Abstract
The lac1 gene encoding an extracellular laccase was isolated from the thermophilic fungus Melanocarpus albomyces. This gene has five introns, and it encodes a protein consisting of 623 amino acids. The deduced amino acid sequence of the laccase was shown to have high homology with laccases from other ascomycetes. In addition to removal of a putative 22-amino-acid signal sequence and a 28-residue propeptide, maturation of the translation product of lac1 was shown to involve cleavage of a C-terminal 14-amino-acid extension. M. albomyces lac1 cDNA was expressed in Saccharomyces cerevisiae under the inducible GAL1 promoter. Extremely low production was obtained with the expression construct containing laccase cDNA with its own signal and propeptide sequences. The activity levels were significantly improved by replacing these sequences with the prepro sequence of the S. cerevisiae α-factor gene. The role of the C-terminal extension in laccase production in S. cerevisiae was also studied. Laccase production was increased sixfold with the modified cDNA that had a stop codon after the native processing site at the C terminus.
Laccases (benzenediol:oxygen oxidoreductases; EC 1.10.3.2) are multicopper enzymes belonging to the group of blue oxidases. They catalyze the oxidation of a variety of phenolic compounds, as well as diamines and aromatic amines, with concomitant reduction of molecular oxygen to water (43). Laccases are widely distributed in higher plants and fungi, and laccase or laccase-like activity has also been demonstrated in some insects and bacteria (18, 31, 12). In fungi, laccases are involved in several physiological functions, such as plant pathogenesis (3, 11), pigment production (2), and degradation of lignocellulosic materials (6). Because of their surprisingly wide variety of substrates, laccases are considered industrially interesting enzymes for various applications, including textile dye bleaching, pulp bleaching, detergents, and enzymatic conversion of chemical intermediates (47).
Despite intensive research, the molecular basis of laccase-catalyzed reactions is still partially unknown. In order to determine the function of laccases and to produce them heterologously in large quantities, several laccase genes have been cloned, especially the genes from basidiomycetous fungi, including Phlebia radiata (39), Cryptococcus neoformans (46), Pleurotus ostreatus (19), Trametes versicolor (25), Trametes villosa (49), Pycnoporus cinnabarinus (13), and Coprinus cinereus (48). Some laccase genes have also been cloned from ascomycetes, including Neurospora crassa (17), Aspergillus nidulans (1), Podospora anserina (14), and Myceliophthora thermophila (5). Generally, the laccase sequences of members of a fungal class exhibit levels of amino acid identity of 50% or more, whereas the levels of identity between sequences of members of different classes are around 30%.
Heterologous expression of laccase genes has been studied in Saccharomyces cerevisiae (27, 10, 29), Trichoderma reesei (38), Aspergillus oryzae (49, 5), Pichia pastoris (24, 16, 34, 7), Aspergillus sojae (22), and Aspergillus niger (36). Especially in S. cerevisiae, reasonable expression levels have proven to be very difficult to achieve. In most previous studies of laccase expression in S. cerevisiae, highly sensitive measurement methods had to be used in order to detect laccase activity. For example, Larsson et al. reported activity measurements that were monitored for 24 h in order to detect satisfactory changes in absorbance, even after optimization of the fermentation conditions for S. cerevisiae expressing a laccase from T. versicolor (29). Reasonable laccase activity levels are essential for using S. cerevisiae in high-throughput screening of directed evolution experiments. P. pastoris has been used as a heterologous host for laccases more often than S. cerevisiae, and higher production levels have been obtained, but the results of use of this organism in high-throughput screening have not been favorable because of a lower transformation frequency and integration of the expression constructs. Successful use of S. cerevisiae expressing M. thermophila laccase in directed evolution was recently reported by Bulter et al., who obtained very promising results (8).
The thermophilic ascomycete Melanocarpus albomyces produces an industrially interesting laccase with substantial thermal stability and activity at alkaline pH values (26). Based on the N-terminal sequence and two internal amino acid sequences of M. albomyces laccase, this enzyme was found to be related to other ascomycete laccases. In this paper, we describe cloning and sequence analysis of the gene encoding M. albomyces laccase. In addition, we expressed the laccase gene in S. cerevisiae so that directed evolution studies can be performed in the future.
MATERIALS AND METHODS
Strains and reagents.
The fungal strains used in this study were M. albomyces VTT D-96490 and P. anserina ATCC 26003. The S. cerevisiae strain used for laccase expression was INVSc1 (MATa his3Δ1 leu2 trp1-289 ura3-52/MATα his3Δ1 leu2 trp1-289 ura3-52), which was obtained from Invitrogen (Carlsbad, Calif.), and the yeast strain used for plasmid construction by the in vivo recombination technique was W3031a (ade2 can1 his3 leu2 trp1 ura3), which was obtained from Hans Ronne, Agricultural University, Uppsala, Sweden. The Escherichia coli strains used were XL1-Blue MR and XL10-Gold from Stratagene (La Jolla, Calif.), TOP10F′ from Invitrogen, and DH5α from Gibco BRL (Gaithersburg, Md.). The enzymes used to manipulate RNA or DNA were obtained from New England Biolabs (Beverly, Mass.) or Boehringer (Mannheim, Germany), unless otherwise stated.
Isolation of the genomic laccase gene.
