Abstract
Forty-four novel strains of Gammaproteobacteria were cultivated from coastal and pelagic regions of the Pacific Ocean using high-throughput culturing methods that rely on dilution to extinction in very low nutrient media. Phylogenetic analysis showed that the isolates fell into five rRNA clades, all of which contained rRNA gene sequences reported previously from seawater environmental gene clone libraries (SAR92, OM60, OM182, BD1-7, and KI89A). Bootstrap analyses of phylogenetic reliability did not support collapsing these five clades into a single clade, and they were therefore named the oligotrophic marine Gammaproteobacteria (OMG) group. Twelve cultures chosen to represent the five clades were successively purified in liquid culture, and their growth characteristics were determined at different temperatures and dissolved organic carbon concentrations. The isolates in the OMG group were physiologically diverse heterotrophs, and their physiological properties generally followed their phylogenetic relationships. None of the isolates in the OMG group formed colonies on low- or high-nutrient agar upon their first isolation from seawater, while 7 of 12 isolates that were propagated for laboratory testing eventually produced colonies on 1/10 R2A agar. The isolates grew relatively slowly in natural seawater media (1.23 to 2.63 day−1), and none of them grew in high-nutrient media (>351 mg of C liter−1). The isolates were psychro- to mesophilic and obligately oligotrophic; many of them were of ultramicrobial size (<0.1 μm3). This cultivation study revealed that sporadically detected Gammaproteobacteria gene clones from seawater are part of a phylogenetically diverse constellation of organisms mainly composed of oligotrophic and ultramicrobial lineages that are culturable under specific cultivation conditions.
Epifluorescence microscopy (41) and direct viable counting methods (33) have shown that only 0.01 to 0.1% of all the microbial cells from marine environments form colonies on standard agar plates (22). Much of the discrepancy between direct counts and plate counts has been explained by measurements of microbial diversity that employed 16S rRNA gene sequencing without cultivation (3, 15, 26, 52). The present consensus is that many of the most abundant marine microbial groups are not yet cultivated, that there is a need to cultivate these groups for genome-enabled studies of physiology, and that cultivation will probably require approaches other than standard plating methods.
Many attempts have recently been made to cultivate previously uncultured microorganisms by the application of novel approaches. These include high-throughput culturing (HTC) using dilution-to-extinction (12, 42), cultivation with a diffusion growth chamber (31), encapsulation of cells in gel microdroplets (59), and modified plating methods (19, 29). One striking success emerging from these efforts was the first cultivation of members of the SAR11 clade (42), but additionally many novel strains among the Proteobacteria, Planctomycetes, Bacteroidetes, Acidobacteria, and Verrucomicrobia were also cultivated. One reason for this success is thought to be the use of growth conditions which closely mimic the chemical composition of natural environments (12, 31, 59). Some of the strains obtained by HTC have already been taxonomically classified as novel genera in a novel order or family and named Parvularcula, Croceibacter, and Fulvimarina (8-10).
The widespread cloning of 16S rRNA genes from marine bacterioplankton has resulted in high numbers of sequences in public databases. Many of the most abundant phylogenetic groups of marine picoplankton, including the clusters SAR86, SAR116, SAR202, SAR324, SAR406, marine Actinobacteria, and Crenarchaeota marine group I (reviewed in reference 25), still remain uncultivated, suggesting that further innovation will be needed for their successful cultivation. However, a number of strains representing marine bacterial clades of lesser abundance have been cultured by the application of new methods. The organisms that we discuss in this report are Gammaproteobacteria (this nomenclature has been suggested by the most recent version of Bergey's Manual of Systematic Bacteriology (24) that belong to previously detected environmental clades (OM60, BD1-7, KI89A, OM182, and SAR92) retrieved from various marine environments, including different geographic areas and ecosystems (2, 3, 5, 6, 12, 18, 19, 36, 43, 51, 54, 58, 60). They are distantly related to the other major marine Gammaproteobacteria lineages, such as the SAR86 cluster, Vibrio, Alteromonas, and Oceanospirillum. Connon and Giovannoni (12) recently cultivated several strains of the OM60/241 and SAR92 clades from the coast of Oregon by HTC methods, which allow large numbers of microbial isolates to be recovered by dilution to extinction in natural seawater media. In addition, Eilers et al. (19) isolated strain KT71 in the NOR5 lineage (a part of the OM60 clade) from a coastal region of the North Sea by plating methods using inorganic N and P compounds at concentrations that were reduced by several orders of magnitude. They also found that microorganisms related to KT71 comprised 8% of total microscopic counts (19).
Here we report on a related set of strains retrieved by applying HTC methods to culture new taxa from seawater. A total of 44 strains were isolated from four different seawater samples over a 1-year period. These strains include the first described representatives of a number of common clades of uncultured marine Gammaproteobacteria and fall into five distinct rRNA gene clades by phylogenetic analysis. We refer to the five clades in this study as the oligotrophic marine Gammaproteobacteria (OMG) group, because all cultured members of the group showed oligotrophic characteristics in culture and were retrieved from exclusively marine environments. Growth characteristics in low-nutrient media were determined for 12 representative cultures.
