inverted cassettes were cloned blunt except for LMO2i and Dδ2i (Table IV). The JH6 heptamer was modified to a perfect consensus by PCR site-directed mutagenesis. The 6131 cassette is a ~400 bp lacZ fragment amplified from the pCR2.1 vector (Invitrogen) after SacI/ NsiI deletion of the MCS, and allows white/blue screening. Cassettes in all constructs were confirmed by sequencing and are illustrated in Fig. 2.
Figure 1.
Recombination substrate (not to scale). The upstream cassette is comprised between M (Mlu1) and S (Sal1). The downstream cassette is comprised between Sp (Spe1) and Sc (Sac2) or N (Not1). The authentic RSS is shown as a triangle (a). Fortuitous RSSs are represented as incomplete triangles: b, fortuitous RSS observed in proto-oncogene translocation breakpoints, c, other potential fortuitous RSS in proto-oncogene flanking sequence, d, other fortuitous RSS in core plasmid sequence. Pathways 1–3: V(D)J recombination; Pathway 4: BR. Horizontal arrows indicate the location of the PCR screen primers.
Table IV.
Primers
| Cassette | Site | Sequence (5′–3′) |
|---|---|---|
| LMO2 | SalI | GGCCGTCGACATCCGTGCACCGAAATTATTGCTGGGTAAGACAATACTGTG |
| MluI | GTCCACGCGTAGGAAAGAGCTTTCCGAAGTTCCAAGGCTATGTAACACACACAGTATTG | |
| Dδ2 | SpeI | CCATACTAGTCACAAACCCCAAGGCAG |
| NotI | CCTCCGCGGCCGCAAAGCAGGGAGGGAAG | |
| Dβ1 | MluI | CTCAACGCGTCCTGAGGACAGTGCCTG |
| SalI | TGTCGTCGACCACAGTCTTGGTCTG | |
| TAL2 | SpeI | CCGGACTAGTGTTTTATTTTTATATCTCCAGTAAGTCACAGTGTGAC |
| NotI | GACCGCGGCCGCTTTATCGAGCAACCACAGTGCAAGTCACACTGTGAC | |
| TAL1 | MluI | AGGAACGCGTCCAAACACCTGCAG |
| SalI | CTTCGTCGACACCGTTTCCACCG | |
| Dδ2i | NotI | CCATAGCGGCCGCCACAAACCCCAAGGCAG |
| SpeI | TACAACTAGTGGAAAGCAGGGAGGGAAG | |
| BCL1 mtc | MluI | CTACACGCGTACTTGTGGGTTG |
| SalI | CAGTGTCGACCAGTGCCCCAG | |
| BCL2 mbr | SpeI | TGAGACTAGTTCAGTTAAAAATCCAG |
| NotI | GCAAGCGGCCGCCATTAAAATG | |
| JH6 | SpeI | TATGTCGACCACTAGTGGTCTGGCTTCTGAGGGGTCAGG |
| NotI | TCCTCGCGGCCGCCCAGTGCCGTCCCCTCTG | |
| DH3.10 | MluI | GTGTCACGCGTGTATTACTATGGTTCGGGGAG |
| LMO2i | SalI | CCATGTCGACTGGCGTTGGGAGGGCAG |
| MluI | GGCCACGCGTATCCGTGCACCGAAATTATTGCTGGGTAATACAATACTGTG | |
| SalI | GTCCGTCGACAGGAAAGAGCTTTCCTAAGTTCCAATGCTATGTAACACACACAGTATTG | |
| 6131 | SalI | CGACTGGTCGACGGGCAGTGAGCG |
| XhoI | GATCCTCGAGCCCGCCGCGCTTAATG | |
| BCL2 mbrf | MluI | TTTTACGCGTCAGGTGTGGAATATGGGGGTTATCTG |
| SalI | TTTTGTCGACGTACAGTTCTGGGGCCAAGAGG | |
| 3′BCL2 | SpeI | TTTTACTAGTGACCGTCATACATGGG |
| SacII | AAATCCGCGGTTCTTAGTATGAGGTTG | |
| Jβ2.7 | NotI | TTTCGCGGCCGCCAGCAGGCTGACCGTGCTGG |
| SacII | CTGACCGCGGACACCCAGCTCCTCCAG | |
| Jδ1 | XhoI | GGTCCTCGAGACTCCTCAGACAACAG |
| SalI | TTATGTCGACCTCCTTAGATGGGAGGATG |
Figure 2.
Schematic representation of the recombination cassettes (to scale). Upstream and downstream cassettes are represented with their authentic and fortuitous RSSs (see legend to Fig. 1). RSSs with 12 bp spacers are white, those with 23 bp spacers are black. For clarity, fortuitous RSSs in cassettes containing authentic RSSs, or fortuitous RSSs without an authentic RSS matching partner are not represented. Cassettes are delimited by lozenges.
Transfection and Harvesting.
18.8 Abelson murine leukemia virus transformed pre-B cells (2 × 107) were transiently transfected with 20 μg of QIAGEN-purified plasmid (QIAGEN) by electroporation with a twin pulse (750 V/25 μF, 128 V/1,500 μF) using an EasyJect device (EquiBio). After 10 min on ice the cells were resuspended in 10 ml of RPMI 1640 supplemented with 10% FCS, 2 mM glutamine, and 1 mM caffeine. After 48 h, plasmids were recovered from the transfected cells by alkaline lysis, and phenol/chloroform extracted. DpnI-digested plasmids were then introduced into chemically competent TOP 10 E. coli (Invitrogen), and plated on ampicillin (50 μg/ml) and chloramphenicol (5 μg/ml). For the lacZ-containing substrates, X-Gal was added to the plates.
Screening of Recombinants.
Clones were screened by PCR using primers located upstream and downstream of the recombination cassettes (CAACTTCTGGTCCGGTAACGTGCTG and CGATGCCATTGGGATATATCAACGGTGG; Fig. 1), followed by direct sequencing as described previously (28). PCR screens were performed from the colonies by resuspending them directly in the PCR mixture, and amplifying for 30 cycles (30 s at 94°C, 30 s at 60°C, and 1 min at 72°C). Colonies negative in the PCR screen were miniprepped and analyzed by BamHI digest. Only minipreps showing at least one BamHI cut were analyzed further. The (6131-DH3i)/Dδ2i construct allowed a first assessment of the recombination site based on blue/white screening. This estimation was confirmed by sequencing 10 blue colonies and all white colonies for each transfection.
DNA Samples.
Thymus samples were obtained from otherwise healthy children undergoing cardiac surgery. This study was approved by the local ethics committee. In all cases informed consent was obtained from the parents. Tissue samples were frozen on dry ice within 1 h from excision. High molecular weight DNA was prepared according to standard procedures. Quality of the DNA preparation was controlled by PCR amplification of the germline LMO2 breakpoint region (CCGTGCACCGA-AATTATTGCTGGGTAA; GTACCCACTTTGCAGGGTT-GTTGAGTGG, same conditions as calibration below) on serial dilutions of thymus DNA (100 ng, 10 ng, 1 ng, 0.1 ng). All preparations gave similar calibration curves.
Thymus PCR Assays.