M. albomyces was cultivated as previously described (26). Total cellular DNA was extracted from the cells by the method of Raeder and Broda (35). The genomic DNA was digested with EcoRI and HindIII in two separate reactions, and the fragments were analyzed by Southern hybridization (40). The laccase-specific probe fragment comprised the portion of the P. anserina lac2 gene that encodes the mature laccase protein (14). The probe fragment was prepared by PCR from P. anserina genomic DNA isolated with an Easy DNA kit (Invitrogen) by using primers 5′-TGCCACACTGCCGCCAACCGTGCT-3′ (forward) and 5′-GTTCTTGATATACCAATCAGGATG-3′ (reverse). The probe was radiolabeled with a random primed DNA labeling kit (Boehringer) by using [α-32P]dCTP. The hybridization solution contained 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7), 1× Denhardt's solution (40), 0.5% sodium dodecyl sulfate (SDS), and 100 μg of herring sperm DNA per ml. To test for optimal hybridization conditions for library screening, hybridization was performed at four different temperatures (48, 50, 55, and 60°C) overnight. Following incubation, the filters were washed with 2× SSC-0.1% SDS twice at room temperature for 5 min and once at the hybridization temperature for 30 min. The hybridization signals were detected by exposing the filters to X-ray film at −70°C.
A genomic library of M. albomyces was constructed in the SuperCos I cosmid (Stratagene). DNA was digested partially with Sau3AI and was dephosphorylated with calf intestinal alkaline phosphatase (Finnzymes, Espoo, Finland). DNA fragments were size fractionated by 15 to 30% sucrose density gradient centrifugation. DNA fragments that were more than 20 kb long were ligated with SuperCos I cosmid vector arms that had been digested with XbaI, dephosphorylated with calf intestinal alkaline phosphatase, and finally digested with BamHI. The ligation mixture was packaged into λ particles with a Gigapack III Gold packaging extract (Stratagene). The synthesized DNA library was amplified in E. coli XL1-Blue MR cells.
Approximately 5 × 105 clones from the genomic DNA library were plated and grown overnight and then transferred to Protran nitrocellulose membranes (Schleicher & Schuell GmbH, Dassel, Germany), and the DNA was fixed by using standard procedures (40). The membranes were hybridized with the labeled P. anserina lac2 gene as described above at 57°C overnight.
To find a suitable fragment for subcloning, the cosmids isolated from strongly hybridizing colonies were digested with 19 different restriction enzymes, and the fragments obtained were analyzed by Southern hybridization with the labeled P. anserina lac2 gene as described above. As a 4.5-kb EcoRI fragment was shown to hybridize with the probe, the cosmid was digested with EcoRI, and the fragment was purified by agarose gel electrophoresis. The fragment was ligated into the vector pBluescriptSK(−) (Stratagene) to obtain plasmid pLLK1. The M. albomyces lac1 gene was sequenced from pLLK1 by using the primer walking technique with a DNA sequencing kit, the dRhodamine terminator cycle sequencing ready reaction (PE Biosystems, Warrington, United Kingdom), and an ABI Prism 3100 Genetic Analyzer automated DNA sequencer (Applied Biosystems, Foster City, Calif.).
cDNA cloning.
The cDNA encoding the laccase was cloned by rapid amplification of cDNA ends (RACE)-PCR. Total RNA was extracted from M. albomyces cells with TRIzol reagent (Life Technologies, Gaithersburg, Md.), dephosphorylated, and reverse transcribed according to the instructions of a FirstChoice RLM-RACE kit (Ambion, Inc., Austin, Tex.). The 5′ and 3′ ends of the lac1 cDNA were amplified in two separate nested PCRs by using two gene-specific primers and two primers specific for adapters ligated into the cDNA. The nested gene-specific primers were as follows: for the 5′ end, 5′-GCCGGTGAGGATGTAGTCGATGAT-3′ (outer primer) and 5′-AGGTGACGTTGAACCAGTAGTTGTC-3′ (inner primer); and for the 3′ end, 5′-CTGGTGCACTTCACGCAGAACAA-3′ (outer primer) and 5′-AGAACCACTTCCAGGTGTCGCT-3′ (inner primer). Thirty five cycles of 94°C for 30 s, 60°C (62°C for the 3′ end) for 30 s, and 72°C for 2 min were performed. The resulting fragments were cloned into the pCR2.1-TOPO vector (TOPO TA cloning kit; Invitrogen). The inserts were sequenced and confirmed to correspond to the laccase gene and to have no PCR mistakes. The two separate cDNA ends were ligated into the same vector in the correct orientation by using a unique AatII restriction site in the overlapping region to obtain plasmid pLLK4.
Southern hybridization of M. albomyces genomic DNA with lac1 cDNA.
In order to demonstrate the presence of other laccase genes in the M. albomyces genome, Southern hybridization of M. albomyces genomic DNA with M. albomyces lac1 cDNA was performed. The DNA was digested with EcoRI, HindIII, and PvuII in three separate reactions, and the fragments were analyzed by Southern hybridization at three different temperatures (50, 55, and 60°C) as described above.
Northern hybridization of M. albomyces RNA with lac1 DNA.
Total RNA was extracted with the TRIzol reagent from a shake flask culture and from a fermentor culture of M. albomyces at a growth stage at which laccase activity was produced. A 594-bp portion of the M. albomyces lac1 gene (bp 218 to 811 from the translation start site) was used as a probe. The hybridization solution contained 50% formamide, 10% dextran sulfate, 1% SDS, 1 M NaCl, and 125 μg of herring sperm DNA per ml. Hybridization was performed at 42°C overnight. Following incubation, the filter was washed once with 5× SSPE and twice with 1× SSPE-0.1% SDS for 15 min at 42°C (1× SSPE is 0.15 M NaCl, 0.01 M sodium phosphate, and 1.3 mM EDTA [pH 7.4]).
Construction of expression vectors for S. cerevisiae.
M. albomyces lac1 cDNA from a SacI site 49 bp upstream of the translation start site to the polyadenylation site was cloned into the EcoRI sites of two yeast expression vectors, pYES2 (Invitrogen) containing the inducible GAL1 promoter and pAJ401 (37) containing the constitutive PGK1 promoter. This created plasmids pLLK10 and pLLK7, respectively.
