SUMMARY
Transforming growth factor beta (TGF-β) signaling, mediated through the transcription factors Smad2 and Smad3 (Smad2/3), directs different responses in different cell types. Here we report that Smad3 co-occupies the genome with cell-type-specific master transcription factors. Thus, Smad3 occupies the genome with Oct4 in embryonic stem (ES) cells, Myod1 in myotubes, and PU.1 in pro-B cells. We find that these master transcription factors are required for Smad3 occupancy and that TGF-β signaling largely affects the genes bound by the master transcription factors. Furthermore, we show that induction of Myod1 in non-muscle cells is sufficient to re-direct Smad3 to Myod1 sites. We conclude that cell-type-specific master transcription factors determine the genes bound by Smad2/3 and are thus responsible for orchestrating the cell-type-specific effects of TGF-β signaling.
INTRODUCTION
Signaling pathways allow cells to respond to their environment and frequently act by regulating gene expression. The terminal components of these pathways tend to occupy the genes they regulate (Darnell et al., 1994; Jarriault et al., 1995; Kim et al., 1997; Molenaar et al., 1996; Pokholok et al., 2006; Sen and Baltimore, 1986) and modulate gene expression through activities that include recruitment of co-activators and chromatin remodeling machinery, modification of transcription factors, and activation of transcription (Clevers, 2006; Guasconi and Puri, 2009; Kopan and Ilagan, 2009; Massague et al., 2005; Natoli, 2009; O’Shea et al., 2002). In this way external signals produce transcriptional responses that allow cells to respond to cues from their environment.
Signaling pathways are required throughout development and play essential roles in numerous disease processes. It is notable that any one signaling pathway can direct very different responses in different cell types (Clevers, 2006; Guasconi and Puri, 2009; Kopan and Ilagan, 2009; Massague et al., 2005; O’Shea et al., 2002). How an extracellular signal produces cell-type-specific responses remains poorly understood, but these diverse responses govern nearly every aspect of cell physiology from growth to differentiation and death.
TGF-β signaling regulates processes that include stem cell maintenance, cell proliferation, differentiation, and apoptosis (Massague et al., 2005; Ross and Hill, 2008). Activation of the TGF-β receptor by TGF-β, Activin or Nodal leads to phosphorylation of the transcription factors Smad2 and Smad3 (Smad2/3). Once phosphorylated, these transcription factors accumulate in the nucleus in association with Smad4 (Massague et al., 2005; Ross and Hill, 2008). Smad3 and the less common isoform of Smad2 can both bind DNA directly through interaction with the Smad binding element (SBE) (Dennler et al., 1998; Shi et al., 1998; Zawel et al., 1998). However, due to the low affinity of this interaction, Smad transcription factors must interact with additional transcription factors in order to form stable complexes with DNA (Massague et al., 2005; Ross and Hill, 2008; Shi et al., 1998).
We mapped genome-wide binding of Smad3 in ES cells, myotubes and pro-B cells and found that a small set of cell-type-specific master transcription factors direct Smad3 to cell-type-specific binding sites and determine cell-type-specific responses to TGF-β signaling. These results are surprising as previous work has suggested that many different transcription factors in a cell are each responsible for directing Smad3 binding to a small number of sites, and it is the sum of these interactions that determines the cell-type-specific response to TGF-β signaling (Massague and Gomis, 2006; Massague et al., 2005; Seoane et al., 2004). Furthermore, we find that master transcription factors help direct Smad3 binding by establishing open chromatin that contains SBEs, allowing Smad3 to bind DNA and form a physical complex with the master transcription factors.
RESULTS
SMAD3 co-occupies the genome with OCT4 in human and murine ES cells
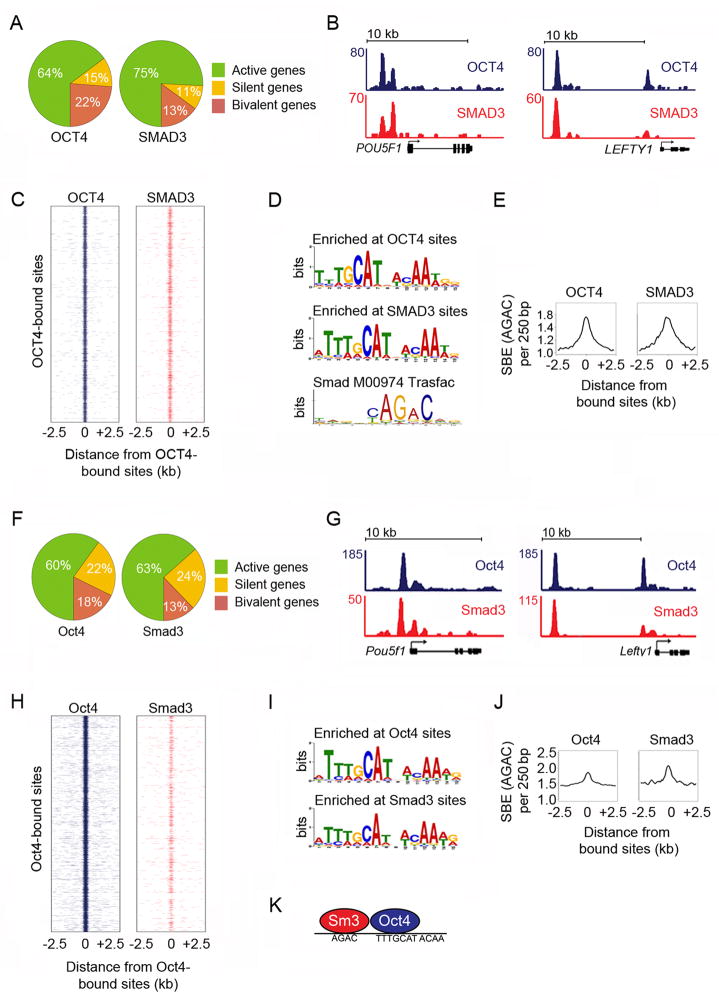
We first investigated whether SMAD3 is directed to DNA sites co-occupied by the master transcription factor OCT4 in human (h) ES cells, where activation of SMAD2/3 is required to maintain hES cell identity (Beattie et al., 2005; James et al., 2005; Vallier et al., 2005; Vallier et al., 2009; Xu et al., 2008). ChIP-seq was performed to determine the genome-wide targets of SMAD3 and OCT4 in hES cells. If SMAD3 is directed to DNA targets by the ES cell-specific master transcription factor OCT4, we would expect to observe that SMAD3 and OCT4 bind to the same sets of genes across the genome. Analysis of the gene targets showed that SMAD3 and OCT4 were predominantly associated with active genes and had a similar distribution to each other across active, silent and bivalent genes (Fig 1A).
Figure 1. SMAD3 and OCT4 co-occupy the genome in ES cells.
(A) Distribution of genes bound by OCT4 (left) and SMAD3 (right) across active, silent and bivalent genes in hES cells (Table S3). In all experiments, hES cells were grown in mTESR1 media, which contains TGF-β. Refer to Extended Experimental Procedures for details of hES cell culture and gene assignments.