MATERIALS AND METHODS
Seawater collection and high-throughput culturing.
Four different seawater samples were collected over a 1-year period from three sampling stations including the surface waters at the southern jetty in Newport, Oreg., a depth of 10 m at a station (NH5) 27.6 km off of the coast of Oregon (44° 39.1′ N, 124° 24.7′ W), and a depth of 10 m at pelagic region of the Pacific Ocean (48° 4.82′ N, 132° 41.03′ W). Seawater samples were immediately stored in the dark at ambient seawater temperatures until further processing. The HTC method (12) was used for cultivating seawater microorganisms in this study. Briefly, the seawater samples were diluted to 10 cells per ml in a low-nutrient heterotrophic medium (LNHM) (0.2-μm-pore-size-filtered and autoclaved seawater amended with 1.0 μM NH4Cl, 0.1 μM KH2PO4, and vitamin mix at a 10−4 dilution of stock [14]) or an LNHM supplemented with 1× mixed carbons (MC) (LNHM plus 1× mixed carbons) (1× concentrations of carbon mixtures were composed of 0.001% [wt/vol] d-glucose, d-ribose, succinic acid, pyruvic acid, glycerol, N-acetyl d-glucosamine, and 0.002% [vol/vol] ethanol) and dispensed into a 48-well microtiter plate. After incubation at 16°C for 3 weeks in the dark or under a 14-h/10-h light/dark regime, 200 μl of samples from each well in the plate was filtered on a 0.2 μm-pore-size polycarbonate membrane (Nuclepore; Whatman, Clifton, N.J.) in a 48-array. Arrays were stained with 4′,6-diamidino-2-phenylindole (DAPI) according to the method of Porter and Feig (41), and cellular growth was determined using an epifluorescence microscope (DMRB; Leica, Wetzlar, Germany). All wells indicating growth were transferred to fresh medium, checked for colony formation, used for DNA extraction, and stored as 10% (vol/vol) glycerol suspensions in liquid nitrogen.
16S rRNA gene sequencing and phylogenetic analyses.
Genomic DNA was extracted from 200 μl of culture using a DNeasy tissue kit (Qiagen, Valencia, Calif.) after three freeze-thaw cycles. A seminested fashion of PCR was used for amplifying 16S rRNA genes. The genes were initially amplified using the slightly modified bacterial universal primers 27F-B (5′-AGRGTTYGATYMTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACTT-3′), and subsequently amplified using the seminested primer set, 27F-B and 1406R (5′-ACGGGCGGTGTGTRC-3′) (34). The PCR mixtures were irradiated with UV (254 nm) for 5 to 10 min before template addition in order to avoid false-positive reactions (38). The PCR products were screened and grouped by restriction fragment length polymorphism (RFLP) analyses employing HaeIII restriction patterns. After purification of the PCR products of at least one culture from each RFLP group using a PCR purification column (QiaQuick, Qiagen), the DNAs were sequenced by the chain termination method on an ABI 377 or ABI 3100 automated sequencer (Applied Biosystems, Foster City, Calif.).
For phylogenetic analyses, nearly full-length 16S rRNA gene sequences were compared with sequences available in the GenBank database by using the BLAST service (1) to determine their approximate phylogenetic affiliations. Sequences were aligned by using the ARB software package (37), and only unambiguously aligned nucleotide positions were used for phylogenetic analyses with PAUP* 4.0 beta 10 (53). The similarity values between sequences were determined from distance matrices by reversing the Jukes-Cantor distance formula (30). Phylogenetic trees were inferred by both neighbor-joining (45) using the Jukes-Cantor model and maximum parsimony with a heuristic search. The resulting neighbor-joining tree and parsimony trees were evaluated by bootstrap analyses based on 1,000 and 100 resamplings, respectively. Short sequences (less than 1,000 bp) were inserted in the tree using the parsimony insertion tool in the ARB database.
Recovery of cultures and isolation as pure cultures.
Twelve representative high-throughput culture collection (HTCC) isolates chosen from five OMG subclades (the clades OM60, BD1-7, KI89A, OM182, and SAR92; see Fig. 1) were revived from frozen 10%-glycerol stocks. Frozen stocks of the isolates were initially thawed in a 20°C water bath, and 200 μl of cultures were transferred to 100 ml of fresh LNHM. The initial HTCC cultures revived from frozen stocks were successively purified using dilution-to-extinction culturing in order to ensure the purity of the cultures. The initial cultures were diluted to 1 to 3 cells per ml in LNHM, and 1 ml of each culture was dispensed into each well of 48-well plates. After 2 weeks of incubation at 16°C, 48-well plates were screened for cellular growth and purity using DAPI-stained epifluorescence microscopy and PCR-RFLP analyses. The positively scored cultures were diluted to fresh medium, and this process was repeated three times. Colony-forming activity was tested by spreading purified HTCC isolates on marine agar 2216 (MA2216; Difco, Detroit, Mich.), R2A agar in aged seawater (R2A) (44, 52), R2A agar diluted in aged seawater 1:10 (vol/vol) (1/10 R2A), 1/10 R2A agar in artificial seawater from Schut et al. (47), 1/10 R2A agar in artificial seawater from Zobell's formula 2 (50), 1/10 R2A agar in artificial seawater from DSMZ medium 246 (http://www.dsmz.de), and LNHM plus 1× MC agar. Colonies grown on solid agar media were purified, transferred to fresh liquid medium, and stored as 10% glycerol stock.