An expression vector in which the native laccase signal and propeptide sequences were replaced with the prepro sequence of the MFα1 gene of S. cerevisiae was constructed. A DNA fragment in which the yeast α-factor prepro region was fused with the 5′ end of the mature laccase was constructed by overlap extension PCR. The 5′ end of this fragment had a 40-bp overlap with the GAL1 promoter, added to the 5′ end primer. The primers used in the PCR were 5′-CTACTAGCAGCTCTAATACGACTCACTATAGGGGAATATTAAGCTTATGAGATTTCCTTCA-3′ (5′ end primer with an overlap with the GAL1 promoter), 5′-AGAAGGGGTATCTTTGGATAAAAGAGAGCCGACGTGCAACACGCCGAGCA-3′ (forward primer for constructing an overlap between α-factor and laccase), 5′-TGCTCGGCGTGTTGCACGTCGGCTCTCTTTTATCCAAAGATACCCCTTCT-3′ (reverse primer for constructing an overlap between α-factor and laccase), and 5′-AGCGGTACGTCCGCTGGCCG-3′ (3′ end primer). The plasmid was constructed by in vivo recombination in S. cerevisiae. Strain W3031a was transformed with the laccase expression plasmid pLLK10 digested with HindIII and BstEII and the PCR fragment described above which had overlaps at both ends with digested pLLK10. Plasmids were rescued into E. coli from the yeast transformants obtained, and plasmid pMS174 was confirmed by restriction enzyme digestion and DNA sequencing.
To study the significance of C-terminal processing for laccase production in yeast, a version of pMS174 was made in which a translation stop codon was added after the C-terminal processing site. This was done by site-directed mutagenesis with a QuickChange mutagenesis kit (Stratagene) and primers 5′-CCCAAGATCGACTCGGGCCTGTAGCGTCGCCGCTGGGTGGAGG-3′ (forward) and 5′-CCTCCACCCAGCGGCGACGCTACAGGCCCGAGTCGATCTTGGG-3′ (reverse) (the mutated bases are indicated by boldface type). The right mutation in the plasmid constructed, pMS175, was confirmed by sequencing.
S. cerevisiae strain INVSc1 was transformed with the expression vectors pLLK7, pLLK10, pMS174, and pMS175 and the corresponding empty control vectors by using the lithium acetate procedure (20). Production of laccase by the transformants was first assayed on SC-Ura plates (41) supplemented with glucose (for transformants carrying pLLK7) or galactose (for other transformants) by soaking the well-grown colonies on plates with 2 ml of 20 mM ABTS [2,2′-azinobis-(3-ethylbenzthiazoline-6-sulfonate] (Roche Diagnostics GmbH, Mannheim, Germany). To study laccase production in liquid cultures, three transformants of each type were grown in parallel in SC-Ura medium (41) buffered to pH 6 with succinate and supplemented with 2% glucose (transformants with pLLK7) or 2% raffinose (transformants with pLLK10, pMS174, or pMS175) and 0.5 mM CuSO4. All the shake flask cultures were inoculated to obtain an initial optical density at 600 nm of 0.2, incubated at 30°C, and shaken at 250 rpm. After 2 days of cultivation, cells from transformants carrying pLLK10, pMS174, or pMS175 were separated by centrifugation (4,000 × g, 3 min) and resuspended in the same volume of induction medium (SC-Ura medium supplemented with 2% galactose and 0.5 mM CuSO4). Extracellular laccase activity was monitored daily by measuring the oxidation of 5 mM ABTS in 25 mM succinate buffer (pH 4.5) at 436 nm with an extinction coefficient of 29,300 M−1 cm−1 (33).
The presence of laccase in yeast culture supernatants and inside the cells was detected by Western blotting with polyclonal antibodies raised in rabbits against the laccase purified from M. albomyces cultures. After 3 days of growth in the induction medium, the cells in 1 ml of culture were collected by centrifugation (4,000 × g, 3 min). The cells were lysed by intensive mixing with glass beads (diameter, 0.45 mm; Sigma) in a 2% SDS solution containing protease inhibitors (Complete Mini protease inhibitor cocktail; Roche). Samples of cell lysates and culture supernatants were separated by SDS-polyacrylamide gel electrophoresis (Mighty Small II SE250; Hoefer Pharmacia Biotech Inc., San Francisco, Calif.) performed as described by Laemmli (28). Proteins were electroblotted onto a polyvinylidene difluoride membrane (Hybond-P; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) by using a Trans-blot cell (Bio-Rad Laboratories Inc., Hercules, Calif.). M. albomyces laccase was detected by using alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G secondary antibody (Bio-Rad).
Nucleotide sequence accession number.
The nucleotide sequence of the M. albomyces lac1 gene has been deposited in the EMBL nucleotide sequence database under accession number AJ571698.
RESULTS
Cloning and characterization of the M. albomyces lac1 gene.