(B) SMAD3 and OCT4 co-occupy DNA sites in hES cells. Gene tracks represent binding of OCT4 (blue) and SMAD3 (red) at POU5F1, the gene encoding OCT4 (left) and LEFTY1 (right). The x-axis represents the linear sequence of genomic DNA and the y-axis represents the total number of mapped reads with the floor set at 2 counts unless specified otherwise. The genomic scale in kilobases (kb) is indicated above each track.
(C) SMAD3 and OCT4 co-occupy the genome. Binding plots show the location of OCT4- (left) and SMAD3- (right) bound sites relative to 7,532 OCT4-bound sites. For each OCT4-bound site (y-axis) the presence of OCT4 (blue) and SMAD3 (red) sites are displayed within a 5 kb window centered on the OCT4-bound site. Intensity at position 0 indicates that sites overlap.
(D) SMAD3 binding sites are enriched for the OCT4 motif. The most enriched motifs at OCT4-bound sites (top) and SMAD3-bound sites (center) were identified using MEME (Bailey and Elkan, 1994) (Fig S1). The canonical Smad Transfac motif (Smad binding element) (Matys et al., 2003) is shown.
(E) The Smad binding element (SBE) is enriched at both OCT4- and SMAD3-bound sites. The histogram shows the average occurrence of the canonical SBE in a 250 bp window (y-axis) relative to the distance from the peak (x-axis) of OCT4- (left) or SMAD3-bound sites (right).
(F) Distribution of genes bound by Oct4 (left) and Smad3 (right) across active, silent and bivalent genes in mES cells. All mES cells analyzed in Fig 1 were grown for 2 passages off feeders without addition of exogenous Activin or TGF-β (see Extended Experimental Procedures). The TGF-β signaling pathway is active under these standard mES cell culture conditions (Fig S1C).
(G) Oct4 and Smad3 co-occupy DNA sites in mES cells. Gene tracks represent binding of Oct4 (blue) and Smad3 (red) at Pou5f1 (left) and at Lefty1 (right).
(H) Smad3 and Oct4 co-occupy the genome. For each of the 15,003 Oct4-bound sites (y-axis) the presence of Oct4 (blue) and Smad3 (red) are displayed within a 5 kb window centered on each Oct4-bound site.
(I) Smad3 binding sites are enriched for the Oct4 motif. Motif discovery was performed using the murine Oct4 and Smad3-bound sites.
(J) The SBE is enriched at both Oct4 and Smad3-bound sites. The histogram of canonical SBE frequency (y-axis) was generated as described in E using murine Oct4 (left) and Smad3 sites (right).
(K) Smad3 (Sm3) and Oct4 co-occupy the genome in ES cells by binding nearby DNA sites. The binding motif for each factor is displayed. See also Figure S1, Table S1–S2.
If SMAD3 is directed to target genes by OCT4, then SMAD3 should co-occupy DNA sites with OCT4, whereas if SMAD3 is directed to target genes by many different transcription factors, the global binding of SMAD3 and OCT4 should not be coincident. Examination of SMAD3 and OCT4 binding at individual hES cell genes revealed that SMAD3 occupies sites with OCT4; for example the two transcription factors bind the same sites at POU5F1 and LEFTY1 (Fig 1B). Furthermore, this binding pattern occurs throughout the genome (Fig 1C, Table S1). Indeed, over 80% of the 1000 highest-confidence SMAD3-bound sites are co-occupied by OCT4 (p < 1e-290, Table S1, S2).
It is possible that SMAD3 co-occupies the genome with many different factors, only one of which is OCT4. If this were true, then many different DNA binding motifs would be present at sites bound by SMAD3. However, if SMAD3 binding is most highly associated with OCT4, the OCT4 motif should be most enriched at SMAD3 sites. De novo motif discovery was performed (Bailey and Elkan, 1994) on sites bound by SMAD3 and sites bound by OCT4. The most enriched motif identifiable at sites bound by each factor was indeed the OCT4 motif (Fig 1D, Fig S1). The canonical SBE was not found by de novo motif discovery at sites bound by SMAD3 (Fig 1D, bottom), which likely reflects the difficulty in determining enrichment of a four-nucleotide motif. However, if SMAD3 and OCT4 co-occupy DNA, then SBEs should be enriched at SMAD3 and OCT4 sites. We scanned the DNA sequence around the sites bound by each factor and found that SBEs are enriched at sites bound by SMAD3 and sites bound by OCT4 (Fig 1E). These results are consistent with the model that in hES cells, SMAD3 predominantly occupies sites with OCT4 throughout the genome.
While human and murine (m) ES cells respond differently to TGF-β signaling (James et al., 2005), both cell types require the master transcription factor Oct4 to maintain cell identity (Chambers and Smith, 2004), so we investigated whether Smad3 also co-occupies sites with Oct4 in mES cells. Global analysis of Smad3 and Oct4 binding in mES cells revealed that these transcription factors occupy the same classes of genes as each other and as their counterparts in hES cells (Fig 1F). Further analysis of Oct4 and Smad3 binding revealed that the two transcription factors bind to the same sites at individual genes (Fig 1G) and throughout the genome (Fig 1H, p <1e-300). Furthermore, the Oct4 DNA motif was the most enriched motif at sites bound by Smad3 and sites bound by Oct4 (Fig 1I) and these sites were also enriched for SBEs (Fig 1J). These results show that Smad3 co-occupies the genome with the master transcription factor Oct4 in both human and murine ES cells (Fig 1K).