The “multiple tube” method has been described previously (29). A two-step nested PCR protocol was performed on 5 μg DNA with appropriate translocus Dδ2/LMO2 (primary: TCCCTGGTCCAGTCAACTTCCTG, GAG-ATGATCAGATCCGTGCACCG, secondary: GCTGTGTT-TGTCTCCTGAGGCATGG, CCGTGCACCGAAATTATT-GCTGGGTAA), and Dδ2/Jβ2.7 (primary: TCCCTGGTCC-AGTCAACTTCCTG, GGCGGGATTCAGGTGGAAGG, secondary: GCTGTGTTTGTCTCCTGAGGCATGG, GAG-CTCGGGGAGCCTTAGAGG) primer pairs. PCR conditions used were as follows: 32 cycles (30 s at 94°C, 30 s at 64°C, and 1 min at 72°C). PCR products were cloned in TA (Invitrogen) and analyzed by sequencing. PCR conditions used for the calibration assay were identical except the number of cycles (20). Serial dilutions of the calibration plasmids (100 pg, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg) were performed in 1 μg DNA from the human lymphoma cell line RL7.
Online Supplemental Material.
Sequence libraries of the junctions. Upstream and downstream sequences of the given recombination are indicated. For recombinants using V(D)J recombination, numbers in parenthesis indicate the status of the coding end processing (0 = precise, –n = deletion, p + n = P nucleotide). P nucleotides are shown in italics. N nucleotides are represented in bold characters. Nucleotides in parenthesis could belong to either upstream or downstream sequence or both using homology-directed recombination. For recombinants using break/repair, the location of the break is indicated. Numbers indicate the location in the core plasmid. Breaks in the cassettes are designed by the name of the cassette. If a break occurred in a cassette containing an authentic RSS (a) or a specific proto-oncogene RSS (b), the location of the break relative to this RSS is indicated (3′ or 5′). Junctions are ordered according to break location (5′ to 3′) in the upstream sequence. The online supplemental sequences are available at http://www.jem.org/cgi/content/full/195/1/85/DC1.
Results
Assessment of V(D)J Targeting of Proto-oncogene Sequences by the Extra-chromosomal Recombination Assay.
To functionally test several proto-oncogene sequences for their ability to target V(D)J recombination, we adapted the extra-chromosomal recombination assay (see general principle in Materials and Methods). This assay typically tests V(D)J recombination between two authentic RSSs derived from immune gene segments, which generally recombine at high frequency. In this case, most if not all recombination events obtained correspond to V(D)J recombination between these two sites. The substitution of one of the two authentic RSSs by a “fortuitous” site containing numerous mismatches from consensus (also called “cryptic” site), results in a decrease in the overall recombination frequency, and in the apparition of other low level recombination events (2). In our assay, we distinguish three distinct recombination events: (a) V(D)J recombination at the “specific” sites, i.e., between the authentic RSS and the fortuitous RSS we wish to assay (illustrated by pathway 1 in Fig. 1); (b) V(D)J recombination at “nonspecific” sites, i.e., between the authentic RSS and other fortuitous RSSs. Such fortuitous RSSs can be located either in the genomic sequence flanking the “specific” RSS (pathway 2 in Fig. 1), or in the plasmid core sequence (pathway 3 in Fig. 1); (c) Unspecific break/repair (BR) recombination events, which are defined here as not mediated by V(D)J recombination, and which constitute the background of the assay (e.g. pathway 4 in Fig. 1, no RSS involved). The potential for V(D)J targeting of the specific RSS tested is therefore assessed relatively to the other 2 “competing” recombination events. Fairly good RSSs (recombining in the range of authentic RSSs) are expected to constitute the vast majority of the readout. In contrast, increasingly poor RSSs should appear at decreasing relative rates in the total readout.
Five representative proto-oncogene breakpoint regions which have been proposed to target V(D)J recombination through fortuitous RSSs were assayed: TAL2, LMO2, TAL1, BCL1 mtc, and BCL2 mbr. For TAL2, LMO2, and TAL1, one particular fortuitous RSS has been observed in vivo in most of the breakpoints characterized so far and proposed to be responsible for V(D)J recombination targeting (Table II). The genomic regions containing the RSSs were therefore PCR-amplified and cloned in recombination substrates (Fig. 2, lines 1–3); at the BCL1 mtc and the BCL2 mbr, however, most breaks observed in vivo cluster in a region of ~400 bp (14, 30) containing several potential cryptic RSSs (Table II; references 31 and 32). In this case, the whole ~400 bp mbr and mtc regions were PCR-amplified and cloned in the recombination substrates (Fig. 2, lines 4 and 5). In the corresponding translocations observed in vivo, the proto-oncogene sequences are all fused to authentic Ig or TCR coding sequences. Furthermore, they preferentially use a specific recombination partner. Such “natural” RSS partners were therefore used as the matching recombination partners in the recombination substrates. Thus, five basic constructs were at first generated and assayed: Dβ1/TAL2, LMO2/Dδ2, TAL1/Dδ2i, BCL1 mtc/JH6, and BCL2 mbr/JH6. As reference, two constructs containing consensus or near-consensus RSSs derived from immune gene segments were also assayed (VκA2.27/Jκ1, and DH3–10/JH6; Fig. 2, lines 6 and 7).
Fortuitous RSSs in TAL2, LMO2, and TAL1 Proto-oncogene Breakpoint Regions Are Able to Target V(D)J Recombination.
Results in Table V show that the control plasmids (VκA2.27/Jκ1, and DH3–10/JH6) gave rise to a high frequency of recombination in our assay (average 250 to 3,700 clones per transfection). As expected with authentic RSSs, most recombination events from these two constructs displayed V(D)J recombination at the specific RSSs (~98–100%). However, and as previously described, large differences in the recombination frequency were found between the two constructs, as the result of variations in the RSSs involved. Similar results were obtained for the Dβ1/TAL2 and LMO2/Dδ2 constructs: a vast majority of the recombination occurred between the specific RSSs assayed, i.e., the authentic immune RSS (Dβ1 or Dδ2) and the TAL2 or LMO2 fortuitous RSS (~96%). Nevertheless, a low level of recombination also occurred between the Dβ1 or Dδ2 RSSs and other fortuitous sites located in the core plasmid, competing therefore with TAL2 (Ψ1200, 2%) and LMO2 (Ψ200, 2%; Ψ250, 2%) (see Table III for sequences of cryptic RSSs). Altogether, these results suggest that both TAL2 and LMO2 sequences contain functional RSSs (see competition substrates analysis below). This is in agreement with recent data from Raghavan et al. (12), who also studied the LMO2 RSS in a similar recombination assay. Sequence analysis of the Dβ1/ TAL2 (n = 38) and LMO2/Dδ2 (n = 26) coding joints confirmed normal features of V(D)J recombination, including break at the coding segment/RSS border, P and N nucleotide additions and deletions (summarized in Fig. 3, A 1, for LMO2/Dδ2; full sequence libraries are shown in the online supplemental sequences). Importantly, these features are also similar to the ones observed in vivo at most derivative chromosome 7 and 14 breakpoints in t(7;9) Dβ1/TAL2 (17) and t(11;14) LMO2/Dδ2 fusion (19) (21), respectively.
Table V.
Recombination Substrates
| Vector | No. Ta | No. clonesb | No. Ac | V(D)J recombination | BRf | |
|---|---|---|---|---|---|---|
| Specific RSSd | Other RSSe | |||||
| VκA2.27/Jκ1 | 8 | ~30,000 (~3,700) | 50 | 50 (100%) | 0 (0%) | 0 (0%) |
| DH3.10/JH6 | 3 | ~750 (~250) | 47 | 46 (98%) | 0 (0%) | 1 (2%) |
| Dβ1/TAL2 | 10 | ~570 (~57) | 110 | 106 (96%) | 2 (2%) | 2 (2%) |
| LMO2/Dδ2 | 6 | ~200 (~34) | 51 | 49 (96%) | 2 (4%) | 0 (0%) |
| TAL1/Dδ2i | 6 | ~110 (~18) | 38 | 2 (5%) | 26 (69%) | 10 (26%) |
| BCL1 mtc/JH6 | 5 | 42 (~8) | 37 | 0 (0%) | 0 (0%) | 37 (100%) |
| BCL2 mbr/JH6 | 4 | 44 (~11) | 37 | 0 (0%) | 0 (0%) | 37 (100%) |
Total number of independent transfections.