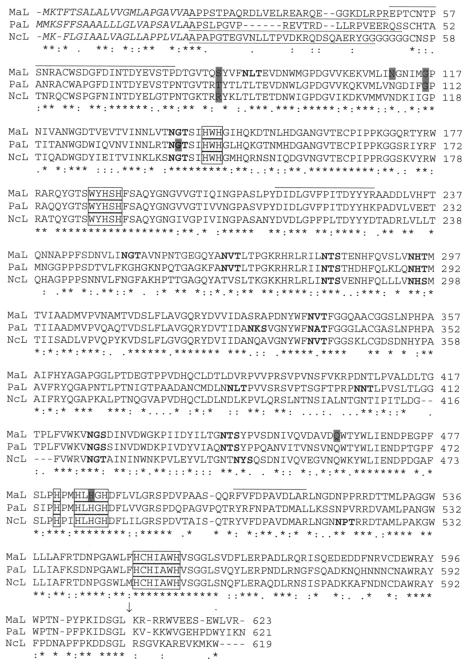
Whether the P. anserina lac2 gene could be used as a laccase probe in heterologous Southern hybridization was tested with M. albomyces total DNA digested separately with EcoRI and HindIII. The results showed that the P. anserina lac2 gene hybridized with a 4.5-kb EcoRI fragment of M. albomyces DNA at all temperatures tested (48 to 60°C) (data not shown), which suggested that M. albomyces had a gene with relatively high conservation with P. anserina lac2. Thus, P. anserina lac2 was used to screen the M. albomyces genomic DNA library that was constructed. Several colonies that gave positive hybridization signals were found, and cosmids from six strongly hybridizing colonies were isolated. The restriction fragment suitable for subcloning the laccase gene from the cosmid was found by mapping the cosmid by digestion with 19 different restriction enzymes, followed by Southern hybridization. A 4.5-kb EcoRI fragment was again shown to hybridize with the labeled P. anserina lac2 gene. This fragment was subcloned into the vector pBluescriptSK(−) to obtain plasmid pLLK1, and the laccase gene region of the insert was sequenced. All three peptide sequences obtained previously for M. albomyces laccase (26) were identified in the amino acid sequence deduced from the gene (Fig. 1), confirming that the right gene was cloned. When the promoter region was sequenced, an EcoRI site was found only 285 bp upstream of the translation start codon. Thus, altogether, 1,619 bp of the promoter region was sequenced from the cosmid instead of pLLK1. A putative TATA box (TATATAAT) was found in the promoter region at position −170 bp with respect to the translation start codon.
FIG. 1.
Alignment of laccase sequences from M. albomyces (MaL), P. anserina (PaL; accession number P78722) (14), and N. crassa (NcL; accession number P06811) (17) as determined with the Clustal W multiple-sequence alignment program (42). An asterisk indicates that the residues at a position are identical in all sequences in the alignment, a colon indicates that conserved substitutions have been observed, and a period indicates semiconserved substitutions. Putative signal sequences are indicated by italics, the propeptides are underlined, and the conserved C-terminal cleavage site is indicated by an arrow. The conserved residues involved in copper binding are enclosed in boxes, and the peptide sequences obtained from purified M. albomyces laccase (26) are overlined. The possible N-glycosylation sites are indicated by boldface type, and the shaded amino acids indicate the locations of introns in the DNA sequence.
In order to determine the intron-exon structure of the M. albomyces lac1 gene and to facilitate heterologous expression, the cDNA copy of the gene was cloned by RACE-PCR by using a method that amplified only capped mRNA molecules from the 5′ end. This enabled determination of the transcription start site based on the 5′ end fragment length. The resulting fragments from the RACE-PCR were 1,194 bp (5′ end) and 1,322 bp (3′ end) long. The positions of five introns (lengths, 78, 73, 87, 86, and 82 bp) (Fig. 1) in the lac1 gene were verified by comparing the cDNA sequence and the sequence of the genomic copy. The 5′ end RACE fragment suggested that there was an ACCAGG transcription start site at bp −120 from the translation start site.
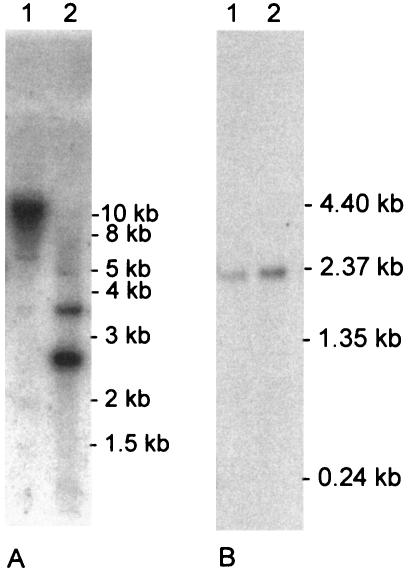
Northern hybridization was performed by using a shake flask culture and a fermentor culture of M. albomyces at a growth stage at which laccase activity was produced. Probing of the Northern blot with a 594-bp portion of the M. albomyces lac1 gene resulted in a 2.0-kb signal, which corresponded well to the size of the cDNA isolated (Fig. 2B).
FIG. 2.
(A) Southern blot analysis of M. albomyces genomic DNA. A 20-μg portion of total DNA was digested with HindIII (lane 1) and with PvuII (lane 2). M. albomyces lac1 cDNA was used as a probe. (B) Northern blot analysis of M. albomyces shake flask (lane 1) and fermentor (lane 2) cultures. Five micrograms of total RNA was hybridized with a portion of the M. albomyces lac1 gene.
The sequence encoded by the open reading frame of the M. albomyces lac1 gene is 623 amino acids long, which is typical for fungal laccases. The regions involved in copper binding are well conserved (14, 48). The amino acid sequence was compared to other known laccase sequences, and it exhibited high levels of identity with other ascomycete laccase sequences (Fig. 1). The levels of amino acid identity were 73% with M. thermophila laccase (4), 68% with P. anserina laccase 2 (14), and 63% with N. crassa laccase (17). On the other hand, the levels of homology with basidiomycete laccases were quite low. For example, the levels of identity of the M. albomyces laccase with laccases from T. versicolor and T. villosa were 27 to 28% (25, 49), and the level of identity with C. cinereus laccase Lcc1 was 31% (48).
