Abstract
Here we report a rapid and sensitive method (using loop-mediated isothermal amplification [LAMP]) for the diagnosis of edwardsiellosis, a fish disease caused by Edwardsiella tarda, in Japanese flounder. A set of four primers was designed, and conditions for the detection were optimized for the detection of E. tarda in 45 min at 65°C. No amplification of the target hemolysin gene was detected in other related bacteria. When the LAMP primers were used, detection of edwardsiellosis in infected Japanese flounder kidney, and spleen and seawater cultures was possible. We have developed a rapid and sensitive diagnostic protocol for edwardsiellosis detection in fish. This is the first report of the application of LAMP for the diagnosis of a fish pathogen.
Edwardsiellosis is a common bacterial disease of fish caused by gram-negative, motile-flagellate, rod-shaped bacteria Edwardsiella tarda. Infection by E. tarda in epidemic proportions has been reported to have a disastrous effect on fish culture. E. tarda has also been recognized as a serious human pathogen causing a variety of clinical syndromes (13). Among several virulence factors studied with E. tarda (5, 12), hemolysis by hemolysins is considered to be a major cause of death in the infected fish. Hemolysin produced by E. tarda has been cloned and extensively characterized (2, 3). Although hemolysis and necrosis of organs are very clear indications of edwardsiellosis, it is difficult to diagnose the infection in its initial stages or during mild infection.
Outbreaks of edwardsiellosis are first seen in the form of infections affecting only a few fish; these fish are able to survive only for a few days. When early detection and proper chemotherapy are not undertaken, the infection spreads to the surrounding areas, creating an edwardsiellosis epidemic. Control of edwardsiellosis is attempted through monitoring and stocking specific-pathogen-free stocks. However, diagnosis of E. tarda is made only on the basis of isolation and biochemical identification and of detection (using enzyme-linked immunosorbent assays) of antigens in infected organs like spleen, kidney, and liver. However, a rapid and sensitive detection technique is required to detect edwardsiellosis in fish at early stages to control the disease at the source.
The loop-mediated isothermal amplification (LAMP) reaction is an autocycling strand displacement DNA synthesis performed using a DNA polymerase with a high level of strand displacement activity and a set of specially designed inner and outer primers (11). First, according to the principles of the procedure, the inner primers (the forward inner primers [FIP]) anneal to hybridize the target DNA and the first strand is synthesized by Bst polymerase; the outer primers (F3), at a lower concentration, hybridize and displace the synthesized first strand linked with the FIP-linked complementary strand, which forms the looped structure at one end. This strand initiates a process in which a backward inner primer (BIP) hybridizes the complementary strand and initiates strand synthesis and, later, strand displacement by B3 inner primers to form a dumbbell shape. This stem-loop structure acts as a template for the mixture of cauliflower-structured products in a LAMP reaction, as thoroughly described by Notomi and coworkers (11). Four highly specific designed primers are known to hybridize with six distinct sequence sites of the template DNA; therefore, the amplification of the target DNA is highly specific. LAMP is a sensitive method which can amplify a few copies of DNA to a magnitude of 109 CFU in less than an hour under isothermal conditions (7-11). This method is promising for rapid diagnosis in cases of infections. LAMP has a wide-ranging applicability in the detection of microorganisms. Recently, Maruyama and coworkers (6) applied a LAMP technique for the in situ detection of an stx2 gene in Escherichia coli. Here we have developed a rapid and sensitive method using LAMP to detect edwardsiellosis in fish and in their environment. This is the first report of the use of a LAMP method for the detection of an environmental isolate.
Development of primers.