Total number of clones obtained (average number of colonies per transfection).
Total number of clones analyzed (PCR screen and/or sequencing).
V(D)J-mediated recombination between two authentic RSSs (VκA2.27/Jκ1, DH3.10/JH6) or between one authentic RSS and the fortuitous RSS identified at the proto-oncogene breakpoints in vivo.
V(D)J-mediated at other fortuitous sites (within the flanking proto-oncogene sequence or in the core plasmid).
BR-mediated recombination (defined as not mediated by V(D)J).
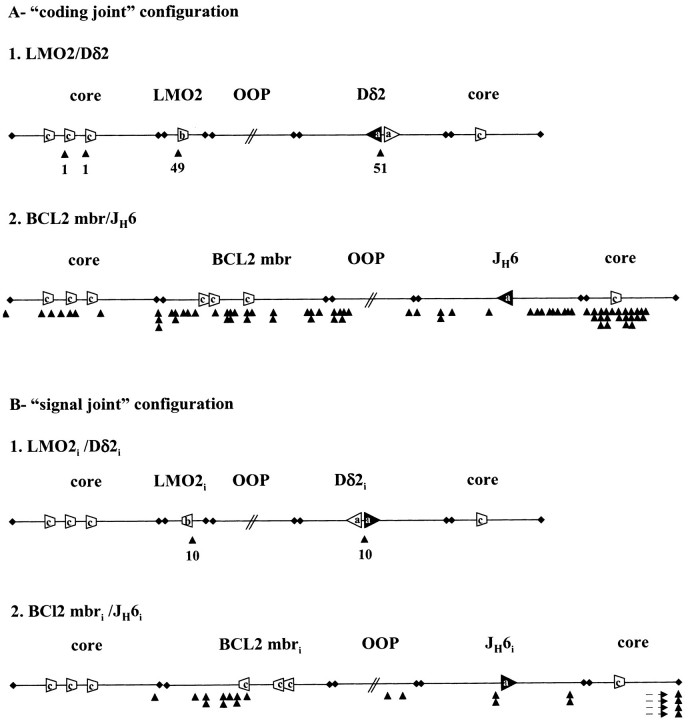
Figure 3.
Schematic representation of the breakpoints distribution (to scale). See also legend to Fig. 1. Black cones indicate the location and frequency of the breaks. Breaks located outside the drawing in BCL2 mbri/JH6i are indicated by dashed arrows.
For the TAL1/Dδ2i construct, recombination rates are much lower than consensus RSSs (Table V). The majority of these events correspond to V(D)J recombination of the Dδ2 3′RSS with cryptic RSSs other than TAL1 (69%). Three different cryptic RSS located in the core plasmid were found recombined: Ψ150, 29%; Ψ6131, 34%; Ψ3490, 5%. Nevertheless, two clones (5%) showed recombination at the specific TAL1 RSS, and sequence analysis of the two coding joints confirmed normal features of V(D)J recombination, including presence of a P nucleotide (see online supplemental sequences). Additionally, cleavages occurred at the immediate border of both the TAL1 RSS (in one case at the exact border, in the other case with 1 nucleotide deletion) and the Dδ2 RSS (in one case with a P nucleotide, in the other case 2 nucleotides deletion). That the two breaks would have occurred by chance at both RSS borders in two independent clones by non-V(D)J–mediated mechanisms is very unlikely. This suggests that TAL1 fortuitous RSS can mediate V(D)J recombination, although at very low levels. Importantly, none of the competitor cryptic sites present within the genomic TAL1 ~400 bp flanking region (Fig. 2, line 3, “c” RSSs) underwent recombination. This observation is in agreement with the identification of this cryptic site as the only RSS mediating the TAL1/TCRδ translocation in this 400 bp region in vivo (22, 23).
In conclusion, these results functionally demonstrate that the fortuitous RSSs of TAL2, LMO2, and TAL1 are able to undergo V(D)J recombination. The data strongly support the view that t(7;9)(q34;q32), t(11;14)(p13;q11), and t(1;14)(p34;q11) translocations can result from bona fide V(D)J recombination between a legitimate TCR locus and an illegitimate proto-oncogene locus bearing a fortuitous but functional RSS. We will refer further to this category of translocations as “type 1.” Our results also suggest that a cryptic site with very low recombinogenic potential such as TAL1 can be involved in such type 1 translocation in vivo.
Fortuitous RSS in BCL2 or BCL1 Breakpoint Regions Are Not Able to Target V(D)J Recombination in Extra-chromosomal Substrates.
At the lower end of the recombination spectrum in Table V, the BCL2 mbr/JH6 and BCL1 mtc/JH6 constructs showed sublevels of recombination events, with an average of ~10 clones per transfection. Sequence analysis of these clones revealed that none of the junctions complies with the criteria of V(D)J recombination (see online supplemental sequences). In particular, breaks are not localized at the immediate border of any of the potential cryptic RSSs in the BCL2 mbr and in the BCL1 mtc, or at the border of the authentic JH6 RSS (summarized in Fig. 3, A 2), in sharp contrast to the other constructs (e.g. LMO2/ Dδ2, Fig. 3, A 1). This is in agreement with the finding of similar break distribution in experiments performed in cells devoid of RAG activity (CHO, not shown). However, junctions generated by BR do not necessarily indicate the precise site of cleavage. To confirm our general interpretation of the breaks, and to exclude the possibility that some of the BCL2 mbr/JH6 and BCL1 mtc/JH6 breaks would occur at unidentified cryptic RSSs, we also tested the formation of signal joints. The presence of the signal joint in addition to the coding joint allows to diagnose unambiguously the use of the V(D)J mechanism in the recombination process. To do so, each of the two cassettes of the BCL2 mbr/JH6 plasmid were inverted so that the retained product would correspond to a SJ instead of a CJ (Fig. 2, line 8, BCL2 mbri/JH6i). As a control, we also inverted the two cassettes of the LMO2/Dδ2 construct (Fig. 2, line 9, LMO2i/Dδ2i). As shown in Fig. 3 B, 100% of the LMO2i/Dδ2i clones (10/10) corresponded to the specific SJ, while none of the BCL2 mbri/JH6i clones (0/10) showed SJ formation. These results confirm our previous break diagnostic based on the CJ, and indicate that the recombined clones obtained for the BCL2 mbr/JH6 and BCL1 mtc/JH6 pairs are indeed not derived from V(D)J recombination, but from unspecific BR. Thus, none of the fortuitous 12-RSSs in the ~400 bp BCL2 mbr and BCL1 mtc were able to target V(D)J recombination with the JH6 RSS as the “natural” recombination partner.