The first 22 N-terminal amino acids of M. albomyces laccase consist of a predicted signal sequence typical of eukaryotic proteins (32). The signal sequence is followed by a cleavable 28-amino-acid propeptide, as shown by comparison of the deduced amino acid sequence to the N-terminal peptide sequence of the purified protein (26). In addition, posttranslational removal of the last 14 predicted C-terminal residues was shown by sequencing the C terminus of the laccase purified from M. albomyces. Ser, Gly, and Leu were identified as the last three amino acids of the mature protein.
Southern blot hybridization of M. albomyces genomic DNA with M. albomyces lac1 cDNA showed the expected strong signals corresponding to the lac1 gene. In addition, weaker but clearly detectable signals were observed with genomic DNA digested with PvuII and with HindIII (Fig. 2A). PvuII digestion resulted in one weaker signal in addition to the expected signals, whereas two weaker signals were detected after HindIII digestion. The signals were seen in hybridization experiments done at all temperatures tested. This indicates that there are at least two laccase genes in the M. albomyces genome.
Heterologous expression.
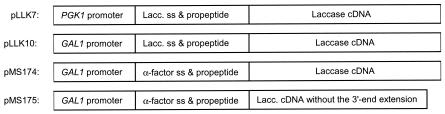
Four different expression vectors were constructed for expression of M. albomyces laccase in S. cerevisiae (Fig. 3). In two of them, the laccase cDNA was alone under the yeast PGK1 promoter or under the GAL1 promoter (pLLL7 and pLLK10, respectively). In the third vector, the region encoding the mature laccase was fused with the yeast α-factor prepro region (pMS174), and the rest of the laccase cDNA was intact. The fourth vector, pMS175, had the α-factor prepro-laccase fusion and a stop codon after the C-terminal processing site of the laccase. All the expression plasmids and the corresponding vectors without an insert were transformed into yeast strain INVSc1. Production of laccase was first assayed with a plate test in which ABTS was the substrate. Formation of a green color around yeast colonies was detected after overnight incubation for transformants carrying pMS175. No color changes were observed for other transformants.
FIG. 3.
Laccase gene expression cassettes created in this study. ss, signal sequence; Lacc., laccase.
Laccase production was then studied in liquid cultures grown in synthetic complete media with 0.5 mM CuSO4 added to support copper incorporation into the laccase. The yeast strains in which laccase production was driven by the GAL1 promoter were first grown in raffinose medium, and the cells were subsequently transferred into a medium with galactose to induce the promoter. Laccase production from the pLLK7 vector with the PGK1 promoter was not detectable with an activity assay in which ABTS was the substrate. In the case of pLLK10, very low laccase activity was detected after 2 days of growth in the galactose medium. Detection of this activity required overnight incubation of the culture supernatant with ABTS. On the other hand, the activity assay was clearly disturbed by some agent present in the yeast culture broth that reduced the color of ABTS at the beginning of the activity measurement. This agent was produced by yeast cells in all the cultures, including controls, since the color of ABTS was not removed by fresh culture medium. Thus, with low laccase activities, the activity assay as such did not give reliable values for production. We therefore attempted to obtain rough estimates for laccase production from pLLK10 by adding known amounts of purified M. albomyces laccase to the activity assay mixture together with the culture supernatant of the control strain. The absorbance changes observed were compared to those of the culture supernatants of the pLLK10 strain. The estimated level of production was around 2 pkat/ml. The higher laccase activities produced by the pMS174 and pMS175 strains could override the effect of the reducing agent.
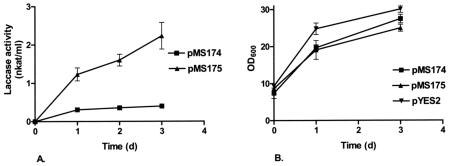
Expression of the M. albomyces laccase from pMS174, in which the mature laccase was fused with the yeast α-factor signal sequence and propeptide, resulted in an approximately 200-fold improvement in production compared to the production with laccase cDNA with its own signal and propeptide sequences in the same vector. The maximum laccase activity in pMS174 cultures was 0.45 nkat/ml. When the C terminus of the laccase was removed from the expression construct (i.e., pMS175), laccase production was further improved about sixfold to the maximum activity, 2.8 nkat/ml. The time course of laccase production by yeast transformants is shown in Fig. 4. Laccase activity for pMS175 strains increased until day 3, whereas for pMS174 the activity was nearly maximal on the first day after induction. Continuing the cultivations for more than 3 days did not markedly increase the laccase activity (data not shown). Figure 4 also shows the growth of yeast transformants and the control strain carrying the empty expression plasmid pYES2 after induction of laccase expression. The control strain carrying pYES2 grew slightly faster than the laccase-producing strains, indicating that laccase production resulted in stress in the cells.
FIG. 4.
(A) Extracellular laccase production by yeast transformants carrying expression plasmid pMS174 or pMS175. (B) Growth of yeast transformants producing laccase (pMS174 and pMS175) and the control strain (pYES2). OD600, optical density at 600 nm.
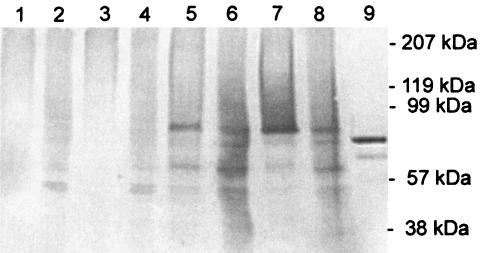
The laccase produced by S. cerevisiae was studied by Western blotting of samples from the culture supernatants and yeast cell lysates with polyclonal antibodies (Fig. 5). No laccase bands were detected in samples from the vector control or transformants carrying pLLK10. A major band at about 95 kDa was observed for the supernatants of transformants carrying pMS174 or pMS175, whereas the native laccase had a molecular mass of about 80 kDa. In addition to the 95-kDa band, there was a smear of larger proteins, which indicates that there was heterogeneous overglycosylation by S. cerevisiae. Two smaller bands which indicated that there was proteolytic degradation were also detected. In the cell lysates of pMS174 and pMS175 strains, the band at 95 kDa, the smear of larger proteins, and a large number of degradation products were detected. The laccase with the C-terminal extension from pMS174 appeared to be more degraded by S. cerevisiae than the product of the truncated laccase gene (pMS175). The supernatant and cell lysate samples used for Western blotting corresponded to the same culture volume. As a major part of the laccase was detected inside the cells, secretion of this enzyme appeared to be one of the steps limiting production.