FIG. 1.

Neighbor-joining 16S rRNA phylogenetic tree showing relationships between the HTCC isolates and environmental sequences in the OMG group. Bootstrap proportions over 50% from the neighbor-joining analysis are shown. Brackets indicate the boundary of each subclade. Names in bold indicate 12 HTCC isolates used for growth characterization. Strain HTCC2149 in italics was not revived from the frozen stock. Seven different sequences in the genus Methylococcus were used as outgroup. Scale bar, 0.1 substitutions per nucleotide position.
Characterization of growth properties.
The growth characteristics of the 12 representative isolates in the OMG group were examined under different culture conditions, including various temperatures and carbon concentrations. For determination of optimum growth temperatures, cell densities were adjusted to approximately 800 cells ml−1 in LNHM plus 1× MC medium using cultures in the exponential growth phase. The cultures were incubated at 4, 10, 16, 20, 25, and 30°C. Cultures in exponential growth phase were also inoculated in LNHM medium amended with different concentrations of MC, including LNHM plus no MC, 0.1× MC, 0.5× MC, 1× MC, 5× MC, 10× MC, and 50× MC, and incubated at 16°C. The average dissolved organic carbon (DOC) concentrations of the LNHM medium (unamended seawater medium) measured by a TOC analyzer, which was made using Oregon coast seawater, were approximately 1.0 mg liter−1 (12), and the calculated DOC concentrations of 1× MC were 35 mg liter−1.
Cell densities, sizes, and morphologies were examined at 1- to 2 day-intervals by DAPI staining using an epifluorescence microscope (DMRB) equipped with a cooled charge-coupled device camera (ORCA-ER, Hamamatsu, Japan) and IPLab v3.5.6 scientific imaging software (Scanalytics, Fairfax, Va.). Cell sizes in exponential and stationary phases were measured from 15 images per sample (ca. 60 cells per image) taken by IPLab v3.5.6. Cell biovolume was calculated from length and width measurements using the following equation: biovolume = (π/4)w2[l − (w/3)], where l is cell length and w is cell width (39). Finally, specific growth rate (μ) was calculated from the exponential phase of the growth curve for every experimental set.
Nucleotide sequence accession numbers.
The sequences of the HTCC isolates used in the phylogenetic analyses have been deposited in GenBank under accession numbers AY386331 to AY386345.
RESULTS
Phylogenetic analyses of the OMG group.
For this study we selected HTCC isolates in the Gammaproteobacteria, which were not closely related to any previously identified bacterial strains. Nearly full 16S rRNA gene sequences were determined for phylogenetic analyses. Our findings led to the definition of the OMG group, which is composed of environmental clone sequences, a few oligotrophic isolates, and the subset of HTCC isolates, all derived from exclusively marine environments (Fig. 1). The OMG group is an association of the five clades, which often appears as a single clade in phylogenetic trees but is not well supported by bootstrap analyses (Fig. 1). The failure of these five clades to consistently form a single larger clade is a consequence of branch swapping with other marine Gammaproteobacteria, particularly “Endobugula” and Teredinibacter; hence, the OMG group appears to loosely define a diverse set of marine Gammaproteobacteria, which may share a common evolutionary origin with “Endobugula” and Teredinibacter, both of which are symbionts of marine vertebrates. Each OMG clade is defined in this study as a 16S rRNA gene lineage of environmental clones or isolates that are reproducibly monophyletic, supported with a high bootstrap value (>75%), and not affiliated with any other OMG clade by significant bootstrap values. Bootstrap support for the clades ranged from 88 to 100% in neighbor-joining trees and from 75 to 100% in maximum-parsimony trees. The clades were named after the first reported clone or isolate from each clade.
Three separate groups of HTCC isolates, represented by HTCC2148, HTCC2246, and HTCC2080, were assigned to the OM60 clade (12, 43), which was composed of environmental clones retrieved from coastal seawater and sediments (3, 18, 43, 54) and seawater isolates (12, 19). Sequence similarities within the OM60 clade ranged from 91.7 to 99.9%, making this a relatively diverse monophyletic clade. Strain HTCC2149 was not recovered from the original frozen stock, so it was removed from further study.
The BD1-7 and KI89A clades each contained only one HTCC isolate. They clustered tightly with deep-sea sediment clones (5, 36), a marine isolate (KI89C, GenBank description), and a seawater clone (KI89A). These two clades were well separated from the other subclades in the OMG group with high bootstrap support (>99%).