In the BCL2 mbr/JH6 and BCL1 mtc/JH6 substrates, however, the complete absence of recombination of the JH6 RSS with other fortuitous cryptic sites in the plasmid core sequence contrasts with the situation in other constructs (Table V, other RSS: 0%). This might be due to the low recombination frequency of the JH6 RSS due to the presence of a 22-bp spacer. Although high enough to target recombination with another authentic RSS such as DH3.10, this frequency might be too low to generate detectable levels of recombination to cryptic sites in the core plasmid or within the BCL2 mbr and BCL1 mtc segments. To test this possibility, a new BCL1 mtc construct was designed in which the JH6 segment was substituted by the Dδ2 segment, in signal joint configuration (Fig. 2, line 10, (BCL1 mtci-6131)/Dδ2i). As an internal control for V(D)J recombination, a known cryptic site was also cloned in the plasmid. We used the “6131” cryptic site described by Lewis and colleagues, which was estimated to recombine at ~1% of the activity of a consensus RSS (2). As a reference, we constructed a similar plasmid, containing the DH3 segment instead of BCL1 mtc (Fig. 2, line 11, (6131-DH3i)/Dδ2i). As shown in Table VI, the substitution of JH6 by Dδ2 as a recombination partner in the (BCL1 mtci-6131)/Dδ2i construct still did not result in targeting V(D)J recombination at the BCL1 mtc fortuitous RSSs (0%). However, in presence of Dδ2, the vast majority of recombination events (98%) consisted of V(D)J recombination between Dδ2 and other fortuitous sites (including 6131, ~79%), leaving only 2% of BR events. The relative recombination rate between 6131 and DH3 in the 6131-DH3i/Dδ2i construct confirmed that 6131 recombines in our hands at ~1–2% of the activity of an authentic RSS. Thus, it can be estimated that the recombinogenic potential of all BCL1 mtc fortuitous sites together is <1% the rate of cryptic sites such as 6131, or <0.01% the rate of a consensus RSS.
Table VI.
Recombination Substrates. Competition with Cryptic Site 6131
| Vector (external-internal) | No. T | No. A | V(D)J recombination | BR | |
|---|---|---|---|---|---|
| Specific RSS | Other RSSa | ||||
| (BCL1 mtci-6131)/Dδ2i | 6 | 82 | 0 (0%) | 80 (98%) | 2 (2%) |
| (6131-DH3i)/Dδ2i | 2 | 293 | 283 (97%) | 7 (2%) | 3 (1%) |
| BCL2 mbrf/Dδ2 | 7 | 23 | 0 (0%) | 16 (70%) | 7 (30%) |
Including 6131.
Importantly, in the BCL2 mbr/JH6 and BCL1 mtc/JH6 constructs, only the appropriately oriented fortuitous 12-RSSs (5′CAC3′) were tested (Fig. 2, lines 4 and 5, white RSSs in BCL2 mbr and BCL1 mtc cassettes). However, in vivo, numerous fortuitous 23-RSS in opposite orientation (3′GTG5′) could target recombination by recombining with the authentic 12-RSSs of the DH segments, and these potential cryptic sites also had to be tested in our assay. In the (BCL1 mtci-6131)/Dδ2i construct, the Dδ2 contains two RSSs (Fig. 2, line 10): one 23-RSS in SJ configuration (black RSS) and one 12-RSS in CJ configuration (white RSS). In this construct, both potential 12- and 23- fortuitous RSSs of the BCL1 mtc in the proper orientation relative to the translocation breakpoints observed in vivo (white and black RSSs, respectively, in the BCL1 mtc cassette), were therefore assayed at once. The inability of the BCL1 mtc to target V(D)J recombination at the sensitivity estimated above takes therefore into account all potential 12- and 23 cryptic sites in the BCL1 mtc. To test in a similar way both the 12- and 23 fortuitous RSSs of the BCL2 mbr, this region (here flanked by an additional ~800 bp of surrounding genomic DNA) was also cloned with the Dδ2 partner (Fig. 2, line 12, BCL2 mbrf/Dδ2). Similar results were found for this construct (Table VI). No recombination of the BCL2 mbr cryptic RSSs occurred (0%), while V(D)J recombination of Dδ2 to other fortuitous sites (Ψ250, 94%; Ψ150, 6%) took place in 70% of the cases. As expected, the absence of the Ψ6131 RSS resulted in a lower V(D)J recombination versus BR ratio than for the (BCL1 mtci-6131)/Dδ2i construct. Thus, as the recombination rates of the cryptic sites above are lower than that of 6131, a recombinogenic potential of <0.01% the rate of a consensus RSS can also be estimated for the 12- and 23 fortuitous RSSs in the BCL2 mbr.
Altogether, these results show that fortuitous RSSs in BCL2 or BCL1 breakpoint regions are not able to target V(D)J recombination in the limit of detection of our extra-chromosomal assay (estimated as at least 1:10,000 the rate of a consensus RSS).
Fortuitous RSSs in the BCL2 mbr Cannot Account for the Relative Rate of the t(14;18)(q32;q21) In Vivo.
What is the significance of the absence of V(D)J recombination at cryptic sites in our ex vivo assay? TAL1/Dδ2i data shows that the frequency of V(D)J recombination can be lower than the frequency of BR (Table V). Nevertheless, this low V(D)J recombination event recapitulates qualitatively the involvement of the V(D)J recombination in the TAL1/ TCRδ translocation process as observed in vivo. Thus, although indicative of the likelihood of usage of the V(D)J recombination, the relative frequency of BR versus V(D)J recombination events does not represent a physiological limit to the potential involvement of V(D)J recombination in the translocation processes. In other words, BCL1 mtc and BCL2 mbr breaks in vivo could still be the result of extremely low levels of V(D)J recombination, but with a frequency under the sensitivity of our ex vivo assay. To set the sensitivity of our assay in perspective of the in vivo process, we sought therefore to compare relative frequencies of V(D)J-mediated breaks occurring at the BCL2 locus in vivo and ex vivo. In rare cases of FL, breaks have been recently proposed to occur through V(D)J recombination at two fortuitous RSSs located outside the BCL2 break clusters (5′ of BCL2 and 3′ of mbr; reference 33). Although not leading to t(14;18) translocation per se, these cases resulted in the transposition of the BCL2 gene into the IgH locus, and in the overexpression of BCL2 in a manner similar to the t(14;18). It was estimated that this transposition event occurs in ~5% of FL cases (2/40). Thus, if one assumes that both the transposition breaks and the BCL2 mbr breaks result from a V(D)J-mediated process at fortuitous RSSs, the relative in vivo rate of V(D)J recombination at the transposition loci and at the BCL2 mbr should be ~5:95% (Table VII). To estimate the sensitivity of our ex vivo assay compared with the in vivo situation, we constructed and assayed a competition substrate containing both the 3′BCL2 region containing the fortuitous RSS involved in the transposition and the large BCL2 mbrf segment, (Fig. 2, line 13, 3′BCL2-BCL2 mbrf)/Dδ2). As shown in Table VII, V(D)J-mediated breaks occurred at high frequency at the 3′BCL2 RSS (100%), but not in the BCL2 mbrf. These data first validates the suggestion of Vaandrager et al. that the transposition process they observed was indeed mediated by V(D)J recombination (33). Most importantly, these results show that the relative recombination rate of ~5% in vivo, corresponds to a relative V(D)J-recombination rate of 100% in our ex vivo assay, and therefore that BCL2 mbr breaks are not generated by V(D)J mis-targeting. No such relative frequencies can be directly assessed for BCL1 mtc in vivo. Nevertheless, considering the similar, very characteristic features of BCL1 mtc and BCL2 mbr translocation breakpoints, and the equal absence of V(D)J recombination at the detection limits of our assay, it is very unlikely that the cryptic sites in BCL1 mtc are involved in the initiation of t(11;14)(q13;q21).