FIG. 5.
Western blot analysis of culture supernatants (lanes 1, 3, 5, and 7) and cell lysates (lanes 2, 4, 6, and 8) of S. cerevisiae transformants after 3 days of induction with galactose. S. cerevisiae was transformed with pYES2 (lanes 1 and 2), pLLK10 (lanes 3 and 4), pMS174 (lanes 5 and 6), and pMS175 (lanes 7 and 8). Fifteen microliters of the supernatant and a corresponding amount of cell lysate were loaded on the gel. Lane 9 contained 30 ng of native M. albomyces laccase.
DISCUSSION
A novel laccase with extremely interesting technical properties was recently isolated from the fungus M. albomyces (26). This laccase showed good thermostability, retaining full activity for 2 h at 60°C, as well as high pH optima at a neutral pH, both of which are unusual properties for most known fungal laccases. In this paper, we describe isolation and heterologous expression of the laccase gene. The amino acid sequence of the laccase was shown to exhibit high levels of homology with the sequences of laccases from other ascomycetes, such as M. thermophila, N. crassa, and P. anserina, and, as expected, low levels of identity with the more widely studied basidiomycete laccases. This finding further strengthens the hypothesis that the fungal laccases can be separated into two divergent classes, ascomycete laccases and basidiomycete laccases (14, 5). The laccase of A. nidulans differs remarkably from all the other fungal laccases, presumably reflecting a functionally different role of the laccase in the formation of conidiophores (2).
The deduced M. albomyces laccase contains nine potential N-linked glycolysation sites (Asn-X-Thr/Ser) (Fig. 1). This protein is, in fact, highly glycosylated, since analysis of the crystal structure of the protein showed that altogether there are seven glycosylated sites (21). In addition to glycosylation, formation of mature M. albomyces laccase requires posttranslational removal of a signal sequence, propeptide cleavage at the N terminus, and removal of a C-terminal extension. A C-terminal truncation was also seen in the crystal structure of MaL as no electron density was observed for the last 14 predicted C-terminal residues (21). This finding was also confirmed in this study by C-terminal sequencing of the purified laccase. Similar C-terminal processing has also been proposed for laccases from the ascomycetes N. crassa (17), P. anserina (14), and M. thermophila (8), and the processing site Asp-Ser-Gly-Leu is conserved in these laccases. Interestingly, the same kind of sequence is found at the C-terminal ends of some other ascomycete laccases with no C-terminal extensions. In the laccases of Cryphonectria parasitica (11), Botrytis cinerea (9), Glomerella lagenarium (44), and Gaeumannomyces graminis var. tritici (30) the last four C-terminal amino acids are Asp, Ser, Gly, and Leu/Ile/Val. We believe that this C-terminal end is conserved because it is related to the catalytic activity of ascomycete laccases. In the crystal structure of M. albomyces laccase, the last four amino acids of the protein were shown to pack tightly inside the protein. In addition, the C-terminal carboxylate group is hydrogen bonded to the side chain of His-140, which is also bound to type 2 copper in the active center (21).
The maximal level of production of M. albomyces laccase in S. cerevisiae was 2.8 nkat/ml. If the enzyme produced by yeast had the same specific activity as the enzyme produced by M. albomyces, yeast cells would produce about 3 mg of laccase per liter. Previously, substantial laccase activities in S. cerevisiae have been detected with the laccase of Coriolus hirsutus (27) or M. thermophila (8). For C. hirsutus laccase no data on how the activity was measured are available. For M. thermophila laccase, the initial level of production obtained with the laccase cDNA alone under a galactose-inducible promoter was low, around 0.6 U/liter, corresponding to 0.01 nkat/ml. Production was improved by directed evolution of the M. thermophila laccase gene. The study of Bulter et al. (8) and our work are the first examples of ascomycete laccase expression in S. cerevisiae. It is possible that laccase production in S. cerevisiae is easier with laccases from the ascomycetous fungi, which are phylogenetically more closely related to the ascomycete S. cerevisiae, than with laccases from basidiomycetes.
The very low laccase activity levels obtained with a construct having the intact native laccase cDNA were significantly improved by replacing the native laccase signal and propeptide sequences with the S. cerevisiae α-factor secretion signal sequence and propeptide. The prepro sequence of the MFα1 gene of S. cerevisiae has been used as a secretion signal in different yeast expression systems (45, 24, 23, 34, 7), but to our knowledge, this is the first time that it has been used in S. cerevisiae expressing a laccase. Use of the α-factor secretion signal and propeptide for laccase expression has previously been reported only in P. pastoris, and studies with T. versicolor lcc1 (24), P. cinnabarinus lac1 (34) and T. versicolor lccIV (7) showed no improvement in laccase production with the α-factor secretion signal and propeptide. Signal sequences are generally interchangeable between eukaryotic species, and therefore it is suspected that the propeptide cleavage step of M. albomyces laccase is the problematic step for production of this enzyme in yeast. In fact, the propeptide of the laccase is cleaved after Pro-Arg, but the KEX2 protease of S. cerevisiae has been shown to cleave the propeptides specifically after two basic amino acids (15).