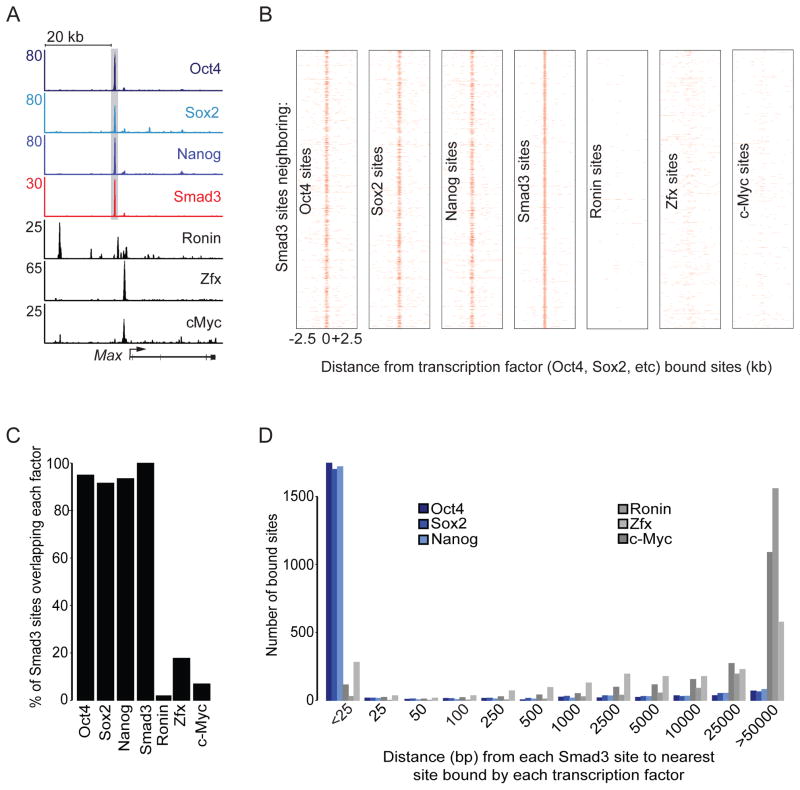
Smad2/3 has been shown to interact with many different transcription factors (Massague et al., 2005; Ross and Hill, 2008), and the analysis described thus far does not eliminate the possibility that Smad3 co-occupies sites with many different transcription factors other than Oct4 in ES cells. To determine if Smad3 co-occupies sites with other characterized transcription factors, we analyzed ChIP-seq data for six different factors in mES cells. Examination of binding data at individual genes such as that encoding Max (Fig 2A) revealed that Smad3 co-occupied sites with Oct4 as well as Sox2 and Nanog, which are key ES cell transcription factors that occupy the genome together (Boyer et al., 2005; Chen et al., 2008; Marson et al., 2008). In contrast, Smad3 did not occupy sites bound by Ronin, Zfx, or c-Myc, despite previous evidence that c-Myc can interact with Smad2/3 in other cell types (Chen et al., 2008; Dejosez et al., 2010; Feng et al., 2002). Genome-wide analysis confirmed that Smad3 tended to co-occupy sites with Oct4, Sox2 and Nanog but rarely co-occupied sites with Ronin, Zfx, or c-Myc (Fig 2B, FigS2A, Table S1). We further quantified the preference for Smad3 to co-occupy sites with these factors by calculating the percent of Smad3 sites co-occupied by each factor (Fig 2C) and determining the distance from each Smad3 site to the nearest binding site for each transcription factor (Fig 2D). Over 90% of the top 1000 Smad3 sites were co-occupied by Oct4, Sox2 and Nanog individually while only a small fraction of Smad3 sites were co-occupied by Ronin, Zfx, or c-Myc (Fig 2C). Furthermore, the majority of sites occupied by Ronin, Zfx or c-Myc are located far from sites occupied by Smad3 (Fig 2D). Thus in mES cells, Smad3 tends to co-occupy the genome specifically at sites bound by Oct4, Sox2 and Nanog.
Figure 2. Smad3 co-occupies the genome with the ES cell core master transcription factors.
(A) Gene tracks represent binding of Oct4, Sox2, Nanog (Marson et al., 2008), Smad3, Ronin (Dejosez et al., 2010), Zfx and c-Myc (Chen et al., 2008) at Max. Gray shading highlights co-occupied sites.
(B) Smad3 and the core master transcription factors co-occupy the genome. The distribution of Smad3-bound sites (red) is shown relative to all bound sites for the indicated transcription factors (y-axis) in a 5 kb window centered on the bound sites for each transcription factor. ChIP-seq performed using an antibody against Smad2/3 showed similar results to Smad3 (Fig S2 B–D).
(C) Smad3 co-occupies the genome with specific transcription factors. The percentage of Smad3 sites (y-axis) co-occupied by each transcription factor (x-axis) is shown. Co-occupancy is defined as greater than or equal to one base pair overlap between sites occupied by each factor. The 1000 strongest Smad3 binding sites were used for this analysis.
(D) Ronin, Zfx, and c-Myc binding is not associated with Smad3. The distance from the center of each Smad3 site to the center of the nearest site bound by the indicated transcription factor was determined. These distances were grouped into bins (x-axis). The sum of bound sites in each bin is shown (y-axis). See also Figure S2.
Oct4 recruits Smad3
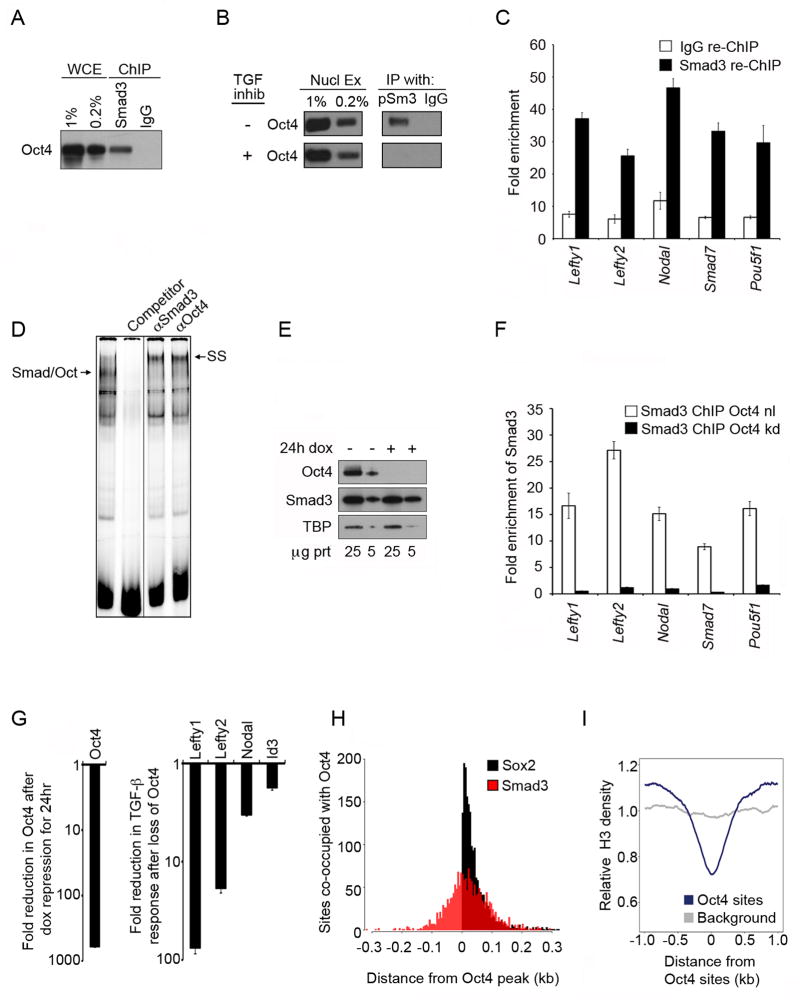
If Oct4 directs Smad3 to sites in ES cells then we would expect Oct4 and Smad3 to co-occupy the same sites at the same time. We performed ChIP for Smad3 in mES cells, followed by Western analysis to detect Oct4, which revealed that Smad3 and Oct4 were contained in the same cross-linked complex (Fig 3A). In addition, co-immunoprecipitation (Co-IP) experiments showed that Smad3 and Oct4 form a physical complex that is dependent on TGF-β signaling (Fig 3B). We then performed ChIP for Oct4 followed by re-ChIP for Smad3 (ChIP-re-ChIP) to determine if Smad3 and Oct4 occupied binding sites in mES cells simultaneously (Fig 3C). The results showed that Oct4 and Smad3 did temporally co-occupy DNA sites. We then asked if Oct4 and Smad3 co-occupy the Lefty1 enhancer by electrophoretic mobility shift analysis (EMSA); antibodies against both Smad3 and Oct4 caused a supershift of the complex suggesting that both Oct4 and Smad3 were part of the complex bound to the Lefty1 enhancer (Fig 3D).
Figure 3. Oct4 recruits Smad3.