The OM182 clade contained several recently obtained marine environmental sequences and three separate groups of HTCC isolates, represented by strains HTCC2151, HTCC2178,and HTCC2188. This clade was clearly separated into two independent lineages with 100% bootstrap support, indicating that these two lineages may represent two separate genera within the class Gammaproteobacteria. Although sequence similarities between the two lineages were very low, in the range of 88.3 to 91.0%, the high level of bootstrap support (88%) between the two lineages made it possible to define this as a single clade. Strains HTCC2151 and HTCC2180 were most closely related to strain HTCC2178 (96.2 to 96.5% similar). Strain HTCC2188 in the other lineage was 94.5 to 97.9% similar to the coastal clones OM182 and OM185 (43), the Arctic Ocean clones (2), and marine sediment clones.
Three groups of HTCC isolates, including strains HTCC2121,HTCC2290, and HTCC2207, together with Sargasso Sea clone SAR92 (6), Pacific coastal clone MB11B11 (51), and North Sea clone ZD0424 (60), comprised the SAR92 clade (12). Sequence similarities of members within the SAR92 clade were very high, from 96.1 to 99.9%, resulting in this clade forming a distinct lineage with high bootstrap support.
Abundance of the OMG group among HTCC isolates.
A total of 185 HTCC isolates were cultivated out of 1,760 wells screened over a 1-year period from four sample collections (Table 1). The culturability of the pelagic seawater sample (15 HTCC isolates per 320 wells screened) was much lower than that of the Oregon coastal samples (33 to 76 HTCC isolates per 480 wells screened). Of the 185 HTCC isolates, 44 HTCC isolates (23.8%) were assigned to the OMG group (Table 1). In addition, 47 isolates in the Roseobacter clade (25.4%) and 24 isolates in SAR11 clade (12.9%) were detected (data not shown). HTCC isolates in the OM60, OM182, and SAR92 clades were recovered from nearly all of the Oregon coast seawater samples, while two isolates in the BD1-7 and KI89A clades were detected only in the jetty sample collected in August 2001. The SAR92 clade was the most abundant subclade among the five clades; its members were recovered from both coastal and Pacific pelagic seawater samples.
TABLE 1.
Numbers of HTCC isolates in the OMG group cultivated by high-throughput culturing over a 1-year period
| Date and location of inoculation samplea | No. of wells screened | No. of bacterial isolates identified | Culture designations | No. of isolates in OMG subclade
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| OMG (total) | OM60 | BD1-7 | KI89A | OM182 | SAR92 | ||||
| 8-20-01, J | 480 | 76 | HTCC2068 to HTCC2144 | 10 | 2 | 1 | 1 | 0 | 6 |
| 9-20-01, OC | 480 | 33 | HTCC2145 to HTCC2180 | 17 | 5 | 0 | 0 | 7 | 5 |
| 8-7-02, Jetty | 480 | 61 | HTCC2181 to HTCC2282 | 15 | 3 | 0 | 0 | 2 | 10 |
| 8-17-02, PO | 320 | 15 | HTCC2283 to HTCC2297 | 2 | 0 | 0 | 0 | 0 | 2 |
| Total | 1,760 | 185 | 44 | 10 | 1 | 1 | 9 | 23 | |
Samples were collected from the jetty, Newport, Oreg. (J), 27.6 km off the coast of Oregon (OC, 44° 39.1′ N, 124° 24.7′ W), and a pelagic region of the Pacific Ocean (PO, 48° 4.82′ N 132° 41.03′ W). Dates are expressed as month-day-year.
Growth characteristics of the HTCC isolates.
Morphology, cell sizes, and biovolumes of isolates in the OMG group were very diverse (Fig. 2). DAPI-stained images of the isolates revealed that they ranged from spherical cocci or rods to straight rods, were generally of small size, and were exclusively unicellular organisms dividing by binary fission. The cell sizes and morphologies of some HTCC isolates were clearly different, even though they were in the same clade. For example, strain HTCC2290 in the SAR92 clade was a straight rod, while strains HTCC2121 and HTCC2207 in the same SAR92 clade were short rods and strain HTCC2290 was three to four times larger in cell length than strains HTCC2121 and HTCC2207 (Fig. 2, panels J, K, and L). All HTCC isolates reduced their sizes during growth (Fig. 2). Nine of the twelve HTCC isolates were less than 1 μm in length in the exponential phase, and all isolates were less than 1 μm in the stationary phase. Due to their small sizes, the biovolume of many isolates was less than 0.1 μm3; they could therefore be defined as “ultramicrocells” (16, 47, 48).
FIG. 2.

Epifluorescence micrographs of DAPI-stained exponential-phase (uppercase panel letters) and stationary-phase (lowercase panel letters) cells of HTCC isolates in the OMG group. Panels: (A, a) HTCC2148, (B, b) HTCC2246, (C, c) HTCC2080, (D, d) HTCC2143, (E, e) HTCC2089, (F, f) HTCC2151, (G, g) HTCC2180, (H, h) HTCC2178, (I, i) HTCC2188, (J, j) HTCC2121, (K, k) HTCC2290, (L, l) HTCC2207. Scale bars, 1 μm.