Based on the production kinetics and the Western analysis of the yeast strains producing M. albomyces laccase, the factors limiting production appear to include inefficient secretion and proteolytic degradation. Analysis of cell lysate samples by Western blotting showed that there were abundant multiple degradation products, and the most abundant products were also detected outside the cells. These data also suggested that the C-terminal extension makes the protein more susceptible to degradation.
The role of the C-terminal extension in heterologous laccase production was studied by producing M. albomyces laccase in yeast in two different forms, with and without the extension. The activity levels were about sixfold higher with the 3′-end-truncated laccase gene. This suggests that S. cerevisiae is not able to process the C terminus correctly or that the cleavage activity is limiting. Our results are in contrast to the results of Bulter et al., who observed a significant decrease in laccase activity with the truncated laccase mutant gene of M. thermophila (8). However, it is worth noting that the M. thermophila laccase gene had already gone through a series of mutations at the time that the C-terminal point mutation was made, and these mutations may have affected the activity levels observed. Germann et al. (17) and Bulter et al. (8) have suggested that the C-terminal extension might be essential for production by inactivating the laccase during early posttranslational processing steps. Our data, however, do not support this theory.
In this work, we isolated the gene encoding a novel ascomycete laccase. The recent determination of the crystal structure of this enzyme (21) makes this laccase very interesting for studies of the basic mechanisms of oxidative enzymes. The crystal structure revealed some very interesting novel features, especially concerning the C-terminal end of the mature protein. This study showed that the C-terminal extension is also of special interest with respect to production of this enzyme. The expression of M. albomyces laccase in yeast established in this work, together with the structure, should facilitate our future studies of the mechanism of action and improvement of this enzyme.
Acknowledgments
We thank Ella Cederlund of the Karolinska Institutet, Stockholm, Sweden, for C-terminal sequencing. The skillful technical assistance of Seija Nordberg and Riitta Nurmi is gratefully acknowledged. This work was part of the VTT Industrial Biotechnology research program (Academy of Finland Finnish Centre of Excellence program 2000-2005, project 64330).
This work was financially supported by Tekes, the National Technology Agency, and the Neste Oy's Foundation.
REFERENCES
- 1.Aramayo, R., and W. E. Timberlake. 1990. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 18:3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aramayo, R., and W. E. Timberlake. 1993. The Aspergillus nidulans yA gene is regulated by abaA. EMBO J. 12:2039-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar Nun, N., A. Tal Lev, E. Harel, and A. M. Mayer. 1988. Repression of laccase formation in Botrytis cinerea and its possible relation to phytopathogenicity. Phytochemistry 27:2505-2509. [Google Scholar]
- 4.Berka, R. M., S. H. Brown, F. Xu, P. Schneider, K. M. Oxenbøll, and D. A. Aaslyng. November 1999. Purified Myceliophthora laccases and nucleic acids encoding same. U. S. patent 5,981,243.
- 5.Berka, R. M., P. Schneider, E. J. Golightly, S. H. Brown, M. Madden, K. M. Brown, T. Halkier, K. Mondorf, and F. Xu. 1997. Characterization of the gene encoding an extracellular laccase of Myceliophtora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 63:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourbonnais, R. E., and M. G. Paice. 1990. Oxidation of non-phenolic substrates—an expanded role for laccase in lignin biodegradation. FEBS Lett. 267:99-102. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. A., Z. Zhao, and A. G. Mauk. 2002. Expression and characterization of a recombinant multi-copper oxidase: laccase IV from Trametes versicolor. Inorg. Chim. Acta 331:232-238. [Google Scholar]
- 8.Bulter, T., M. Alcalde, V. Sieber, P. Meinhold, C. Schlachtbauer, and F. H. Arnold. 2003. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 69:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantone, F. A., and R. C. Staples. 1993. A laccase cDNA from Botrytis cinerea. Phytopathology 83:1383. [Google Scholar]
- 10.Cassland, P., and L. J. Jönsson. 1999. Characterization of a gene encoding Trametes versicolor laccase A and improved heterologous expression in Saccharomyces cerevisiae by decreased cultivation temperature. Appl. Microbiol. Biotechnol. 52:393-400. [DOI] [PubMed] [Google Scholar]
- 11.Choi, G. H., T. G. Larson, and D. L. Nuss. 1992. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol. Plant-Microbe Interact. 5:119-128. [DOI] [PubMed] [Google Scholar]
- 12.Claus, H. 2003. Laccases and their occurrence in prokaryotes. Arch. Microbiol. 179:145-150. [DOI] [PubMed] [Google Scholar]
- 13.Eggert, C., P. R. LaFayette, U. Temp, K.-E. L. Eriksson, and J. F. D. Dean. 1998. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 64:1766-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Larrea, J., and U. Stahl. 1996. Isolation and characterization of a laccase gene from Podospora anserina. Mol. Gen. Genet. 252:539-551. [DOI] [PubMed] [Google Scholar]
- 15.Fuller, R. S., A. Brake, and J. Thorner. 1989. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. 86:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelo-Pujic, M., H.-H. Kim, N. G. Butlin, and G. T. R. Palmore. 1999. Electrochemical studies of a truncated laccase produced in Pichia pastoris. Appl. Environ. Microbiol. 65:5515-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germann, U. A., G. Müller, P. E. Hunziker, and K. Lerch. 1988. Characterization of two allelic forms of Neurospora crassa laccase. Amino- and carboxyl-terminal processing of a precursor. J. Biol. Chem. 263:885-896. [PubMed] [Google Scholar]