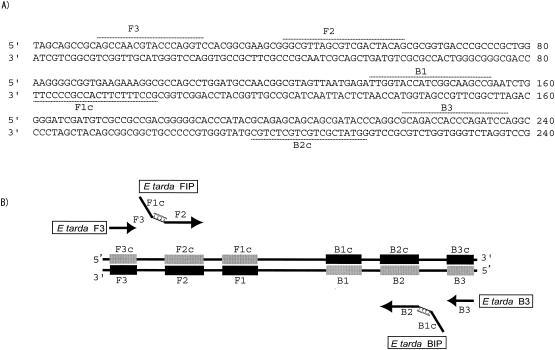
A highly specific set of primers was designed, and these were efficient in amplifying an E. tarda hemolysin gene (Fig. 1). Primers for LAMP were designed to target the hemolysin gene (ethA) from E. tarda (accession no. BAA21097), by using primer software at the Net Laboratory website (http://www.venus.netlaboratory.com/partner/lamp/index.html), as the gene had low homology to the same gene from other bacteria. For efficient stem-loop formation, the size of the target DNA should be 130 to 200 bp. The BIP for the E. tarda hemolysin gene consisted of B1 (20 nucleotides [nt]), a TTTT linker, and B2 (19 nt) (5′-TTGGTACCATCGGCAAGCCG-TTTT-GGTATCGCTGCTGCTCTGC-3′); the FIP consisted of F1c (20 nt), a TTTT linker, and a complementary sequence of F2c (19 nt) (5′-GCCTTTCTTCACCGCCCCTT-TTTT-GGCGTTAGCGTCGACTACAG-3′). Primers B3 and F3 were 5′-TGGATCTGGGTGGTCGTC-3′ and 5′-AGCCAACGTACCCAGGTC-3′ (Fig. 1A). The PCR primers (Fw, 5′-AAGTCGAGCGGTAGCAGG-3′; Rv, 5′-GGTGAGCCATTACCTCACCT-3′) targeted 290 bp of 16S rRNA from E. tarda.
FIG. 1.
(A) Nucleotide sequence of partial hemolysin gene used to design inner and outer primers for LAMP. The nucleotide sequences and the positions used to design the primers are represented by dashed lines and labels. (B) A schematic diagram showing the positions at which the primers attach for amplification of the target gene. The picture has been adapted from data provided by Eiken Chemical Co. Ltd.
DNA extraction and LAMP.
The DNA template was prepared by growing a 1-ml culture of E. tarda (FPC 498; isolated from a Japanese flounder, Nagasaki, Japan) in Luria-Bertani broth to late logarithmic phase (12 h postinoculation) and by centrifugation at 5,000 × g for 3 min to form a pellet. The pellet was then resuspended in 500 μl of Tris-EDTA buffer, boiled for 5 min at 95°C, and centrifuged to collect the supernatant. This supernatant was used for LAMP and PCRs. LAMP reactions were carried out using a Loopamp kit (Eiken Chemical Co. Ltd., Tokyo, Japan). In brief, LAMP was carried out in a total of 25 μl of reaction mixture containing 40 pmol of FIP and BIP, 5 pmol of F3 and B3, 12.5 μl of 2× reaction mixture (40 mM Tris-HCl [pH 8.8], 20 mM KCl, 16 mM MgSO4, 20 mM [NH]4SO4, 0.2% Tween 20, 1.6 M betaine, deoxynucleotide triphosphates [dNTPs] [2.8 mM each]), Bst DNA polymerase, and template DNA. The reaction mixture was incubated at 65°C for 45 min and then at 80°C for 2 min to terminate the reaction. A total of 2 μl of the product was analyzed in 2.0% agarose gel by electrophoresis. The reaction produced many bands of various sizes, ranging from approximately 100 bp to the capacity of the loading well (Fig. 1).
Optimization of LAMP for E. tarda detection.
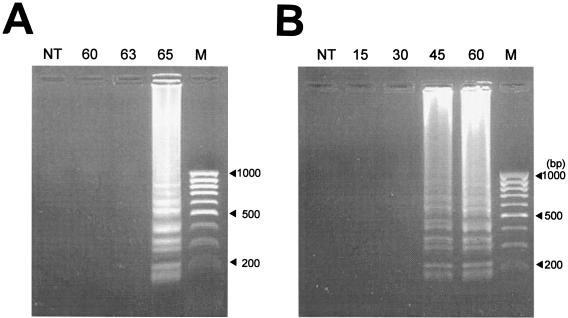
Detection (using LAMP) of E. tarda in diseased fish was standardized in this study. To optimize the conditions needed for the specific amplification of the hemolysin gene of E. tarda, the DNA template from strain FPC 498 (Table 1) was used. The product was formed at 65°C with an amplification time of 60 min. No amplification was detected at 60 and 63°C (Fig. 2A). When time-specific amplification for 15, 30, 45, and 60 min was carried out, amplification of the E. tarda DNA template at 65°C was detected at as early as 45 min (Fig. 2B). On the basis of the above analyses, the conditions of amplification were optimized as 45 min at 65°C.
TABLE 1.