When initial extinction cultures obtained from seawater samples were spread on solid agar surfaces, none of 44 HTCC isolates in the OMG group formed colonies on either 1/10 R2A agar or LNHM plus 1× MC agar. Seven of the 12 purified HTCC isolates, however, produced colonies on 1/10 R2A agar after the 12 isolates were reinoculated into fresh LNHM medium at a dilution of 1 cell ml−1 three times (Table 2). Growth curves of the 12 HTCC isolates were determined under various growth conditions, and maximum specific growth rates were calculated from exponential growth kinetics. These isolates were generally psychro- to mesophilic bacteria that grew best at 16 to 20°C in LNHM to LNHM plus 1× MC media. The μ values of the isolates at 16°C in LNHM and LNHM plus 1× MC ranged from 1.23 to 2.63 day−1 and 1.57 to 2.32 day−1, respectively (Table 2), and these values were much lower than μ value of standard laboratory marine organisms, such as Vibrio sp. and Alteromonas sp.
TABLE 2.
Growth characteristics of the representative HTCC isolates in the OMG group
| Clade | Strain | Specific growth rate at 16°C (day−1)
|
Maximum cell density at 16°C (106 cells ml−1)
|
Growth on agar mediaa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LNHM | LNHM + 1× MC | LNHM | LNHM + 1× MC | 1/10 R2A | R2A | MA 2216 | ASWS | ASWZ | DSMZ246 | ||
| OM60 | HTCC2148 | 1.57 | 2.19 | 1.05 | 3.03 | G | − | − | − | G | G |
| HTCC2246 | 2.09 | 2.11 | 0.89 | 0.87 | G | − | − | − | − | − | |
| HTCC2080 | 2.21 | 2.22 | 1.29 | 5.71 | G | − | − | − | − | − | |
| BD1-7 | HTCC2143 | 2.11 | 2.32 | 0.27 | 2.25 | G | − | − | G | G | G |
| KI89A | HTCC2089 | 1.56 | 2.01 | 0.68 | 1.47 | − | − | − | − | − | − |
| OM182 | HTCC2151 | 1.35 | 1.75 | 1.28 | 4.00 | − | − | − | − | − | − |
| HTCC2180 | 1.23 | 1.57 | 0.83 | 3.49 | − | − | − | − | − | − | |
| HTCC2178 | 1.94 | 1.65 | 0.64 | 1.20 | G | G | G | − | G | G | |
| HTCC2188 | 2.63 | 1.83 | 1.32 | 5.00 | G | G | − | − | − | − | |
| SAR92 | HTCC2121 | 1.18 | 1.61 | 1.26 | 9.93 | − | − | − | − | − | − |
| HTCC2290 | 1.36 | 1.65 | 0.73 | 3.99 | − | − | − | − | − | − | |
| HTCC2207 | 2.05 | 2.27 | 1.34 | 15.7 | G | − | − | − | − | − | |
Growth on agar media was determined at 16°C. The inocula were the HTCC cultures adapted to laboratory conditions after successive purifications. The original HTCC extinction cultures obtained from natural seawaters did not form colonies on LNHM plus 1 × MC and 1/10 R2A agar media. ASWS, artificial seawater from Schut et al. (47); ASWZ, 1/10 R2A agar in artificial seawater from Zobell's formula 2 (50); DSMZ246, 1/10 R2A agar in artificial seawater from DSMZ medium 246; G, growth; −, no growth.