(A) Smad3 and Oct4 are part of the same complex. ChIP was performed for Smad3 and IgG in mES cells followed by Western to detect Oct4. Whole cell extract (WCE) was used as a loading control. mES cells were grown under standard culture conditions unless otherwise specified.
(B) pSmad3 interacts with Oct4, and this interaction is dependent on TGF-β signaling. mES cells were grown without (−) or with (+) SB431542 (TGF inhib) for 24 hr. Co-IPs with antibodies against pSmad3 and IgG were performed on nuclear lysates. Precipitated complexes were probed for Oct4.
(C) Smad3 and Oct4 bind DNA sites at the same time. Oct4 ChIP was performed followed by re-ChIP using antibodies against Smad3 and IgG. qPCR was performed in triplicate to quantify the fold enrichment (y-axis) of Smad3 and IgG at the indicated genes (x-axis) relative to an unbound control region. Error bars indicate standard deviation.
(D) Smad3 and Oct4 simultaneously occupy the Lefty1 enhancer. A 40 bp probe from the Lefty1 enhancer was incubated with nuclear extracts from mES cells (left lane). Nuclear extracts and probe were also incubated with cold competitor, antibody against Smad3 or antibody against Oct4. The complex formed by Smad3 and Oct4 (Smad/Oct) is supershifted (SS) by both antibodies. mES cells were cultured with SB431542 for 24 hr and then washed to remove inhibitor before treating with Activin for 1 hr to activate TGF-β signaling.
(E) Smad3 levels are not affected by loss of Oct4. ZHBTc4 mES cells were cultured in the absence or presence of dox for 24 hr. Western blot was performed to quantify levels of Smad3 and Oct4. 25μg and 5μg of cell lysates were loaded. TATA binding protein (TBP) was used as a loading control.
(F) Oct4 is required for Smad3 binding. ChIP was performed for Smad3 in ZHBTc4 mES cells without dox (Smad3 ChIP Oct4 nl) and with dox for 24 hr (Smad3 ChIP Oct4 kd). qPCR was performed to quantify the fold enrichment of Smad3 (y-axis) at the indicated genes (x-axis). Fold enrichment was normalized to IgG.
(G) Oct4 is required for genes to respond to TGF-β signaling. ZHBTc4 mES cells were cultured without or with dox for 24 hr. Cells were then washed and treated with Activin for 1 hr prior to analysis of gene expression by qPCR. The fold reduction of Oct4 expression after 24 hr of dox (left) and the fold reduction in TGF-β response for genes co-occupied by Oct4 and Smad3 (right) are shown.
(H) Oct4 sites are more tightly associated with Sox2 than Smad3. The distances from the Oct4 peak to the peaks of Sox2 (black) and Smad3 (red) were calculated for each region bound by Oct4, Sox2 and Smad3 (1849 regions). The distances between peaks were organized into 5-base pair bins, and the number of peaks in each bin (y-axis) is shown over a 0.6 kb window (x-axis) centered on the Oct4 peak at position 0. The distance between Oct4 and Sox2 sites is defined as positive.
(I) Oct4 sites are depleted of nucleosomes. ChIP-seq was performed to map genome-wide H3 occupancy. The relative H3 density (y-axis) is shown across a 2kb window (x-axis) centered on sites occupied by Oct4.
To determine if Oct4 is required for recruiting Smad3 to DNA sites bound by Oct4 in ES cells, we inhibited Oct4 expression in ZHBTc4 mES cells with doxycycline (dox) treatment for 24 hr (Niwa et al., 2000). We found that loss of Oct4 did not affect the levels of Smad3 protein (Fig 3E). However, loss of Oct4 expression did result in a dramatic reduction in Smad3 occupancy at key genes normally co-occupied by Oct4 and Smad3 (Fig 3F). Furthermore, loss of Oct4 resulted in reduced responsiveness to TGF-β signaling at genes normally co-occupied by Oct4 and Smad3 (Fig 3G).
Oct4 might recruit Smad3 through a direct interaction and/or by inducing a more open chromatin state to make Smad binding elements (SBEs) available for Smad3 binding. Our evidence suggests that Oct4 and Smad3 are associated in some fashion (Fig 3A–F), although analysis of the positions of Oct4 and Smad3 binding peaks indicates that these two proteins do not interact in a unique and direct manner such as that observed for Oct4 and Sox2 (Fig 3H). Analysis of nucleosome occupancy at sites bound by Oct4 revealed that these sites are relatively depleted of nucleosomes (Fig3I). These results are consistent with the possibility that Oct4 recruits Smad3 to adjacent SBEs that are available due to nucleosome depletion.
Smad3 co-occupies the genome with master transcription factors in various cell types
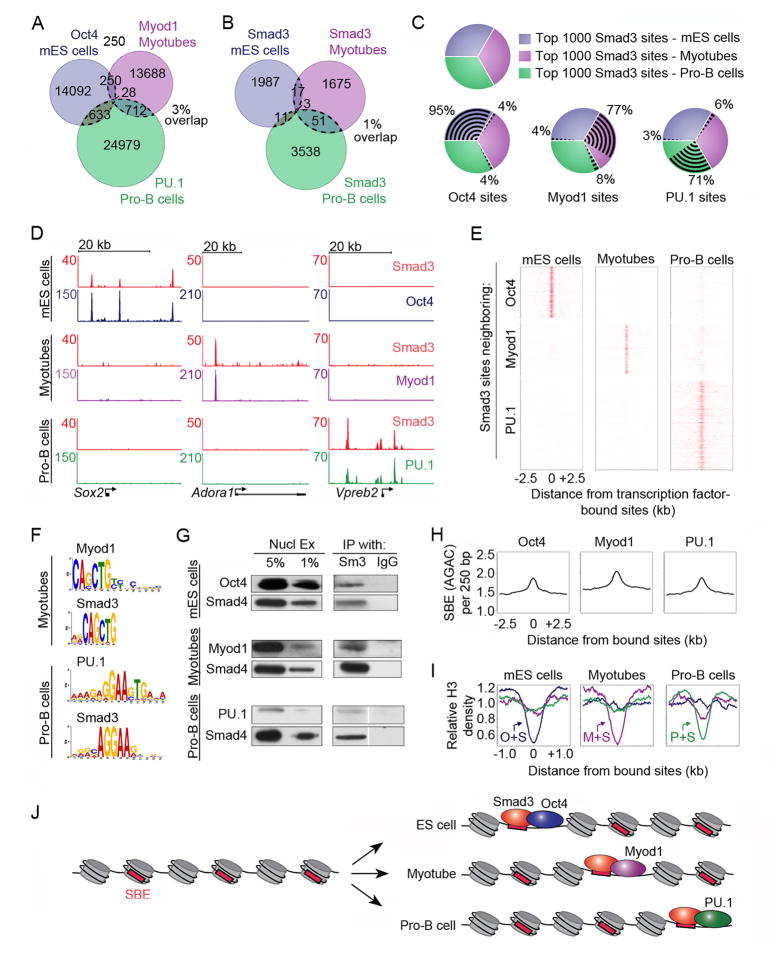
We next investigated whether Smad3 co-occupies the genome of additional cell types with the master transcription factors of those cell types. We performed ChIP-seq analysis for Myod1 in myotubes (Davis et al., 1987; Tapscott, 2005) and PU.1 in pro-B cells (DeKoter and Singh, 2000; Nutt and Kee, 2007) and found that sites bound by Oct4, Myod1 and PU.1 were largely unique (Fig 4A, Table S1). We also performed ChIP-seq analysis of Smad3 binding in myotubes and pro-B cells and found that Smad3 also tends to bind unique sites in these different cell types (Fig 4B).
