Abstract
The multianalyte array biosensor (MAAB) is a rapid analysis instrument capable of detecting multiple analytes simultaneously. Rapid (15-min), single-analyte sandwich immunoassays were developed for the detection of Salmonella enterica serovar Typhimurium, with a detection limit of 8 × 104 CFU/ml; the limit of detection was improved 10-fold by lengthening the assay protocol to 1 h. S. enterica serovar Typhimurium was also detected in the following spiked foodstuffs, with minimal sample preparation: sausage, cantaloupe, whole liquid egg, alfalfa sprouts, and chicken carcass rinse. Cross-reactivity tests were performed with Escherichia coli and Campylobacter jejuni. To determine whether the MAAB has potential as a screening tool for the diagnosis of asymptomatic Salmonella infection of poultry, chicken excretal samples from a private, noncommercial farm and from university poultry facilities were tested. While the private farm excreta gave rise to signals significantly above the buffer blanks, none of the university samples tested positive for S. enterica serovar Typhimurium without spiking; dose-response curves of spiked excretal samples from university-raised poultry gave limits of detection of 8 × 103 CFU/g.
Salmonellosis is a serious health concern and is responsible for approximately 30% of all reported cases of food poisoning in the Unites States for which the etiology is determined (42). While forty to fifty thousand cases of salmonellosis and 500 deaths due to salmonellosis are reported in the United States each year (11, 42, 66), estimates of the true incidence of salmonellosis cases are much higher, with estimates of economic costs ranging from $1.3 to $4.0 billion each year (19, 65).
Current Food and Drug Administration methods for detecting contamination of foodstuffs with Salmonella consist of conventional culture techniques, immunoassays, and biochemical analyses (3, 47). Microbiological culture methods typically involve pre-enrichment of food samples by incubation in a nonselective medium, followed by one or more selective enrichments. Because each enrichment cycle may take up to 24 h, these methods therefore require up to 36 h before results are known. Validated immunoassays and biochemical analyses (Gene-Trak, SalmonellaTek, Assurance, 1-2 Test, TECHRA, and VIDAS), as well as other nonvalidated “rapid” analytical methods, also require pre-enrichment of samples, resulting in a turnaround time of at least 24 h (2). Furthermore, many of these methods require significant sample preparation and/or manipulation and lengthy assay procedures. Thus, the need remains for additional methods capable of detecting Salmonella contamination rapidly and with a minimum of manipulations.
In this report, we describe the rapid detection of Salmonella enterica serovar Typhimurium in a variety of foodstuffs with minimal sample preparation by use of the multianalyte array biosensor (MAAB) (48, 49, 52). This system consists of an array of biological recognition elements, a light source for fluorescence excitation, an optical detector, and a fluidics component. The recognition elements are immobilized onto an optical waveguide in patterned arrays, either as a microarray (15, 16) or as a series of parallel stripes. Typically, these recognition elements are antibodies, although other recognition species have been used (50). Multiple samples are applied to the waveguide such that each sample encounters multiple capture elements. Analytes present within each sample bind to appropriate capture elements and are then detected by fluorescent tracer antibodies. With cocktails of tracer antibodies and capture elements with different specificities, up to nine different analytes have been detected on a single waveguide (61).
The MAAB uses the evanescent wave, an electromagnetic component of the light launched into the wave guide, to selectively excite fluorophores present in the array of surface-bound immunocomplexes. The surface specificity of the evanescent wave allows real-time measurements of binding reactions (54) as well as analysis of turbid or nonhomogeneous samples, with little interference from the sample matrix (49, 52). The use of this method and other evanescent wave techniques for biosensing has been well established (36, 46, 55, 67). This report documents the first use of the MAAB to detect a pathogen in food samples. Also demonstrated is the potential for use in preharvest testing of chickens for Salmonella infection.
MATERIALS AND METHODS
Antibodies, antigens, and biochemicals.
Antibodies and antigens were purchased from the following sources: monoclonal anti-S. enterica serovar Typhimurium lipopolysaccharide (clone M32242), Fitzgerald Industries International (Concord, Mass.); heat-killed S. enterica serovar Typhimurium, Escherichia coli O157:H7, and Campylobacter jejuni, KPL (Gaithersburg, Md.); and rabbit anti-Salmonella sp. immunoglobulin G, Biodesign International (Saco, Maine). Low-biotin-content bovine serum albumin (BSA) was purchased from Sigma-Aldrich (St. Louis, Mo.). EZ-Link biotin-LC-NHS ester, NeutrAvidin biotin-binding protein, and N-succinimidyl 4-maleimidobutyrate (GMBS) were obtained from Pierce (Rockland, Ill.).