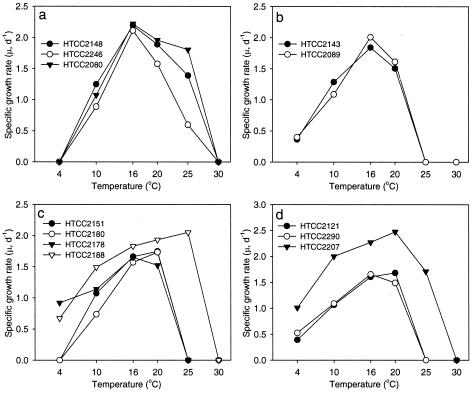
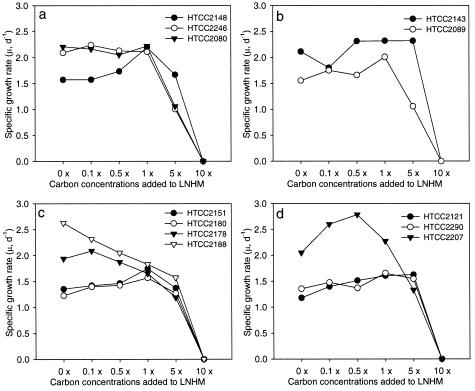
The specific growth rates of the HTCC isolates under various growth conditions were summarized in graphs and compared to each other (Fig. 3 and 4). None of the HTCC isolates grew at 30°C, only 5 of the 12 HTCC isolates grew at 25°C, and 7 isolates grew at 4°C. All five strains in the OM60, BD1-7, and KI89A clades showed a maximum μ value at 16°C, while most strains in the OM182 and SAR92 clades (five of seven) showed a maximum μ value at 20 to 25°C (Fig. 3). From this result, temperature range and optima for growth could be one of the criteria for differentiating clades or strains within the OMG group. None of the strains grew in LNHM plus 10× MC medium (ca. 350 mg of DOC liter−1), and the μ value for 10 HTCC isolates was reduced dramatically in LNHM plus 5× MC medium (Fig. 4). This indicated that all of the HTCC isolates in the OMG group have typical characteristics of marine obligately oligotrophic bacteria. Strains HTCC2188 and HTCC2178 in the OM182 clade were typical examples of oligotrophic bacteria; HTCC2188 and HTCC2178 grew best in LNHM medium (ca. 1.0 mg of DOC liter−1) and LNHM plus 0.1× MC (ca. 4.5 mg of DOC liter−1), respectively, and high concentrations of DOC reduced their specific growth rates (Fig. 4c). Interestingly, growth characteristics of the HTCC isolates were generally related to their phylogenetic positions. Strains HTCC2151 and HTCC2180 in the OM182 clade showed 99.6% 16S ribosomal DNA sequence similarity and had very similar growth patterns. Strain HTCC2188, which was the most distantly related to strains HTCC2151 and HTC2180, however, had growth characteristics that were quite different from those of the two strains (Fig. 3c and 4c). Similar results were found for members of the SAR92 clade; strain HTCC2207, the most phylogenetically distant of the branches within the SAR92 clade, was distinct from strains HTCC2121 and HTCC2290 in characteristics such as temperature range and optimum, maximum cell densities, and optimum concentrations of DOC (Fig. 3d and 4d).
FIG. 3.
Variation of specific growth rates (μ) of HTCC isolates in the OMG group at different temperatures. Panels: (a) HTCC isolates in the OM60 clade; (b) HTCC isolates in the BD1-7 and KI89A clades; (c) HTCC isolates in the OM182 clade; (d) HTCC isolates in the SAR92 clade. d, day.
FIG. 4.
Variation of specific growth rates (μ) of HTCC isolates in the OMG group at different DOC concentrations. Panels: (a) HTCC isolates in the OM60 clade; (b) HTCC isolates in the BD1-7 and KI89A clades; (c) HTCC isolates in the OM182 clade; (d) HTCC isolates in the SAR92 clade. d, day.
DISCUSSION
The recovery of 16S rRNA genes and isolates of the OMG group from different geographic regions and different types of seawater suggests that this is a cosmopolitan group of Gammaproteobacteria. Members of the group have been found in clone libraries, denaturing gradient gel electrophoresis bands, and isolates retrieved from the Pacific Ocean (3, 12, 13, 51), Atlantic Ocean (6, 27, 32, 43), North Sea (19, 60), Arctic Ocean (2), Antarctic Sea (5), Mediterranean Sea (46), Gulf of Elat (58), Tokyo Bay (54), Suruga Bay (36), and Aegean Sea (AEGEAN-133; GenBank description). The majority of OMG representatives have been retrieved from the surface layer of coastal seawaters (3, 12, 13, 19, 32, 43, 46, 51), while other members have been found in diverse marine ecological settings, such as surface or deep layers of the open ocean (2, 6, 18, 60), shallow or deep marine sediments (5, 36, 54), and even in sea grass (58). Most cultured members of the OMG group have been obtained from temperate environments (12, 19), but tropical strains affiliated with the OMG group were very recently isolated from the Sargasso Sea, Atlantic Ocean (M. S. Rappé and S. J. Giovannoni, unpublished results). Thus, biogeographic information based on 16S rRNA genes indicates that OMG members are cosmopolitan, autochthonous marine microorganisms which inhabit diverse marine habitats.
There have not been any comprehensive phylogenetic analyses of the OMG group, even though members of this group have been repeatedly found in seawater clone libraries. This might be explained by the relatively low abundance of this group in clone libraries. Since members of the OMG group comprise less than 3% of the total number of clones in seawater clone libraries typically, only one or two of the OMG clades are represented in each library (43, 60). Although members of the OMG group are infrequent in clone libraries, a recent fluorescence in situ hybridization (FISH) study (19) showed that the number of bacteria in the OM60 clade accounted for as many as 1.5 × 105 cells ml−1 (8% of DAPI counts) in surface waters of the North Sea and 33 to 66% of FISH-detectable Gammaproteobacteria. Similarly, FISH counts of bacteria related to OM182 and OM185 in the OM182 clade comprised 1.3 to 3.7% of total DAPI counts at NH5 station off the Oregon coast over a 1-year period (C. S. Alexander and S. J. Giovannoni, unpublished results). Therefore, overall in situ abundance of members of the OMG group may be higher than indicated by clone libraries. Further studies are required for determining the in situ abundance of the entire OMG group.