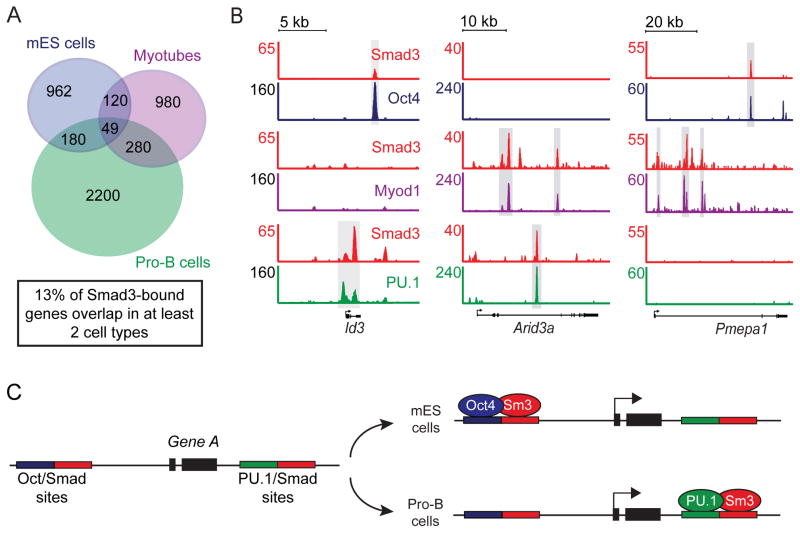
Figure 4. Smad3 co-occupies DNA with cell-type-specific master transcription factors.
(A) Master transcription factors bind unique sites in different cell types. The Venn diagram shows the overlap of sites bound by Oct4 in mES cells (blue), Myod1 in myotubes (purple) and PU.1 in pro-B cells (green) (Table S1). The total number of bound sites is indicated for each shaded area. 3% of all sites overlap in at least two cell types (indicated by dotted lines).
(B) Smad3 binds unique sites in different cell types. The Venn diagram shows the overlap of Smad3-bound regions between mES cells (blue), myotubes (purple) and pro-B cells (green). 1% of Smad3-bound sites overlap in at least two cell types. Myotubes and Pro-B cells were treated with TGF-β prior to analysis of Smad3 binding.
(C) Smad3 co-occupies sites with master transcription factors that are cell-type specific. The 1000 strongest Smad3 binding sites (by peak height) were chosen from each cell type for analysis (top left). The co-occupancy of Oct4 (bottom left), Myod1 (bottom center) and PU.1 (bottom right) with Smad3 in each cell type is shown.
(D) Smad3 co-occupies cell-type-specific sites with master transcription factors at individual genes. Gene tracks represent binding of Smad3 and Oct4 in mES cells (top), Smad3 and Myod1 in myotubes (center) and Smad3 and PU.1 in pro-B cells (bottom) for the genes encoding Sox2, Adora1 and Vpreb2. The floor is set at 3 counts. See also Figure S3.
(E) Smad3 co-occupies the genome with cell-type-specific master transcription factors. Binding plots show the location of Smad3-bound sites in mES cells (left), myotubes (center) and pro-B cells (right) relative to sites bound by Oct4 in mES cells (top), Myod1 in myotubes (middle) and PU.1 in pro-B cells (bottom).
(F) Smad3 binding sites are enriched for the motif of the cell-type-specific master transcription factor. Motif discovery was performed using Myod1 and Smad3-bound sites identified in myotubes (top) and PU.1 and Smad3-bound sites in pro-B cells (bottom). The most enriched motifs are shown.
(G) Smad3 interacts with master transcription factors. Co-IPs with antibodies against Smad3 (Sm3) and IgG were performed using nuclear lysates from mES cells (top), myotubes (center), and pro-B cells (bottom). Precipitated complexes were probed for Oct4 in mES cells, Myod1 in myotubes and PU.1 in pro-B cells. Smad4 was used as a positive control for immunoprecipitation.
(H) SBEs are enriched at sites occupied by master transcription factors. The average frequency of SBEs in a 250 bp window across a 5kb region centered on the binding site of each transcription factor is indicated.
(I) Nucleosomes are depleted at sites co-occupied by Smad3 and master transcription factors. Relative H3 density centered on sites co-occupied by Oct4 and Smad3 (O+S) in mES cells (left), Myod1 and Smad3 (M+S) in myotubes (center) and PU.1 and Smad3 (P+S) in pro-B cells (right) is shown.
(J) Model for cell-type specific genome occupancy by Smad3. Cell-type specific Smad3 binding may be determined by interactions with master transcription factors, which occupy nucleosome-depleted regions and recruit Smad3 to cell-type-specific sites. Red boxes indicate SBEs and gray cylinders represent nucleosomes.
If Smad3 is recruited to DNA sites by master transcription factors, then we would expect Smad3 sites in each cell type to be occupied by the master transcription factor found in that cell type. We initially analyzed the top 1000 bound sites for Smad3 in mES cells, myotubes and pro-B cells and asked if those sites were occupied by master transcription factors (Fig 4C, S3A). Indeed, Oct4 occupied sites with Smad3 in mES cells but did not occupy sites bound by Smad3 in myotubes or pro-B cells. Myod1 occupied sites with Smad3 in myotubes, and PU.1 occupied sites with Smad3 in pro-B cells. We next analyzed all Smad3 sites in each cell type, which confirmed that Smad3 tends to co-occupy sites with the cell-type-specific master transcription factors (Fig S3B).
The cell-type-specific association of master transcription factors and Smad3 was striking at individual genes (Fig 4D). For example, Smad3 and Oct4 co-occupied sites at the gene encoding Sox2 in mES cells, but these sites were not occupied by Smad3 or Myod1 in myotubes or Smad3 or PU.1 in pro-B cells. The gene encoding Adora1 was uniquely co-occupied by Smad3 and Myod1 in myotubes, and the gene encoding Vpreb2 was uniquely occupied by Smad3 and PU.1 in proB cells. Genome-wide analysis confirmed that Smad3 occupied unique sites with Oct4 in mES cells, Myod1 in myotubes and PU.1 in pro-B cells (Fig 4E). In addition, motif discovery revealed enrichment of the Myod1 motif in Smad3-bound regions of myotubes and the PU.1 motif in Smad3-bound regions of pro-B cells (Fig 4F), further supporting the conclusion that Smad3 co-occupies the genome with cell-type-specific master transcription factors.