Preparation of antibodies.
The capture antibody, rabbit anti-Salmonella spp., was biotinylated by 30 min of incubation with a fivefold molar excess of EZ-Link biotin-LC-NHS ester in 0.1 M borate, pH 8.5. Unincorporated biotin was then separated from biotinylated antibody by gel chromatography with Bio-Gel P-10 (Bio-Rad, Hercules, Calif.), and the antibody was equilibrated in phosphate-buffered saline, pH 7.4 (PBS). The tracer antibody, monoclonal anti-S. enterica serovar Typhimurium lipopolysaccharide (clone M32242), was labeled with Cy5 bisfunctional NHS ester (Amersham, Arlington Heights, Ill.) by incubation of 1 packet of dye with 3 mg of protein in 0.1 M borate, pH 8.5, for 30 min. Labeled protein was separated from unbound dye by gel chromatography, as described above. Dye-to-protein ratios of 2:1 to 4:1 were obtained; labeling efficiencies in this range have been shown to be optimal for this fluorophore (1).
Preparation of assay substrates.
NeutrAvidin biotin-binding protein was covalently immobilized onto sensor substrates essentially as described previously (49). Briefly, cleaned soda lime slides (Daigger, Wheeling, Ill.) (13) were treated with a 2% solution of 3-mercaptopropyl triethoxysilane (Fluka, Ronkonkoma, N.Y.) in toluene for 1 h under nitrogen, rinsed thrice with toluene, and dried. The silanized slides were subsequently incubated with 1 mM GMBS in anhydrous ethanol for 30 min, rinsed with deionized water, and incubated overnight with 30 μg of NeutrAvidin per ml in PBS. After rinsing in PBS, NeutrAvidin-coated slides were stored in PBS until patterning was performed.
Biotinylated capture antibodies were immobilized in patterned stripes on the NeutrAvidin-treated slides by use of 12-channel poly(dimethyl)siloxane patterning templates; the flow channels were oriented along the short (1 in.) axis of the slide (dimensions of each channel, 21 by 1 by 2.5 mm3 [length by width by height]). Approximately 60 μl of biotinylated capture antibody (15 μg/ml in PBS) was loaded into each channel and was incubated overnight at 4°C. After the capture antibody solution was removed, each lane was rinsed with 1.0 ml of PBS containing 0.05% Tween 20 and 1 mg of BSA/ml (PBSTB), and the patterning template was removed. The slides were then immersed in a blocking and drying solution of 10 mg of BSA per ml in 10 mM sodium phosphate buffer, pH 7.4, dried under a stream of nitrogen without further rinsing, and stored in the dark at 4°C until use; dried, patterned slides processed in this manner have been stored up to 17 weeks without a significant loss in activity (62).
Biochemical assays.
Sample analysis was performed on the antibody-patterned slides by use of poly(dimethyl)siloxane assay flow guides similar to those used for patterning, except that the channels (dimensions, 40 by 1 by 2.5 mm3 [length by width by height]) were oriented orthogonal to those molded in the patterning template. After placement of the flow guide onto the patterned slide, the ends of each channel were connected to a peristaltic pump (outlet) or reservoir (inlet), and each lane was washed with 0.8 ml of PBSTB at a flow rate of 0.8 ml/min. Samples (0.8 ml/lane, diluted in PBSTB) were loaded into the reservoirs and allowed to flow over the slide surface at a rate of 0.1 ml/min. Following a buffer wash (0.8 ml PBSTB, 0.8 ml/min), tracer antibody solution (15 μg/ml in PBSTB, 0.45 ml/lane) was allowed to flow over the wave guide at a rate of 0.1 ml/min. After a final rinse with PBSTB, the assay template was removed from the slide, and the slide was rinsed with deionized water, dried, and imaged.
To determine whether increased incubation times would improve the assay sensitivity, the standard 15-min assay protocol was extended to approximately 1 h. The sample incubation time was increased to 40 min, with recirculation of the sample, and the tracer antibody incubation time was extended to 20 min, with recirculation of the tracer solution.
Biosensor optics.