Although the definitions of the oligotroph, obligate oligotroph, and facultative oligotroph have been the subject of debate for several decades (reviewed in reference 49), it is widely accepted that a general characteristic of oligotrophic bacteria is the ability for growth in low-nutrient (0.5 to 15 mg of C liter−1) media, irrespective of whether they grow in high nutrient media or not. We observed that all 12 representative isolates in the OMG group grew well in carbon-unamended natural seawater medium (LNHM, 1.0 mg of C liter−1) to 1× MC-amended LNHM (36.0 mg of C liter−1) and that addition of MC (176 mg of C liter−1) significantly reduced specific growth rates. No isolates grew in the medium containing more than 351 mg of DOC liter−1. According to the definition that obligately oligotrophic bacteria cannot grow at substrate concentrations above 0.3 g of C liter−1 (28) or 6 g of C liter−1 (23) and the fact that they were unable to grow in nutrient-rich media upon first cultivation of organisms from natural habitats (7, 47, 48), the HTCC isolates in the OMG group are obligately oligotrophic bacteria.
HTCC isolates in the OMG group, therefore, can be considered model oligotrophic bacteria, similar to well-studied oligotrophic species, such as Sphingomonas alaskensis (17, 55) and “Candidatus Pelagibacter ubique” (42) in the SAR11 clade. Since S. alaskensis strain RB2256T was first isolated from Resurrection Bay, Alaska (7, 47), many studies have defined this species as a model oligotrophic marine ultramicrobium for the following reasons. It was able to grow in very low nutrient media (<1 mg of C liter−1) but not in 5 mg of DOC liter−1, and it has an ultramicrosize of <0.1 μm3, a relatively low growth rate (μ = <0.2 h−1), and high-affinity substrate uptake systems (7, 16, 17, 20, 21, 40, 47, 48). After prolonged storage at 4 to 5°C and laboratory adaptation, strains RB2256T and AFO1 formed colonies on nutrient-rich media and became facultative oligotrophs (17, 47, 48). The oligotrophic characteristics of the HTCC isolates in the OMG group were comparable to those of S. alaskensis. None of the isolates formed colonies on low-nutrient media (LNHM plus 1× MC agar, 1/10 R2A) or relatively high-nutrient medium (marine agar 2216) upon their first isolation from seawater. The specific growth rate of the isolates (0.05 to 0.11 h−1) was generally lower than that of strain RB2256T (0.13 to 0.16 h−1). None of the isolates grew in the medium containing 351 mg of DOC liter−1, while strain RB2256T grew well in the medium containing 800 mg of DOC liter−1 (16). Most OMG isolates, therefore, still possess the obligately oligotrophic properties of marine bacteria rather than the facultatively oligotrophic features of S. alaskensis. Similar to S. alaskensis, the biovolume of many isolates in the group (5 of 12 in exponential phase, 9 of 12 in stationary phase) was less than 0.1 μm3. At least five isolates in the group retained a constant ultramicrosize (<0.1 μm3), irrespective of growth stage or nutrient conditions. Because ultramicrobial cells dominate ocean bacterioplankton communities (4, 35) and because very few ultramicrobial strains have been isolated from oceanic ecosystems, the cultivation of various ultramicrocells in the OMG group provides the first evidence that many of ultramicrobial cells are readily culturable when provided with appropriate culture conditions.
Based on tree topologies and 16S ribosomal DNA sequence similarities between representative isolates, the OMG group appears to be comprised of five to seven novel genera. If we used the practical 3% cutoff value for 16S rRNA divergence to demarcate species (57), HTCC isolates in the group could be divided into a maximum of 10 species. However, it has been suggested that distinctions between environmental clades in tree topologies might be a better basis than sequence similarity for drawing taxonomic distinctions, particularly when the clades correlate with their phenotypic properties (11). Since the different physiological responses to environmental factors, such as DOC and temperature, were observed to correlate with phylogenetic positions of the HTCC isolates in the OMG group, it may be that this group consists of several ecologically differentiated species, or ecotypes (11). For example, strain HTCC2207 in the SAR92 clade, which is closely related (>96%) to the other two HTCC strains in the same clade, showed clear differences in its response to temperature and DOC. Therefore, strain HTCC2207 can probably be differentiated as a separate species from the other strains. It is premature to conclude the taxonomy of the HTCC isolates in the OMG group before assessing such information as DNA-DNA relatedness, DNA base composition, mode of energy metabolism, and biochemical properties required for classification. Future polyphasic taxonomy (56) of the OMG isolates based on genotypic and phenotypic characteristics will provide expanded information regarding diversity of the OMG group. Finally, future physiological and genomic research on the isolates will elucidate their role in oceanic biogeochemical cycling and the biochemical features which render them oligotrophic.
Acknowledgments
We acknowledge Stephanie Connon, who set up most of the high-throughput culturing and provided advice throughout this study. We thank Michael Rappé and Christopher Alexander for sharing their unpublished results. We also thank Kevin Vergin, Carol DiMeo, Markus Moseneder, Robert Morris, and Jim Tripp for critical review. Special thanks are given to Eun Jung Shin for her technical assistance with preparing cell arrays and media.