Table VII.
Relative Rates of V(D)J Recombination at BCL2 mbr and 3′BCL2 Loci
| Vector (internal-external) | No. T | No. clones | No. A | V(D)J recombination |
|---|---|---|---|---|
| 3′BCL2 : BCL2 mbr | ||||
| In vivoa | NA | NA | 40 | 2 (5%) : 38 (95%) |
| (3′CBL20BCL2 mbrf)/D′2 | 3 | 105 | 41 | 29 (100%)b : 0 (0%) |
Reference 33.
The 12 remaining clones consist of BR located outside BCL2 mbrf.
This strongly supports the view that both t(14;18)(q32; q21) and t(11;14)(q13;q21) translocations belong to a category of translocation distinct from type 1, in which only the breaks at the immune locus are mediated by V(D)J recombination. Breaks at the nonimmune locus bearing the proto-oncogene are therefore initiated by other mechanisms, and could subsequently invade the V(D)J synaptic complex during the rearrangement process. Such translocation mechanism will be referred to as “type 2” in this manuscript.
Precise Quantification of the Recombinogenic Potentials of the Proto-oncogene Breakpoint Regions in Type 1 Translocations.
To determine if the recombination rates of the functional cryptic RSSs in type 1 translocations are sufficient to compete with physiological TCR RSSs, we performed competition assays in which the proto-oncogene breakpoint region competes for recombination with the normal partner of the immune locus to which it translocates. In these competition assays, the presence of the two competing segments on the same plasmid allows a direct and very precise estimation of the relative rates of recombination (10).
This is demonstrated here by the (1:1) relative ratio obtained for a control substrate in which the two competing segments are identical (Dβ1.1/(TAL2-TAL2), Table VIII). Thus, to estimate the recombinogenic potential of the TAL2 sequence, the TAL2 segment was set to compete with Jβ2.7 for rearrangement to Dβ1.1 (Fig. 2, line 15, Dβ1.1/(TAL2-Jβ2.7)). As shown in Table VIII, the rate of recombination of TAL2 RSS is ~50 times lower than its Jβ2.7 physiological competitor (2 vs. 98%). A similar type of experiment was performed for LMO2. The Jδ1 segment was used as competitor for rearrangement to Dδ2 (Fig. 2, line 16, LMO2i-Jδ1i)/Dδ2i). Strikingly, results in Table VIII indicate that the recombination frequency of the LMO2 RSS (25%) displayed only a threefold decrease relatively to the Jδ1 RSS (75%). This is slightly different from the estimation of Raghavan et al. (12) who found a 27-fold lower recombinogenic potential relative to a consensus 12-signal. However, their estimation is based on comparison with a standard consensus RSS, while we used the natural Jδ1 competitor of the t(11;14)(p13;q11) translocation which diverges from consensus at several positions (Table I).
Table VIII.
Competition Substrates
| Vector (internal-external) | No. T | No. clones | No. A | Internal : External |
|---|---|---|---|---|
| Dδ1/(TAL2-TAL2) | 4 | ~418 | 70 | 33a (49%) : 34 (51%) |
| Dδ1/(TAL2-Jδ2.7) | 4 | ~1,600 | 89 | 2 (2%) : 87 (98%) |
| Dδ2/(Jδ1-LMO2) | 2 | ~279 | 71 | 53 (75%) : 18 (25%) |
The three remaining clones consist of recombination to cryptic sites or of BR.
Thus, precise quantification of the recombinogenic potentials of the TAL2 and LMO2 proto-oncogene breakpoint regions show that they are essentially in the same range as the physiologic recombination partners of the TCR Dβ and Dδ gene segments. Importantly, if the potency of a fortuitous RSS largely contributes to the final frequency of translocation in vivo, recombinogenic potentials in this range should constitute a considerable risk of genomic instability and cancer through mis-targeting by the V(D)J recombination.
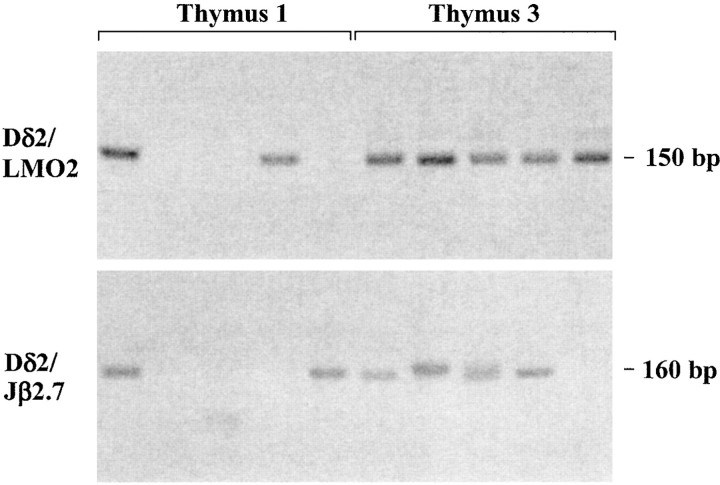
The Recombinogenic Potential of the LMO2 Breakpoint Region Is Directly Predictive of the t(11;14)(p13;q11) Translocation Frequency In Vivo. To test the contribution of the re-combinogenic potential of the LMO2 proto-oncogene breakpoint region in the final frequency of the t(11;14) (p13;q11) translocation, we sought to detect the Dδ2/ LMO2 rearrangement in vivo, and to assess if the relative frequency of translocation matches the relative frequency of recombination determined ex vivo. To minimize possible bias from subsequent selection and proliferation of malignant cells carrying the specific translocation in vivo, we used thymus from healthy individuals as the source of DNA. In thymus DNA, however, the frequency of Dδ2/ LMO2 translocation cannot be assessed relatively to the Dδ/Jδ rearrangements for two main reasons: (a) Dδ/Jδ rearrangements are subject to differentiation and selection in normal thymus; (b) Dδ/Jδ rearrangements occur in cis, i.e., by deletion on the same chromosome, while Dδ2/LMO2 rearrangements occur in trans, i.e., by translocation between two chromosomes. Trans-chromosomal V(D)J recombination has been estimated to be at least 1,000 times less abundant than standard V(D)J recombination within a given receptor locus (34, 35). We reasoned therefore that since the LMO2 recombinogenic potential is comparable to those of authentic TCR RSSs, its rate of translocation with TCR gene segments should be comparable to the rates of TCR trans-chromosomal V(D)J recombination (e.g., TCRδ/β). We thus compared the relative in vivo frequencies of t(11;14)(p13;q11) (Dδ2/LMO2) and t(7;14) (q34;q11) (Dδ2/Jβ2.7) V(D)J-mediated translocations. To detect Dδ2/LMO2 and Dδ2/Jβ2.7 rearrangements, which were anticipated to be rare in thymus DNA of healthy individuals, we used a sensitive double nested PCR assay, in a “multiple tube” procedure (29). Detection of rare events by such sensitive PCR assay gives rise to fluctuation, probably depending on the presence or not of the event in the aliquot taken from the sample, and on how early in the PCR cycling the event is first amplified. To circumvent this fluctuation, each individual estimate is based on the sum of PCR products from 20–40 separate reactions, analyzed in parallel lanes. The results obtained from 3 independent thymuses are summarized in Table IX, and representative PCR signals are shown in Fig. 4. As can be seen, Dδ2/LMO2 signal joints could be detected at a strikingly high rate (55/100 [55%] reactions in 2/3 thymuses), which can be roughly estimated at ~1 translocation event every 1–10 × 105 cells (34). To confirm the identity of the Dδ2/LMO2 rearrangements, the PCR bands were cloned and sequenced. All bands sequenced showed the presence of a proper Dδ2/LMO2 signal joint (see online supplemental sequences). Importantly, these breakpoints are also qualitatively similar to the two Dδ2/ LMO2 derivative chromosome 11 breakpoints described so far at that site in T-ALL (19), and to the junctions generated ex vivo in this study. In addition, comparison between Dδ2/LMO2 and Dδ2/Jβ2.7 recombination rates shows that the appearance of Dδ2/LMO2 rearrangements strikingly parallels the one of Dδ2/Jβ2.7 rearrangements (Table IX, 25/60 [42%] reactions in 2/3 thymuses). The absence of detection of both translocations in thymus 2 is not due to differences in DNA quality (see Materials and Methods) and could be related to age difference. To exclude possible bias due to differences in the efficiency of the PCR primer combinations used, each PCR amplification was calibrated. Serial dilutions of a plasmid containing either a Dδ2/ LMO2, or a Dδ2/Jβ2.7 rearrangement were PCR-amplified using the same sets of nested primers as above. Both rearrangements could be detected with a comparable sensitivity (not shown), indicating that there is no large bias in the PCR detection of the rearrangements. This suggests that the t(11;14)(p13;q11) involving Dδ2/LMO2 recombination, and the t(7;14)(q34;q11) involving TCR Dδ2/Jβ recombination, occur at similar frequencies in the thymus of healthy individuals.