The optical components of the MAAB consisted of a stage for holding the slide, a source of illumination for evanescent excitation (635-nm, 12 mW diode laser; Lasermax, Rochester, N.Y.), a GRIN lens array (Nippon Sheet Glass, Somerset, N.J.) (24), a series of filters, and a Peltier-cooled charge-coupled device (SpectraSource Instruments, Westlake Village, Calif.) for imaging; more complete details are presented elsewhere (20, 25, 67, 68). The angle of incidence of excitation light (approximately 36 degrees) was such that it allowed evanescent excitation of the fluorescent immunocomplexes in the patterned region of the slide.
Data analysis.
Data were extracted from the digitized images by use of a custom automated data analysis program (54). After control spots were manually located with a “spot-pick” tool, the program automatically located the remaining spots in the array by a Sobel edge detection algorithm. For each array element, the mean fluorescence intensity within each spot and the background fluorescence from both sides of the spot were determined. The net mean fluorescence was then calculated by subtracting the mean local background value from the mean fluorescence within the spot. Limits of detection (LODs) were calculated as the lowest tested concentrations at which the average fluorescent signal (number of squares; n = 10) was at least 3 standard deviations above the mean fluorescence intensity of the buffer blank (number of blank squares; n = 10). To account for interslide variability and effects of sample matrices, positive controls (107 CFU of S. enterica serovar Typhimurium per ml in buffer) and negative controls (unspiked buffer) were analyzed in parallel with the samples. All sample data shown were normalized to the positive control for direct comparison of multiple slides.
Unpaired Student's t tests were used to compare unspiked food samples with negative controls (unspiked buffer). Signals from different samples of excreta were compared by analysis of variance, with randomized blocks. Analysis of covariance was used to test for differences in regression lines calculated from dose-response curves for spiked foodstuffs and spiked excreta. Since saturation was observed at high concentrations, only the ranges of concentrations at which regression (R2) values were >0.98 were included in the analyses of covariance. These ranges of concentrations corresponded to the following: ≤2 × 106 CFU/ml for all spiked foodstuffs, E. coli-spiked samples, and C. jejuni-spiked samples; ≤4 × 104 CFU/ml for excreta from university-raised poultry; and ≤4 × 106 CFU/ml for excreta from leisure farm-raised chickens.
Preparation of foodstuffs.
Cubed whole cantaloupe was mixed with an equal volume of 2× concentrated stock of PBS containing 2 mg of BSA per ml (2× PBS-BSA) and was spiked with various concentrations of S. enterica serovar Typhimurium. The samples were homogenized on high for 2 min in a Waring blender (Torrington, Conn.) and centrifuged at 3,000 × g, and the liquid fractions were analyzed.
Whole liquid eggs were diluted with equal volumes of 2× PBS-BSA containing various concentrations of S. enterica serovar Typhimurium and mixed on high for 2 min in a Waring blender. The diluted egg homogenates were used in the MAAB assays without further treatment.
The chicken carcass rinse was prepared by incubating a fresh chicken (approximately 2.7 kg) with 100 ml of 1× PBS-BSA in a large zip-lock bag for 2 h on a rocker. The rinse was removed from the bag and immediately frozen for later analysis. After thawing, aliquots were spiked with S. enterica serovar Typhimurium and analyzed without further treatment.
Samples of a national brand pork sausage, with a fat content of approximately 35%, were combined with 2 volumes of 2× PBS-BSA containing various concentrations of S. enterica serovar Typhimurium. The sausage-buffer mix was homogenized in a Waring blender for 2 min. After centrifugation at 3,000 × g, the liquid fraction immediately under the uppermost layer of fat was removed and analyzed immediately without further purification.
For preparation of the sprout rinse, fresh alfalfa sprouts were combined with an equal volume of PBSTB containing various concentrations of S. enterica serovar Typhimurium. After swirling for 15 s, the particulates were allowed to settle. The liquid supernatant (sprout rinse) was removed with a pipette and analyzed immediately, without filtration or further clarification. Sprout homogenates were prepared separately by mixing 9 volumes of PBSTB (containing various concentrations of S. enterica serovar Typhimurium) with fresh alfalfa sprouts, mixing them on high in a Waring blender for 30 s (60), and filtering them through Whatman 1 filter paper.
Poultry excreta.
Chicken fecal samples were obtained from two poultry facilities owned by the University of Maryland and a noncommercial, leisure farm in New York. Chickens housed in university facilities were kept in separate cages; thus, the excreta from each bird could be collected as an individual sample. Excreta from chickens housed at the leisure farm were collected as a pool, as birds were housed in a communal coop. Excretal samples were weighed and mixed with 2 volumes of 2× PBSTB containing various concentrations of S. enterica serovar Typhimurium. After vortexing for 30 s, samples were incubated at room temperature for 1 h. Particulates were removed by filtration through Whatman 1 filter paper and samples were immediately analyzed.