We next investigated whether interactions could be detected between Smad3 and the master transcription factors of each cell type and whether Smad3 binding depends on Myod1 in myotubes and PU.1 in pro-B cells. We found that Smad3 immunoprecipitated with Oct4 in mES cells, Myod1 in myotubes and PU.1 in pro-B cells (Fig 4G). We next asked if knockdown of Myod1 affected binding of Smad3 in myotubes. Myoblasts were transfected with Myod1 siRNA, and a 52% knockdown of Myod1 was observed after 48 hr of myotube differentiation. The reduction in Myod1 was associated with an intermediate level of myotube differentiation (data not shown) and resulted in decreased Smad3 occupancy at sites normally co-occupied with Myod1 (Fig S3C). Similarly, deletion of PU.1 and the functionally redundant protein spib (DeKoter et al., 2002) in pro-B cells resulted in decreased Smad3 occupancy at sites co-occupied by PU.1 (Fig S3D). Together these results suggest that master transcription factors are required for wild-type levels of Smad3 recruitment.
If Smad3 binds DNA at sites with cell-type-specific master transcription factors, then SBEs should be enriched at these sites. We scanned Oct4, Myod1 and PU.1 sites and found that these sites were each enriched for SBEs (Fig 4H). This result indicates that Smad3 binds a unique subset of SBEs in each cell type, and that this subset is associated with sites bound by the cell-type-specific master transcription factors.
Finally, we investigated whether nucleosome depletion occurs at sites bound by cell-type-specific master transcription factors and Smad3, as increased accessibility of SBEs in these regions may contribute to Smad3 binding. Genome-wide ChIP-seq analysis of histone H3 occupancy revealed that master transcription factors occupied cell-type-specific regions that were relatively depleted of nucleosomes (Fig S3E). In addition, Smad3 and the master transcription factors co-occupied unique nucleosome-depleted regions in each cell type (Fig 4I). For example, in mES cells Oct4 and Smad3 co-occupied sites that were relatively depleted of nucleosomes (Fig 4I, left, blue), while sites occupied by Smad3 and Myod1 in myotubes (purple), or Smad3 and PU.1 in pro-B cells (green) were not associated with significant nucleosome depletion in mES cells. Nucleosome depletion was unchanged when TGF-β signaling was inhibited (Fig S3F) suggesting that Smad3 did not affect nucleosome occupancy but was directed to sites that were already depleted of nucleosomes. Thus, Smad3 appears to bind unique SBEs in nucleosome-depleted regions adjacent to master transcription factors in each cell type (Fig 4J).
Smad3 can occupy different cell-type-specific enhancers at the same gene
Master transcription factors can bind cell-type-specific enhancers (Heintzman et al., 2009). Thus, we would expect that Smad3 could occupy different enhancers of the same gene in different cell types. We analyzed the intersection of genes bound by Smad3 in mES cells, myotubes and pro-B cells. While only 1% of the Smad3 binding sites were occupied in more than one cell type (Fig 4B), 13% of genes bound by Smad3 were occupied by Smad3 in more than one cell type (Fig 5A). Analysis of individual genes showed that Smad3 could occupy the same gene in different cell types but usually did so by co-occupying cell-type-specific enhancers with the cell-type-specific master transcription factor (Fig 5B). For example, Id3 was occupied by Oct4 in mES cells and PU.1 in pro-B cells. However, Oct4 and PU.1 occupied Id3 at different sites, and Smad3 co-occupied sites with the cell-type-specific master regulators. Thus, for the small fraction of genes that appear to be targeted by TGF-β signaling in multiple cell types, these genes tend to be bound by Smad3 at different enhancers (Fig 5C).
Figure 5. Smad3 can bind the same gene at different sites in different cell types.
(A) Smad3 binds a small number of genes in common between different cell types. The Venn diagram shows the overlap of genes bound by Smad3 in mES cells, myotubes and pro-B cells (Table S3). The numbers represent the total number of bound genes in each shaded area.
(B) Smad3 co-occupies the same gene but at cell-type-specific sites. Gene tracks show binding of Smad3 and Oct4 in mES cells (top), Smad3 and Myod1 in myotubes (center) and Smad3 and PU.1 in pro-B cells (bottom) for Id3, Arid3a and Pmepa1. Gray boxes highlight sites co-occupied by Smad3 and master transcription factors in each cell type. The floor is set at 3 counts.
(C) Smad3 co-occupies a fraction of genes with different master transcription factors by binding at different sites. At hypothetical Gene A one SBE (red box) is adjacent to an Oct4 site and another is adjacent to a PU.1 site. In mES cells Smad3 (Sm3) binds with Oct4 while in pro-B cells Smad3 binds with PU.1. See also Table S3.
TGF-β signaling regulates genes bound by master transcription factors
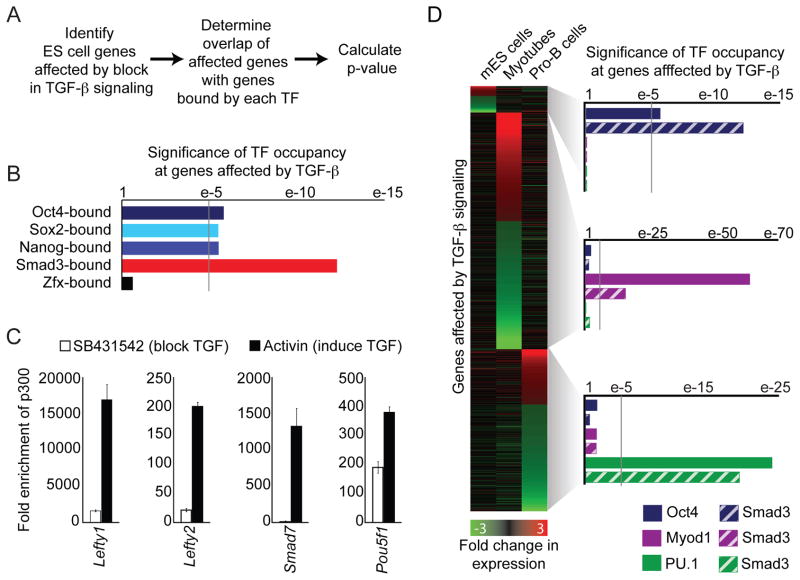
If Smad3 is directed to sites occupied by Oct4 then genes bound by Oct4 should be modulated by TGF-β signaling. Genome wide expression analysis was performed on mES cells under standard conditions and after treatment with the TGF-β inhibitor SB431542 for 24 hr (James et al., 2005; Ross et al., 2006; Vallier et al., 2005; Xu et al., 2008) to identify genes that are modulated by TGF-β signaling. We found that genes bound by Oct4, Sox2, Nanog, or Smad3 were affected by a block in TGF-β signaling (Fig 6A, B, Table S3, S4). As a control we also analyzed genes bound by Zfx, a transcription factor that binds a similar number of genes to Oct4 (Table S1, S3) but does not co-occupy sites with Smad3. In contrast to genes bound by Oct4, genes bound by Zfx were not affected by TGF-β signaling (Fig 6B, bottom).
Figure 6. TGF-β signaling regulates genes bound by master transcription factors.
(A) Schematic of analysis. Genes affected by loss of TGF-β signaling were identified by genome-wide microarray analysis. Next, the overlap of genes affected by TGF-β signaling and genes bound by each transcription factor (TF) were determined. The p-value was calculated using the hypogeometric distribution.