- 18.Gianfreda, L., F. Xu, and J.-M. Bollag. 1999. Laccases: a useful group of oxidoreductive enzymes. Bioremed. J. 3:1-25. [Google Scholar]
- 19.Giardina, P., R. Cannio, L. Martitrani, L. Marzullo, G. Palmieri, and G. Sannia. 1995. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 61:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakulinen, N., L.-L. Kiiskinen, K. Kruus, M. Saloheimo, A. Paananen, A. Koivula, and J. Rouvinen. 2002. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 9:601-605. [DOI] [PubMed] [Google Scholar]
- 22.Hatamoto, O., H. Sekine, E. Nakano, and K. Abe. 1999. Cloning and expression of a cDNA encoding the laccase from Schizophyllum commune. Biosci. Biotechnol. Biochem. 63:58-64. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh, H. P., and N. A. Da Silva. 1998. Partial-pKD1 plasmids provide enhanced structural stability for heterologous protein production in Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 49:411-416. [DOI] [PubMed] [Google Scholar]
- 24.Jönsson, L. J., M. Saloheimo, and M. Penttilä. 1997. Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr. Genet. 32:425-430. [DOI] [PubMed] [Google Scholar]
- 25.Jönsson, L., K. Sjöström, I. Häggström, and P. O. Nyman. 1995. Characterization of a laccase gene from the white-rot fungus Trametes versicolor and structural features of basidiomycete laccases. Biochim. Biophys. Acta 1251:210-215. [DOI] [PubMed] [Google Scholar]
- 26.Kiiskinen, L.-L., L. Viikari, and K. Kruus. 2002. Purification and characterisation of a novel laccase from the ascomycete Melanocarpus albomyces. Appl. Microbiol. Biotechnol. 59:198-204. [DOI] [PubMed] [Google Scholar]
- 27.Kojima, Y., Y. Kita, and Y. Tsukuda. March 1990. DNA for expression and secretion. European patent EP0388166.
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Larsson, S., P. Cassland, and L. J. Jönsson. 2001. Development of a Saccharomyces cerevisiae strain with enhanced resistance to phenolic fermentation inhibitors in lignocellulose hydrolysates by heterologous expression of laccase. Appl. Environ. Microbiol. 67:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litvintseva, A. P., and J. M. Henson. 2002. Cloning, characterization, and transcription of three laccase genes from Gaeumannomyces graminis var. tritici, the take-all fungus. Appl. Environ. Microbiol. 68:1305-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins, L. O., C. M. Soares, M. M. Pereira, M. Teixeira, T. Costa, G. H. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 33.Niku-Paavola, M.-L., E. Karhunen, P. Salola, and V. Raunio. 1988. Ligninolytic enzymes of the white-rot fungus Phlebia radiata. Biochem. J. 254:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otterbein, L., E. Record, S. Longhi, M. Asther, and S. Moukha. 2000. Molecular cloning of the cDNA encoding laccase from Pycnoporus cinnabarinus I-937 and expression in Pichia pastoris. Eur. J. Biochem. 267:1619-1625. [DOI] [PubMed] [Google Scholar]
- 35.Raeder, U. A., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 36.Record, E., P. J. Punt, M. Chamkha, M. Labat, C. A. M. J. J. van Den Hondel, and M. Asther. 2002. Expression of the Pycnoporus cinnabarinus laccase gene in Aspergillus niger and characterization of the recombinant enzyme. Eur. J. Biochem. 269:602-609. [DOI] [PubMed] [Google Scholar]
- 37.Saloheimo, A., B. Henrissat, A.-M. Hoffrén, O. Teleman, and M. Penttilä. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 38.Saloheimo, M., and M.-L. Niku-Paavola. 1991. Heterologous production of a ligninolytic enzyme: expression of the Phlebia radiata laccase gene in Trichoderma reesei. Bio/Technology 9:987-990. [Google Scholar]
- 39.Saloheimo, M., M.-L. Niku-Paavola, and J. K. Knowles. 1991. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J. Gen. Microbiol. 37:1537-1544. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thurston, C. 1994. The structure and function of fungal laccases. Microbiology 140:19-26. [Google Scholar]
- 44.Tsuji, G., J. Fujikawa, H. Ishisa, O. Horino, and Y. Kubo. 2001. Laccase gene LAC1 of Colletotrichum lagenarium is not essential for melanin biosynthesis and pathogenicity. J. Gen. Plant Pathol. 67:182-190. [Google Scholar]
- 45.Weydemann, U., P. Keup, M. Piontek, A. W. M. Strasser, J. Schweden, G. Gellissen, and Z. A. Janowicz. 1995. High-level secretion of hirudin by Hansenula polymorpha—authentic processing of three different preprohirudins. Appl. Microbiol. Biotechnol. 44:377-385. [DOI] [PubMed] [Google Scholar]
- 46.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, F. 1999. Recent progress in laccase study: properties, enzymology, production, and applications, p. 1545-1554. In M. C. Flickinger and S. W. Drew (ed.), The encyclopedia of bioprocessing technology: fermentation, biocatalysis and bioseparation. John Wiley & Sons, New York, N.Y.
- 48.Yaver, D. S., M. J. Overjero, F. Xu, B. A. Nelson, K. M. Brown, T. Halkier, S. Bernauer, S. H. Brown, and S. Kauppinen. 1999. Molecular characterization of laccase genes from the basidiomycete Coprinus cinereus and heterologous expression of the laccase Lcc1. Appl. Environ. Microbiol. 65:4943-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaver, D. S., F. Xu, E. J. Golightly, K. M. Brown, S. H. Brown, M. W. Rey, P. Schneider, T. Halkier, K. Mondorf, and H. Dalboge. 1996. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl. Environ. Microbiol. 62:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]