Table IX.
In Vivo Frequencies of Translocation
| Thymus (age) | Dδ2/LMO2 | Dδ2/Jβ2.7 |
|---|---|---|
| 1 (6 yr) | 16/40 | 12/20 |
| 2 (2 d) | 0/20 | 0/20 |
| 3 (1 yr) | 39/40 | 13/20 |
| Total | 55/100 (~55%) | 25/60 (~42%) |
Figure 4.
Detection of t(11;14)(p13;q11) (Dδ2/LMO2) and t(7;14) (q34;q11) (Dδ2/Jβ2.7) V(D)J-mediated translocations by the multiple tube procedure. Reactions 1–5 from thymus 1 and 3 are shown.
Remarkably, these data show that: (a) the t(11;14)(p13;q11) involving Dδ2/LMO2 recombination previously found only in T-ALL patients, can also be detected at high frequency in thymus from healthy individuals; and (b) the recombinogenic potential of LMO2, as assessed in recombination assays ex vivo, is directly predictive of the actual in vivo rate of translocation.
Discussion
Lymphoid neoplasms (NHL and ALL) are among the most frequent malignancies. In industrial countries, their incidence is increasing more rapidly than that of most other tumors, but the reasons for this progression are largely unexplained. Multiple factors are likely to be involved, and act over the lifetime of an individual through many pathogenic pathways, rendering causal relationships complex. It is therefore of considerable importance to gain further understanding on the initial molecular mechanisms involved in lymphomagenesis, and on the role played by genetic and exogenous factors in this process. The recombination assay described here provides a functional mean by which such studies can be initiated.
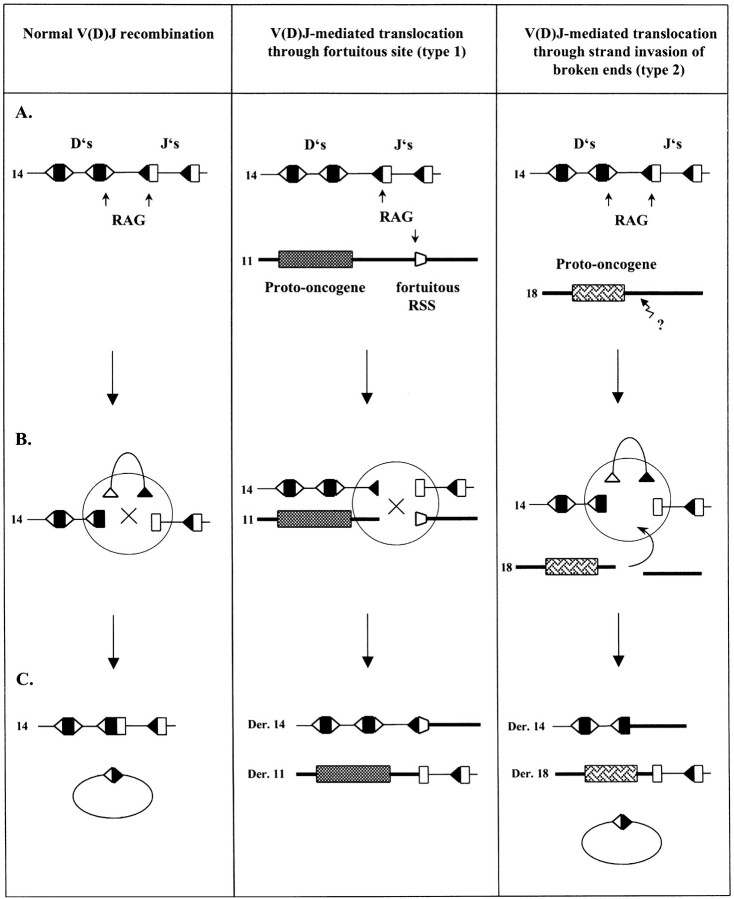
Our data indicate that at least two distinct mechanisms involving V(D)J recombination can lead to translocation, as previously anticipated on the basis of in vivo translocation breakpoints analysis (Fig. 5): in a first category (type 1), illegitimate V(D)J recombination would occur between a legitimate Ig or TCR locus and an illegitimate proto-oncogene locus bearing a fortuitous but functional RSS (Fig. 5, middle panel). Together with the breakpoint features previously observed in vivo in T-ALL, our results strongly support the possibility that most t(7;9)(q34;q32), t(11;14) (p13;q11), and t(1;14)(p34;q11) are mediated by such type 1 mechanism. In a second category of translocation (type 2), only the Ig/TCR loci breaks are mediated by V(D)J recombination (Fig. 5, right panel). Breaks at the nonimmune locus bearing the proto-oncogene, are therefore initiated by other mechanisms, and could subsequently invade the V(D)J synaptic complex during the rearrangement process (also called “strand donation,” [35;36]). Our data indicate that the t(11;14)(q13;q32), and t(14;18)(q32;q21) translocations in NHL are very likely to belong to this second category, in full agreement with the predictions based on the most recent in vivo observations (28, 37). Interestingly, the three translocations identified above as type 1 correspond to the translocations associated with the development of T cell malignancies, while the two translocations identified as type 2 are the ones associated with the development of B cell neoplasms. This could reflect a more general difference between B and T cells toward regulation of locus accessibility. Indeed, several lines of evidence suggest that the accessibility to the V(D)J recombinase is more tightly restricted to the Ig loci in B cells than it is to the TCR loci in T cells (38–40). In this scenario, the accessibility of the recombinase to nonimmune loci such as proto-oncogenes would be inhibited in B cells, and other mechanisms would then be necessary to generate the breaks near the various proto-oncogene loci.
Figure 5.