RESULTS
Detection of S. enterica serovar Typhimurium in buffer.
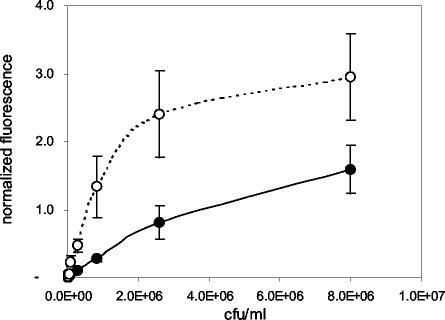
Dose-response curves were determined for heat-killed S. enterica serovar Typhimurium spiked into laboratory buffer, using standard (15-min) and extended (1-h) assay formats (Fig. 1). The LOD for the standard, 15-min assay was 8 × 104 CFU/ml, although samples containing 2.6 × 104 CFU/ml occasionally gave signals above those of the negative controls (P < 0.05). When sample and tracer incubations were increased to 40 and 20 min, respectively, the net background fluorescence and signals from negative controls did not change (P > 0.05), but fluorescence signals generated from positive samples increased, resulting in a 10-fold improvement in the LOD (8 × 103 CFU/ml).
FIG. 1.
Dose-response curves for S. enterica serovar Typhimurium in buffer. Closed circles, results obtained using the standard, 15-min assay format; open circles, results from the extended, 1-h assay. Values shown on the ordinate are mean normalized fluorescence values (± standard errors of the means [SEM]; n = 10) from duplicate slides.
Detection of S. enterica serovar Typhimurium in spiked foodstuffs.
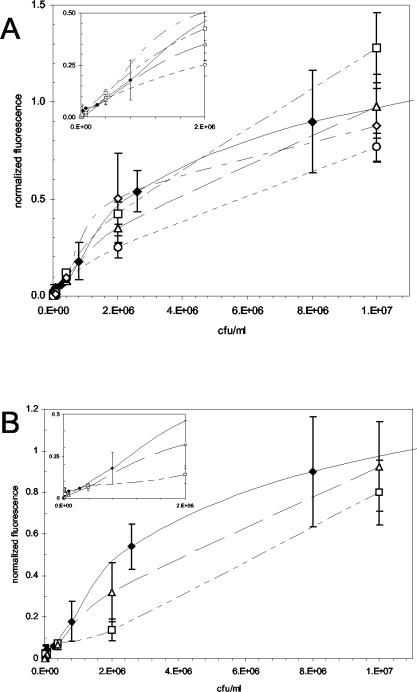
Various foodstuffs were spiked with S. enterica serovar Typhimurium and tested by the standard 15-min assay protocol (Fig. 2). With the exception of the chicken carcass rinse, spiking of the foodstuffs was performed before processing to assess the combined effects of the sample matrix and the processing procedures. The concentrations of S. enterica serovar Typhimurium in the buffer used to spike the solid foodstuffs are shown on the abscissa; for liquid samples, the value shown is the final concentration of S. enterica serovar Typhimurium after spiking. These values (CFU per milliliter) are shown in place of CFU per gram to facilitate comparison between the various matrices and buffer samples (shown in black diamonds in both panels). LODs in spiked foodstuffs were as follows: 8 × 104 CFU/ml in cantaloupe and chicken carcass rinse, 4 × 105 CFU/ml in sausage, and 1.6 × 104 CFU/ml in whole liquid egg (panel A); and 4 × 105 CFU/ml in both sprout rinse and sprout homogenate (panel B). These concentrations correspond to 8 × 104 CFU/g of cantaloupe, 8 × 105 CFU/g of sausage, 1.6 × 104 CFU/g of egg, 4 × 105 CFU/g of sprouts for sprout rinse, and 3.6 × 106 CFU/g for sprout homogenate. A detection limit of 2.7 × 103 CFU/cm2 for the chicken carcass rinse was determined by using Thomas's (64) formula for converting carcass weight to total surface area.
FIG. 2.