This study was supported by grants from Diversa Corp. and by National Science Foundation grant DEB-0207085.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 4.Børsheim, K. Y., G. Bratbak, and M. Heldal. 1990. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hilds, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 6.Britschgi, T. B., and S. J. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Button, D. K., F. Schut, P. Quang, R. Martin, and B. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, J.-C., and S. J. Giovannoni. 2003. Croceibacter atlanticus gen. nov., sp. nov., a novel marine bacterium in the family Flavobacteriaceae. Syst. Appl. Microbiol. 26:76-83. [DOI] [PubMed] [Google Scholar]
- 9.Cho, J.-C., and S. J. Giovannoni. 2003. Fulvimarina pelagi gen. nov., sp. nov., a marine bacterium that forms a deep evolutionary lineage of descent in the order “Rhizobiales.” Int. J. Syst. Evol. Microbiol. 53:1853-1859. [DOI] [PubMed] [Google Scholar]
- 10.Cho, J.-C., and S. J. Giovannoni. 2003. Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the α-Proteobacteria. Int. J. Syst. Evol. Microbiol. 53:1031-1036. [DOI] [PubMed] [Google Scholar]
- 11.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 12.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, H. C., and R. R. L. Guillard. 1958. Relative value of ten genera of micro-organisms as food for oyster and clam larvae. USFWS Fish Bull. 58:293-304. [Google Scholar]
- 15.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eguchi, M., T. Nishikawa, K. MacDonald, R. Cavicchioli, J. C. Gottschal, and S. Kjelleberg. 1996. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 62:1287-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contribution to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fegatella, F., and R. Cavicchioli. 2000. Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 66:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry, J. C. 1990. Oligotrophs, p. 93-116. In C. Edwards (ed.), Microbiology of extreme environments. Open University Press, Milton Keynes, United Kingdom.
- 24.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-166. In G. M. Garrity (ed.), Bergey's manual of systemic bacteriology, 2nd ed., vol. 1. Springer, New York, New York.
- 25.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, Inc., New York, N.Y.
- 26.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 27.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida, Y., M. Eguchi, and H. Kadota. 1986. Existence of obligately oligotrophic bacteria as a dominant population in the south China Sea and the west Pacific Ocean. Mar. Ecol. Prog. Ser. 30:197-203. [Google Scholar]
- 29.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 31.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 32.Kelly, K. M., and A. Y. Chistoserdov. 2001. Phylogenetic analysis of the succession of bacterial communities in the Great South Bay (Long Island). FEMS Microbiol. Ecol. 35:85-95. [DOI] [PubMed] [Google Scholar]
- 33.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 34.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 35.Lee, S., and J. A. Fuhrman. 1987. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 37.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 38.Moore, J. L., C.-Y. Ou, and G. Schochetman. 1991. Use of UV irradiation to reduce false positivity in polymerase chain reaction. BioTechniques 10:442-446. [PubMed] [Google Scholar]
- 39.Norland, S., M. Heldal, and O. Tumyr. 1987. On the relation between dry matter and volume of bacteria. Microb. Ecol. 12:95-101. [DOI] [PubMed] [Google Scholar]
- 40.Ostrowski, M., R. Cavicchioli, M. Blaauw, and J. C. Gottschal. 2001. Specific growth rate plays a critical role in hydrogen peroxide resistance of the marine oligotrophic ultramicrobacterium Sphingomonas alaskensis strain RB2256. Appl. Environ. Microbiol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 42.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 43.Rappé, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811-826. [Google Scholar]
- 44.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Schäfer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 47.Schut, F., E. J. DeVries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schut, F., J. C. Gottschal, and P. A. Rudolf. 1997. Isolation and characterisation of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20:363-369. [Google Scholar]
- 49.Schut, F., R. A. Prins, and J. C. Gottschal. 1997. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat. Microb. Ecol. 12:177-202. [Google Scholar]
- 50.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 611-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular microbiology. American Society for Microbiology, Washington, D.C.
- 51.Suzuki, M. T., O. Beja, L. T. Taylor, and E. F. DeLong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among SSU rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford, D. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4b8 ed. Sinauer Associates, Sunderland, Mass.
- 54.Urakawa, H., T. Yoshida, M. Nishimura, and K. Ohwada. 2000. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ. Microbiol. 2:542-554. [DOI] [PubMed] [Google Scholar]
- 55.Vancanneyt, M., F. Schut, C. Snauwaert, J. Goris, J. Swings, and J. C. Gottschal. 2001. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. Evol. Microbiol. 51:73-79. [DOI] [PubMed] [Google Scholar]
- 56.Vandamme, P., B. Pot, M. Gillis, P. De Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 58.Weidner, S., W. Arnold, and A. Pühler. 1996. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microbiol. 62:766-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zengler, K., G. Toledo, M. S. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]