(B) Inhibition of the TGF-β pathway affects genes bound by the mES cell core regulatory transcription factors. Genome-wide expression was performed in mES cells that were cultured under standard conditions or in the presence of SB431542 for 24 hr. The association of genes bound by each transcription factor with genes affected by TGF-β signaling (significance of transcription factor occupancy at genes affected by TGF-β signaling, x-axis) was calculated for genes bound by Oct4, Sox2, Nanog, Smad3 and Zfx (y-axis). A factor was considered to bind genes affected by TGF-β signaling at a p value < 1e-5 (gray line).
(C) TGF-β signaling leads to recruitment of p300 at genes co-occupied by Oct4 and Smad3. mES cells were treated with SB431542 for 24 hr before being washed and treated with fresh SB431542 or 10 ng/ml Activin for 1 hr. ChIP was performed for p300 and IgG, and qPCR was performed to quantify the fold enrichment of p300 relative to IgG (y-axis) at the indicated genes.
(D) TGF-β signaling regulates different genes in different cell types. Genome-wide expression analysis was performed after 24 hr treatment with SB431542 in mES cells and 12 hr after activation of TGF-β signaling in myotubes and pro-B cells. The fold change in expression for each affected gene is indicated by color (bottom) and is shown for mES cells (left), myotubes (center) and pro-B cells (right). All genes that change in only one cell type are shown. Statistical analysis was then performed (as described in A) to determine if there was an association between genes bound by each transcription factor and genes affected by TGF-β signaling for each cell type. Analysis was performed for genes affected by TGF-β signaling in mES cells (top right), myotubes (middle right) and pro-B cells (bottom right). See also Table S4.
To determine if key ES cell genes bound by Smad3 and Oct4 are direct targets of TGF-β signaling, we next asked if activation of TGF-β signaling resulted in rapid recruitment of transcriptional co-activators (Ross et al., 2006). mES cells were treated with SB431542 for 24 hr before they were washed and re-treated with fresh SB431542 or Activin to activate the TGF-β pathway. ChIP was performed for the histone acetyltransferase p300 one hr after activation and showed that TGF-β signaling led to rapid recruitment of p300 to genes co-occupied by Oct4 and Smad3 (Fig 6C).
If master transcription factors direct Smad3 to different targets in different cell types we would expect that the genes regulated by TGF-β signaling are different in different cell types. Genome wide expression analysis was performed on myotubes and pro-B cells to identify genes that changed in expression after treatment with TGF-β for 12 hr. Expression changes in myotubes and pro-B cells were also compared to changes in mES cells after inhibition of TGF-β signaling for 24 hr. As expected, a largely unique set of genes was found to be affected in each cell type (Fig 6D, left). We next asked if the set of genes bound by Smad3 or the master transcription factors in each cell type were associated with the set of genes affected by TGF-β signaling in each cell type. In each case we found that only genes bound by the cell-type-specific master transcription factor or cell-type-specific Smad3 were affected by TGF-β signaling (Fig 6D, right), indicating that TGF-β signaling regulates genes bound by cell-type-specific master transcription factors.
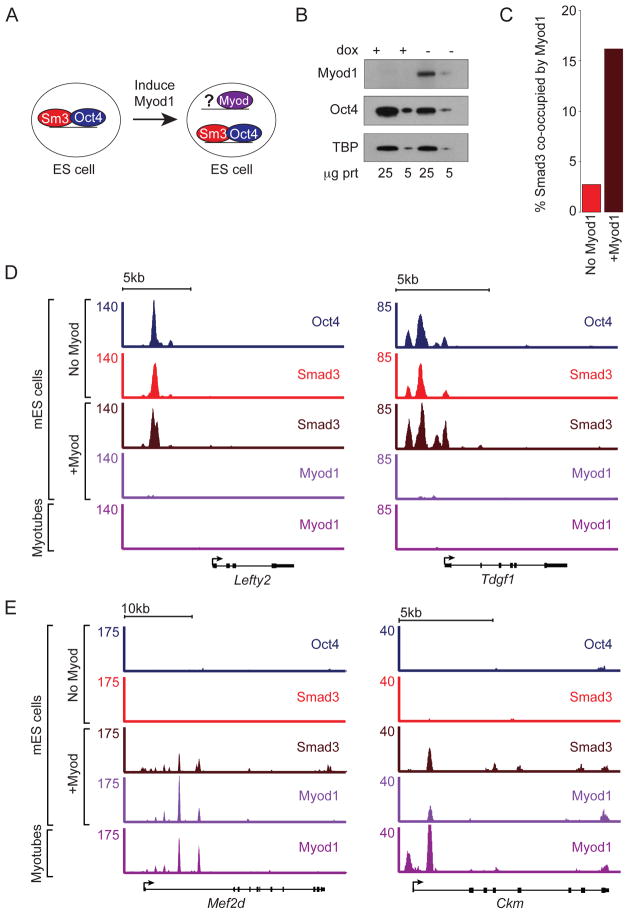
Induction of Myod1 redirects Smad3 binding in mES cells
If master transcription factors direct Smad3 to their sites of occupancy, then induction of a master transcription factor that is not normally expressed in a specific cell type should direct Smad3 to the unique sites occupied by the induced master transcription factor. To test this idea, we induced expression of Myod1 in mES cells (Fig 7A) (Nishiyama et al., 2009). ChIP-seq was performed for Smad3 in mES cells that were maintained for 5 days in mES cell culture conditions with and without induction of Myod1. Despite expression of Myod1, mES cells maintained expression of Oct4 (Fig 7B). However, expression of Myod1 was sufficient to direct a fraction of Smad3 to sites occupied by Myod1 (Fig 7C). Inspection of ChIP-seq profiles showed that Smad3 continued to occupy sites with Oct4 in the setting of Myod1 expression (Fig 7D), while also occupying new sites with Myod1 (Fig 7E).
Figure 7. Myod1 expression redirects Smad3 occupancy in mES cells.
(A) Experimental model. Smad3 co-occupies the genome with Oct4 in mES cells. Myod1 expression was induced in mES cells for 5 days in standard mES cell culture conditions. ChIP-seq was performed to determine if Smad3 was directed to new sites occupied by Myod1.
(B) Oct4 expression is maintained despite induction of Myod1. mES cells containing dox-repressible Myod1 (Nishiyama et al., 2009) were cultured in standard mES cell conditions for 5 days with and without dox. Western analysis was performed to detect expression of Myod1 (top) and Oct4 (middle). TBP was used as a loading control (bottom).
(C) Smad3 occupies new sites with Myod1. The percentage of Smad3 sites in mES cells that are also occupied by Myod1 in myotubes (y-axis) is shown for mES cells without induction of Myod1 (No Myod1) and with induction of Myod1 (+Myod1). The 1000 strongest Smad3 binding sites in each condition were used for this calculation.
(D) Smad3 continues to occupy sites bound by Oct4. Gene tracks show binding of Oct4, Smad3 without induction of Myod1 (red), Smad3 with induction of Myod1 (brown) and Myod1 (after induction) in mES cells at Lefty2 and Tdgf1. Myod1 binding in myotubes is shown at the bottom. The floor is set at 3 counts.
(E). Smad3 occupies new sites bound by Myod1. Gene tracks show binding of Oct4, Smad3 without induction of Myod1 (red), Smad3 with induction of Myod1 (brown) and Myod1 (after induction) in mES cells at the muscle specific genes Mef2d and Ckm. Myod1 binding in myotubes is shown at the bottom.