E. tarda strains used in this study
| Strain | Origin | Date | Location |
|---|---|---|---|
| E 22 | Blood (eel) | 1972 | Shizuoka, Japan |
| E 381 | Kidney (tilapia) | 1979 | Niigata, Japan |
| FPC 498 | Ascites (flatfish) | 1978 | Nagasaki, Japan |
| SU 226 | Eel pond water | 1981 | Shizuoka, Japan |
| 158 | Sea bream | 1995 | Kagoshima, Japan |
FIG. 2.
Determination of LAMP conditions for E. tarda detection. (A) Effect of temperature on the amplification of a LAMP product. Amplification (using FPC 498 as the DNA template) at 60, 63, and 65°C was carried out for 60 min. (B) Effect of time on amplification of E. tarda by LAMP. Amplification using FPC 498 was carried out at 65°C for 15, 30, 45, and 60 min. A 100-bp molecular mass marker is shown in the extreme right lane, and the results of a control reaction with no template (NT) are shown in the first lane.
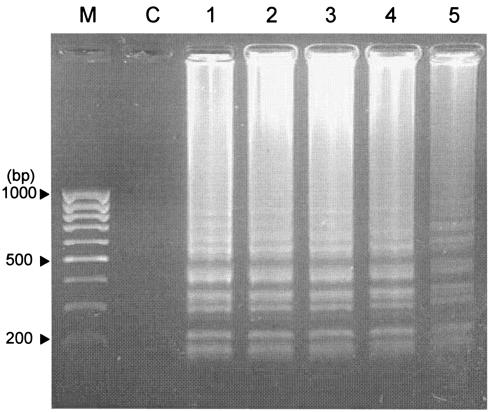
To determine the specificity of detection, LAMP reactions were carried out (using E. tarda hemolysin-specific LAMP primers) in experiments with E. tarda, Salmonella enterica serovar Typhi, E. coli, Shigella sonnei, Proteus mirabilis (kindly provided by Yoshitaka Goto, Miyazaki University, Miyazaki, Japan), Vibrio sp., Aeromonas hydrophila, Enterococcus sp., and Photobacterium sp. After 45 min at 65°C, using templates of DNA from other enteric bacteria did not produce any amplification. Some of these isolates are known to produce enterotoxins and have been isolated from fish-rearing facilities. Five strains of E. tarda (Table 1) obtained from farms in Japan and tested using E. tarda LAMP primers gave specific amplification products (Fig. 3).
FIG. 3.
Specificity for hemolysin gene amplification using a LAMP reaction and different strains of E. tarda. Lanes 1 to 5, strains E 381, FPC 498, SU 226, E 22, and 158. A 100-bp molecular-mass marker (M) is shown in the extreme left lane. C, control reaction with distilled water.
Previous reports have shown that LAMP can detect target DNA at levels as low as 6 copies (10). In this study, using a LAMP method we detected the hemolysin gene of E. tarda from DNA obtained from 10 to 109 CFU of E. tarda. This was achieved by a 10-fold serial dilution of 2.9 ×109 CFU E. tarda in phosphate-buffered saline. The colonies were enumerated on a Casamino Acids (0.5%)-supplemented Trypticase soy agar plate. E. tarda detection (targeting 16S rRNA) by PCR was carried out to compare the levels of sensitivity of LAMP. All PCRs were performed according to the following protocol. A total of 1 μl of cDNA was mixed with 5 μl of dNTPs (10 μM concentrations of each dNTP), 0.5 μl of Taq polymerase (5 U/μl), 5 μl of each gene-specific primer (5 pmol/μl), and 27.5 μl of water. The PCR was performed using a PCR apparatus (MJ Research Inc., Waltham, Mass.) under conditions of 0.5 min at 94°C, 0.5 min at 60°C, and 1 min at 72°C for 30 cycles. However, PCR amplified the target gene at a dilution of bacterial DNA of up to 103 CFU. The sensitivity of the PCR detection also depends (among several other factors) upon the efficiency of the primers, and the use of nested PCR can increase the sensitivity of detection. The need for a specialized machine, and the time required for the detection by PCR, makes LAMP-mediated detection advantageous. However, until recently, a fluorescent antibody technique (4) and PCR of the hemolysin gene (1) were used to diagnose edwardsiellosis. Thus, these observations show that E. tarda detection using LAMP is highly specific and sensitive.
Diagnosis of edwardsiellosis by LAMP.