Legitimate and illegitimate V(D)J recombination. Two models of V(D)J-mediated translocation are shown: type 1 translocation (middle panel) results from V(D)J recombination between a fortuitous but functional RSS in the proto-oncogene sequence; in type 2 translocations (right panel) DSBs at the proto-oncogene locus are created by a yet unknown mechanism and the resulting DNA broken ends subsequently invade the V(D)J synapse at the Ig/TCR locus. The D gene segments (D’s) are represented as black boxes with their corresponding RSSs as white triangles. The J gene segments (J’s) are shown as white boxes with their RSSs as black triangles. (A) Genomic configuration. Putative chromosomes are indicated. Arrows indicate the sites of V(D)J-mediated cleavage during the attempted rearrangement. The broken arrow with question mark shows the site of breakage by a mechanism distinct from V(D)J-mediated cleavage. (B) The cleaved signal complex stage of the rearrangement and end-joining. (C) Rearranged configuration.
For type 1 translocations, mis-targeting by the V(D)J recombination in developing T cells should present a considerable risk of genomic instability and cancer: (a) even sublevels of V(D)J mis-targeting can lead to chromosomal alterations, as previously observed for chromosomal deletion (2), and as shown in this report for chromosomal translocation with TAL1/Dδ; (b) an estimated 10 million fortuitous cryptic sites in the genome could potentially mis-target the V(D)J recombination process at ~1% the range of canonical frequency (2), a frequency well above that of TAL1; and (c) the genomic instability conferred by V(D)J mis-targeting can be considerable, as demonstrated here by the high recombinogenic potential of the LMO2 sequence tested and correspondingly high rates of Dδ2/ LMO2 translocation in healthy individuals. This major impact of the recombinogenic potential of a RSS is surprising in the view that several additional factors are expected to play an important role in the outcome of the translocation: the accessibility of the recombinase to a given locus; the inhibition of trans-chromosomal V(D)J recombination; the recently described post-cleavage specificity of the V(D)J machinery (41); and the selection and proliferation of cells carrying the translocation due to the ectopic expression of the proto-oncogene. Some of these factors, such as the one responsible for the inhibition of trans-locus recombination, are likely to be general mechanisms which should be operating equally in all translocations, regardless of the involvement of an immune or nonimmune locus. However, some others, such as accessibility, should be more locus dependent (42). We cannot exclude the possibility that the LMO2 locus is accidentally relatively accessible to the recombinase in developing T cells, but that this situation might be different for other proto-oncogene loci. Interestingly, trans-V(D)J recombination between immune loci has been previously described as a potent bio-marker for genomic instability and cancer (43). It will be of importance to determine in prospective studies if the rate of type 1 translocations and the development of the corresponding associated neoplasms, is also upmodulated in the situations of genomic instability identified by trans-locus V(D)J recombination. The understanding of the regulating forces involved in type 1 translocation, and the identification of the possible genetic or exogenous factors involved in their modulation, might prove to be important steps in cancer prevention.
For type 2 translocations, the identification of the additional mechanisms involved in the translocation processes is the present challenge in the further understanding of this type of illegitimate recombination. Numerous mechanisms have previously been proposed as potential candidates, including CHI sequences (44, 45), 2-ended transposition (46, 47), and the somatic hypermutation mechanism (28). Interestingly, the possible involvement of the somatic hypermu-tation as the additional mechanism generating the breaks at the proto-oncogene locus, would be in line with the association between type 2 translocations and mature B cell malignancies. On the other hand, the lack of obvious consensus motifs found so far between the BCL1 mtc and the BCL2 mbr despite the striking similarities in the t(14;18)(q32;q21) and t(11;14)(q13;q32) junctions, might suggest a difference in the origin of the specific breakage, but a common mechanism of invasion of the V(D)J synapse by the broken ends. One could conceive that various causes of breakage could all eventually lead to broken ends trapped in the same DNA/protein repair complex (e.g. through recruitment of Ku70/80). In support of this scenario, it would be of interest to determine if “naked” broken ends can invade the specific V(D)J synaptic complex, in absence of mechanisms other than the ubiquitous DNA repair process. If so, the contribution of V(D)J recombination to genomic instability could be the result of more diverse and complex processes than previously anticipated.
Acknowledgments
We are grateful to Dr. C. Mannhalter for technical expertise, and to Dr. P. Ferrier for helpful comments on the manuscript. This work was supported by a grant for the ICP Program of the Austrian Federal Ministry for Education, Science and Culture, and by a grant from the Fonds zur Foerderung der Wissenschaftlichen Forschung (FWF P-13984GEN).
Footnotes
Abbreviations used in this paper: BR, break/repair; CJ, coding joint; FL, follicular lymphoma; mbr, major breakpoint region; mtc, major translocation cluster; NHL, non-Hodgkin’s lymphoma; RSS, recombination signal sequence; SJ, signal joint; T-ALL, T cell acute lymphoblastic leukemia.
References
- Lieber M.R., Chang C.P., Gallo M., Gauss G., Gerstein R., Islas A. The mechanism of V(D)J recombination: site-specificity, reaction fidelity and immunologic diversity. Semin. Immunol. 1994;6:143–153. doi: 10.1006/smim.1994.1020. [DOI] [PubMed] [Google Scholar]
- Lewis S.M., Agard E., Suh S., Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko B., Sklar J. Chromosomal translocations in lymphoid neoplasia: a reappraisal of the recombinase model. Cancer Cells. 1990;2:1–8. [PubMed] [Google Scholar]
- Lieber M.R. Warner-Lambert/Parke-Davis Award Lecture. Pathological and physiological double-strand breaks: roles in cancer, aging, and the immune system. Am. J. Pathol. 1998;153:1323–1332. doi: 10.1016/s0002-9440(10)65716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney A.J., Victor K.D., Vu K., Nadel B., Chuk-wuocha R.U. Influence of the V(D)J recombination mechanism on the formation of the primary T and B cell repertoires. Semin. Immunol. 1994;6:155–163. doi: 10.1006/smim.1994.1021. [DOI] [PubMed] [Google Scholar]
- Akira S., Okazaki K., Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- Hesse J.E., Lieber M.R., Mizuuchi K., Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y., Tsurushita N., Nagawa F., Matsuoka M., Okazaki K., Imai M., Sakano H. Essential residues in V(D)J recombination signals. J. Immunol. 1994;153:4520–4529. [PubMed] [Google Scholar]
- Fanning L., Connor A., Baetz K., Ramsden D., Wu G.E. Mouse RSS spacer sequences affect the rate of V(D)J recombination. Immunogenetics. 1996;44:146–150. doi: 10.1007/s002510050103. [DOI] [PubMed] [Google Scholar]
- Nadel B., Tang A., Escuro G., Lugo G., Feeney A.J. Sequence of the spacer in the recombination signal sequence affects V(D)J rearrangement frequency and correlates with nonrandom Vkappa usage in vivo. J. Exp. Med. 1998;187:1495–1503. doi: 10.1084/jem.187.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel B., Tang A., Lugo G., Love V., Escuro G., Feeney A.J. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J. Immunol. 1998;161:6068–6073. [PubMed] [Google Scholar]