DISCUSSION
Transcription factors bind specific DNA sequences and regulate gene expression (Ptashne, 1988), and master transcription factors are required for establishment, maintenance and even reprogramming of cell identity (Takahashi and Yamanaka, 2006; Feng et al., 2008; Lassar et al., 1986; Seale et al., 2008; Zhou et al., 2008; Graf and Enver, 2009). Here we report that master transcription factors also are responsible for directing the gene targets of TGF-β signaling and thus determine the cell-type-specific effects of TGF-β signaling.
This conclusion is supported by the finding that Smad3 co-occupies the genome with Oct4 in ES cells, Myod1 in myotubes and PU.1 in pro-B cells. In addition, Smad3 interacts with these master transcription factors and binds accessible DNA sites adjacent to those bound by master transcription factors. Furthermore, the expression of genes bound by master transcription factors is modulated by TGF-β signaling. Finally, expression of Myod1 in non-muscle cells can re-direct Smad3 to the new sites occupied by Myod1.
If many different transcription factors can interact with Smad2/3 (Chen et al., 2002; Chen et al., 1996; Chen et al., 1997; Germain et al., 2000; Liu et al., 1997; Massague et al., 2005; Seoane, 2004), why do master transcription factors have such a profound effect in determining the genes regulated by TGF-β signaling in each cell type? The answer may lie in the relative concentration of different transcription factors, as master transcription factors tend to be expressed at high levels compared to other transcription factors (Young, 2011). Thus, the abundance of cell-type-specific master transcription factors may allow them to dominate the competition for interactions with Smad2/3. Recent work in mES cells has also suggested that the Wnt, LIF and BMP pathways target sites bound by mES cell master transcription factors (Chen et al., 2008; Cole et al., 2008; Young, 2011), and our findings now suggest that cell-type-specific master transcription factors may operate in many different cell types to determine the gene targets of signaling pathways.
TGF-β signaling, through activation of Smad2 and Smad3, plays an essential role in normal development and tissue homeostasis as well as in human diseases from cancer to autoimmunity to cirrhosis (Friedman, 2008; Li and Flavell, 2008; Massague et al., 2005; Padua and Massague, 2009; Wandzioch and Zaret, 2009). It is therefore critical to understand how activation of Smad2/3 can lead to such diverse cellular responses. Our findings reveal that the cell-type-specific effects of TGF-β signaling are determined in large part by the interaction of Smad2/3 proteins with master transcription factors that specify and maintain cell identity. It is through this mechanism that TGF-β signaling is tailored to modulate genes that are most relevant to cell identity, which may explain why aberrations in this pathway can have such profound effects in a range of human diseases.
EXPERIMENTAL PROCEDURES
Cell culture
hES (BGO3) cells were maintained as previously described (Ludwig et al., 2006) using mTESR1 media (Stem Cell Technologies), which contains TGF-β. mES cells were cultured as previously described (Marson et al., 2008). mES cells were maintained on murine embryonic fibroblast (MEF) feeder cells and then passaged 2x off feeders prior to analysis. Exogenous Activin was not added to mES cell cultures unless stated. When indicated, mES cells were treated with SB431542 (10 μM) for 24 hr to inhibit TGF-β signaling. ZHBTc4 mES cells were treated with dox for 24 hr to repress Oct4 expression as previously described (Niwa et al., 2000). Myod1 was induced in ES cells by dox withdrawal as previously described (Nishiyama et al., 2009). Analysis of Myod1 and PU.1 binding was performed under standard culture conditions and analysis of Smad3 binding was performed after treatment with TGF-β in myotubes and pro-B cells. See Extended Experimental Procedures for additional details.
Chromatin Immunprecipitation (ChIP)
ChIP and ChIP coupled with massively parallel sequencing (ChIP-seq) was performed as previously described (Marson et al., 2008). Analysis of H3 occupancy was performed by normalizing the average H3 density across all sites co-occupied by the indicated transcription factors. Antibodies and additional information can be found in the Extended Experimental Procedures.
ChIP-seq Analysis
Analysis methods were derived from previously published methods (Marson et al., 2008). Briefly, the number of extended reads were calculated in bins across the genome and bins that contained statistically significant ChIP-seq enrichment were identified by comparison to a Poissonian background model. A p-value cutoff of 1e-9 was used for all datasets except for c-Myc and Ronin where a cutoff of 1e-5 was used in order to analyze comparable numbers of bound sites. Refer to the Extended Experimental Procedures for details.
Mircroarray Analysis
Analysis was performed using Agilent Whole-Mouse Genome Microarrays (Agilent, G4122F) as previously described (Cole et al., 2008). A gene was determined to be significantly affected by TGF-β signaling if its expression level changed by at least 1.5 fold with a p-value less than or equal to 0.05. Refer to the Extended Experimental Procedures for details.
Co-immunoprecipitation (Co-IP)
Co-IP experiments using nuclear extracts were performed as previously described (Kagey et al., 2010). Refer to the Extended Experimental Procedures for details.
Electrophoretic mobility shift (EMSA)
A 40 bp sequence containing adjacent Smad and Oct4 binding sites in the murine Lefty1 enhancer was labeled with [γ-32P]-ATP and incubated with nuclear extract from mES cells treated with Activin for 1 hr. Competitor DNA was used at 100-fold excess to labeled DNA. Supershift was performed by incubating the assembled complex with antibodies against Smad3 and Oct4. Refer to Extended Experimental Procedures for additional details.
Previously published ChIP-seq Datasets used in this study
The following previously published datasets were used: Oct4, Nanog, Sox2 in mES cells (Marson et al., 2008), c-Myc and Zfx in mES cells (Chen et al., 2008) and Ronin in mES cells (Dejosez et al., 2010).
Supplementary Material
Acknowledgments
We are grateful to M. S. H. Ko for the gift of ES[MC1R(20)]:tetMyod1 cells, D. Wotton for the gift of Smad2/3 antibody and H. Singh for providing the 38B9 cells with permission from N. Rosenberg. We thank T. Lee and L. Lawton for helpful discussions, J. Love, J-A. Kwon, V. Dhanapal, S. Gupta and T. Volkert for Illumina sequencing and microarray preparation, G. Frampton, and B. Yuan for help with computational analysis, and T. DiCesare for help with graphics. We also thank A. Blais for assistance with siRNA transfection. This work was supported by fellowships from the American Gastroenterological Association and by NIH grant DK090122 (A.C.M), the Canadian Institutes of Health Research (S.B.) and by NIH grant HG002668 (R.A.Y.).
Footnotes
ACCESSION NUMBERS
The ChIP-seq and microarray data are deposited in GEO under accession numbers GSE21621 and GSE23830.
Supplemental Information includes Extended Experimental Procedures, three figures and four tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24:1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. Embo J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B) Mol Cell. 2002;9:133–143. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109(Suppl):S121–131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. Embo J. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Seoane J. p21(WAF1/CIP1) at the switch between the anti-oncogenic and oncogenic faces of TGFbeta. Cancer Biol Ther. 2004;3:226–227. doi: 10.4161/cbt.3.2.717. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandzioch E, Zaret KS. Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science. 2009;324:1707–1710. doi: 10.1126/science.1174497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.