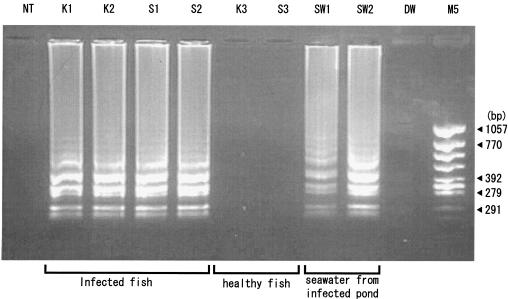
Fish diseases are a major problem in the aquaculture industry because of the economic losses incurred. The rapidity and sensitivity of LAMP make it a suitable technique for the detection of pathogens in aquaculture facilities and for management of disease. Healthy and E. tarda-infected samples of Japanese flounder (Paralichthys olivaceus) were obtained from Ehime Prefectural Chuyo Fisheries Experimental Station, Iyo, Japan. The kidney and spleen were removed aseptically, and DNA was extracted using a DNA extraction kit (ISOHAIR; Nippon Gene, Toyama, Japan). E. tarda was detected in the infected kidney (n = 5) and spleen (n = 5) samples. Healthy spleen and kidney samples did not produce amplification when specific primers for E. tarda detection by LAMP were used (Fig. 4). Seawater samples from E. tarda-infected Japanese flounder culture ponds contained 3.8 × 102 CFU. A total of 1 ml of the seawater was aliquoted and centrifuged at 16,000 × g to obtain a pellet, which was dissolved in 10 μl of distilled water and tested (using LAMP) for the presence of E. tarda. Seawater samples were positive for the presence of E. tarda.
FIG. 4.
Detection of E. tarda hemolysin gene in samples from healthy and infected Japanese flounder and in samples of seawater from an infected culture facility. K1 and K2, E. tarda-infected kidney samples; S1 and S2, E. tarda-infected spleen samples; K3 and S3, healthy kidney and spleen samples. SW1 and SW2 are seawater samples from E. tarda-infected ponds. Molecular mass marker 5 (M5) (φ/X174/HincII digest) is shown in the last lane on the right. All the products were electrophoresed on 2.0% agarose gels and stained with ethidium bromide. NT, no template; DW, distilled water.
In conclusion, LAMP, a rapid and highly sensitive system for detecting edwardsiellosis, has been designed. This is the first report of the use of a LAMP technique, which has applications for the detection of environmental isolate. This method can be effectively used for diagnosis of edwardsiellosis in fish and in a culture environment.
REFERENCES
- 1.Chen, J. D., and S. Y. Lai. 1998. PCR for direct detection of Edwardsiella tarda from infected fish and environmental water by application of the hemolysin gene. Zool. Stud. 37:169-176. [Google Scholar]
- 2.Chen, J. D., S. Y. Lai, and S. L. Huang. 1996. Molecular cloning, characterization, and sequencing of the hemolysin gene from Edwardsiella tarda. Arch. Microbiol. 165:9-17. [DOI] [PubMed] [Google Scholar]
- 3.Hirono, I., N. Tange, and T. Aoki. 1997. Iron-regulated haemolysin gene from Edwardsiella tarda. Mol. Microbiol. 24:851-856. [DOI] [PubMed] [Google Scholar]
- 4.Kawahara, E., and R. Kusuda. 1987. Direct fluorescent antibody technique for diagnosis of bacterial disease in eel. Bull. Jpn. Soc. Sci. Fish. 53:395-399. [Google Scholar]
- 5.Kusuda, R., and N. Kitadai. 1993. Hemolysin production by Edwardsiella tarda isolated from eel, Anguilla japonica. Jpn. Soc. Aquacult. Res. 41:251-255.
- 6.Maruyama, F., T. Kenzaka, N. Yamaguchi, K. Tani, and M. Nasu. 2003. Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification. Appl. Environ. Microbiol. 69:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 8.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Nagamine, K., Y. Kuzuhara, and T. Notomi. 2002. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochem. Biophys. Res. Commun. 290:1195-1198. [DOI] [PubMed] [Google Scholar]
- 10.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 11.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [Online.] [DOI] [PMC free article] [PubMed]
- 12.Ullah, M. A., and T. Arai. 1983. Pathological activities of naturally occurring strains of Edwardsiella tarda. Fish. Pathol. 18:65-70. [Google Scholar]
- 13.Wilson, J. P., R. Rebecca, M. D. Waterer, J. D. Wofford, M. D. Stanley, and M. D. Chapman. 1989. Serious infections with Edwardsiella tarda. Arch. Intern. Med. 149:208-210. [PubMed] [Google Scholar]