- Raghavan S.C., Kirsch I.R., Lieber M.R. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J. Biol. Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Gorham J., Cossman J., Jaffe E., Croce C.M. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- Cleary M.L., Sklar J. Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc. Natl. Acad. Sci. USA. 1985;82:7439–7443. doi: 10.1073/pnas.82.21.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi A., Wright J.J., Graninger W., Seto M., Owens J., Cossman J., Jensen J.P., Goldman P., Korsmeyer S.J. Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc. Natl. Acad. Sci. USA. 1987;84:2396–2400. doi: 10.1073/pnas.84.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Louie E., Bashir M.M., Croce C.M. The reciprocal partners of both the t(14; 18) and the t(11; 14) translocations involved in B-cell neoplasms are rearranged by the same mechanism. Oncogene. 1988;2:347–351. [PubMed] [Google Scholar]
- Tycko B., Reynolds T.C., Smith S.D., Sklar J. Consistent breakage between consensus recombinase heptamers of chromosome 9 DNA in a recurrent chromosomal translocation of human T cell leukemia. J. Exp. Med. 1989;169:369–377. doi: 10.1084/jem.169.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne E., Takihara Y., Sagman U., de Sousa J., Burrow S., Lewis W.H., Mak T.W., Minden M.D. The T-cell receptor delta chain locus is disrupted in the T-ALL associated t(11;14)(p13;q11) translocation. Blood. 1989;73:1672–1676. [PubMed] [Google Scholar]
- Cheng J.T., Yang C.Y., Hernandez J., Embrey J., Baer R. The chromosome translocation (11;14)(p13;q11) associated with T cell acute leukemia. Asymmetric diversification of the translocational junctions. J. Exp. Med. 1990;171:489–501. doi: 10.1084/jem.171.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T., Buluwela L., Williams D., White L., Rabbitts T.H. A cluster of chromosome 11p13 translocations found via distinct D-D and D-D-J rearrangements of the human T cell receptor delta chain gene. EMBO J. 1988;7:2011–2017. doi: 10.1002/j.1460-2075.1988.tb03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I.S., Kaneko Y., Gonzalez-Sarmiento R., Campbell K., White L., Boehm T., Rabbitts T.H. A study of chromosome 11p13 translocations involving TCR beta and TCR delta in human T cell leukaemia. Oncogene. 1991;6:577–582. [PubMed] [Google Scholar]
- Chen Q., Cheng J.T., Tasi L.H., Schneider N., Buchanan G., Carroll A., Crist W., Ozanne B., Siciliano M.J., Baer R. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 1990;9:415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Yang C.Y., Tsan J.T., Xia Y., Ragab A.H., Peiper S.C., Carroll A., Baer R. Coding sequences of the tal-1 gene are disrupted by chromosome translocation in human T cell leukemia. J. Exp. Med. 1990;172:1403–1408. doi: 10.1084/jem.172.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley C.G., Aplan P.D., Denning S.M., Haynes B.F., Waldmann T.A., Kirsch I.R. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc. Natl. Acad. Sci. USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger L.R., Kagan J., Christopher G., Kurtzberg J., Hershfield M.S., Nowell P.C., Croce C.M. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc. Natl. Acad. Sci. USA. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L.Y., Baer R.J. The role of chromosome translocations in T cell acute leukemia. Curr. Opin. Immunol. 1995;7:659–664. doi: 10.1016/0952-7915(95)80074-3. [DOI] [PubMed] [Google Scholar]
- Nadel B., Feeney A.J. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol. Cell. Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger U., Boecskoer S., Le T., Mitterbauer G., Bolz I., Chott A., Kneba M., Mannhalter C., Nadel B. Follicular lymphomas’ BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- Fuscoe J.C., Setzer R.W., Collard D.D., Moore M.M. Quantification of t(14;18) in the lymphocytes of healthy adult humans as a possible biomarker for environmental exposures to carcinogens. Carcinogenesis. 1996;17:1013–1020. doi: 10.1093/carcin/17.5.1013. [DOI] [PubMed] [Google Scholar]
- Williams M.E., Swerdlow S.H., Meeker T.C. Chromosome t(11;14)(q13;q32) breakpoints in centrocytic lymphoma are highly localized at the bcl-1 major translocation cluster. Leukemia. 1993;7:1437–1440. [PubMed] [Google Scholar]
- Stamatopoulos K., Kosmas C., Belessi C., Stavroyianni N., Kyriazopoulos P., Papadaki T. Molecular insights into the immunopathogenesis of follicular lymphoma. Immunol. Today. 2000;21:298–305. doi: 10.1016/S0167-5699(00)01650-9. [DOI] [PubMed] [Google Scholar]
- Stamatopoulos K., Kosmas C., Belessi C., Kyriazopoulos P., Papadaki T., Anagnostou D., Loukopoulos D. Molecular analysis of bcl-1/IgH junctional sequences in mantle cell lymphoma: potential mechanism of the t(11;14) chromosomal translocation. Br. J. Haematol. 1999;105:190–197. doi: 10.1111/j.1365-2141.1999.01314.x. [DOI] [PubMed] [Google Scholar]
- Vaandrager J.W., Schuuring E., Philippo K., Kluin P.M. V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood. 2000;96:1947–1952. [PubMed] [Google Scholar]
- Tycko B., Palmer J.D., Sklar J. T cell receptor gene trans-rearrangements: chimeric gamma-delta genes in normal lymphoid tissues. Science. 1989;245:1242–1246. doi: 10.1126/science.2551037. [DOI] [PubMed] [Google Scholar]
- Bailey S.N., Rosenberg N. Assessing the pathogenic potential of the V(D)J recombinase by interlocus immunoglobulin light-chain gene rearrangement. Mol. Cell. Biol. 1997;17:887–894. doi: 10.1128/mcb.17.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel B., Marculescu R., Le T., Rudnicki M., Bocskor S., Jaeger U. Novel insights into the mechanism of t(14;18)(q32;q21) translocation in follicular lymphoma. Leuk. Lymphoma. 2001 doi: 10.3109/10428190109097743. In press. [DOI] [PubMed] [Google Scholar]
- Welzel N., Le T., Marculescu R., Mitterbauer G., Chott A., Pott C., Kneba M., Du M.Q., Kusec R., Drach J., et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–1636. [PubMed] [Google Scholar]
- Born W., White J., Kappler J., Marrack P. Rearrangement of IgH genes in normal thymocyte development. J. Immunol. 1988;140:3228–3232. [PubMed] [Google Scholar]
- Ferrier P., Krippl B., Blackwell T.K., Furley A.J., Suh H., Winoto A., Cook W.D., Hood L., Costantini F., Alt F.W. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bories J.C., Demengeot J., Davidson L., Alt F.W. Gene-targeted deletion and replacement mutations of the T-cell receptor beta-chain enhancer: the role of enhancer elements in controlling V(D)J recombination accessibility. Proc. Natl. Acad. Sci. USA. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard E.A., Lewis S.M. Postcleavage sequence specificity in V(D)J recombination. Mol. Cell. Biol. 2000;20:5032–5040. doi: 10.1128/mcb.20.14.5032-5040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel W.M., Leduc I., Mathieu N., Tripathi R.K., Ferrier P. Accessibility control of V(D)J recombination: lessons from gene targeting. Adv. Immunol. 1998;69:309–52. doi: 10.1016/s0065-2776(08)60610-0. [DOI] [PubMed] [Google Scholar]
- Kirsch I.R. Molecular genetics of lymphogenesis: an overview. In: Magrath I.T., editor. The Non-Hodgkin’s Lymphomas. Second Edition. E. Arnold; London, UK: 1997. pp. 253–276. [Google Scholar]
- Wyatt R.T., Rudders R.A., Zelenetz A., Delellis R.A., Krontiris T.G. BCL2 oncogene translocation is mediated by a chi-like consensus. J. Exp. Med. 1992;175:1575–1588. doi: 10.1084/jem.175.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger U., Purtscher B., Karth G.D., Knapp S., Mannhalter C., Lechner K. Mechanism of the chromosomal translocation t(14;18) in lymphoma: detection of a 45-Kd breakpoint binding protein. Blood. 1993;81:1833–1840. [PubMed] [Google Scholar]
- Hiom K., Melek M., Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/S0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]