Dose-response curves for S. enterica serovar Typhimurium spiked into foodstuffs and food extracts. Values shown on the abscissa indicate the concentrations of S. enterica serovar Typhimurium in the spiking solution (for sausage, egg, cantaloupe, and sprout homogenate) or the final concentrations of S. enterica serovar Typhimurium (for carcass rinse and sprout rinse). Values shown on the ordinate are mean normalized fluorescence values (± SEM; n = 10) from duplicate slides. (A) Open diamonds, spiked sausage; squares, cantaloupe; triangles, chicken carcass rinse; circles, whole liquid egg. (B) Triangles, spiked sprout rinse; squares, sprout homogenate. The dose-response curve for S. enterica serovar Typhimurium in buffer is shown for comparison (closed diamonds). Insets show sensor responses for the linear portion of the dose-response profiles.
None of the unspiked food matrices gave rise to signals significantly above that of the buffer blank (P > 0.05). While dose-response regressions for the spiked cantaloupe, sprout rinse, sausage, and chicken carcass rinse were not significantly different from dose-response curves for buffer (P > 0.05), the MAAB response was significantly inhibited by the presence of egg or sprout homogenate (P < 0.005).
Cross-reactivity.
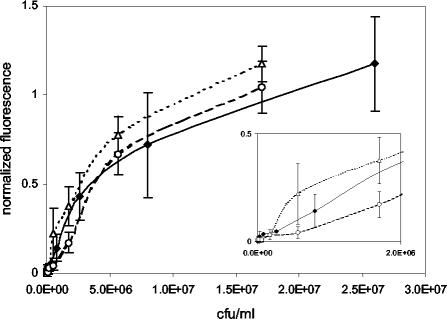
Dose-response curves for S. enterica serovar Typhimurium were constructed in the presence of two potentially co-occurring bacteria. Various concentrations of S. enterica serovar Typhimurium were spiked into solutions containing either 108 CFU of E. coli O15:H7 per ml or 12 μg of C. jejuni per ml and tested by the standard 15-min assay protocol. Although samples containing C. jejuni or E. coli alone did not give rise to signals significantly above those for negative control values (buffer alone, P > 0.2), the LODs for mixed assays were 1.7 × 105 CFU/ml (E. coli) and 5.6 × 105 CFU/ml (C. jejuni), which are 3 to 10 times higher than the LOD determined for S. enterica serovar Typhimurium alone (Fig. 3). Furthermore, while dose-response curves for E. coli-spiked samples did not differ from those for controls (P > 0.25), the sensor response was dampened in samples spiked with C. jejuni (P < 0.05).
FIG. 3.
Dose-response curves for S. enterica serovar Typhimurium in the presence of E. coli and C. jejuni. Samples containing S. enterica serovar Typhimurium were spiked with either E. coli O15:H7 (108 CFU/ml) (triangles) or C. jejuni (12 μg/ml) (circles). The dose-response curve for S. enterica serovar Typhimurium alone is shown for comparison (diamonds). Values shown on the abscissa indicate the concentrations of S. enterica serovar Typhimurium. Values shown on the ordinate are mean normalized fluorescence values (± SEM; n = 10) from duplicate slides. The inset shows sensor responses for concentrations in the linear range.
Detection of S. enterica serovar Typhimurium in chicken excreta.
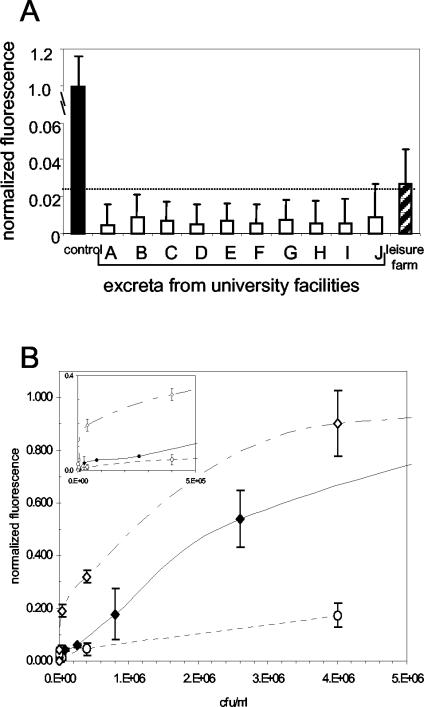
Standard 15-min assay protocols were used to test samples of chicken excreta obtained from birds raised either in university facilities or on a noncommercial, leisure farm (Fig. 4). None of the excretal samples from university facilities had fluorescent signals that were significantly above that of the negative control (P > 0.25) or above the threshold for LOD determination (panel A). On the other hand, pooled excreta from the leisure farm chickens gave rise to fluorescent signals that were significantly above the background (P < 0.01), above those from the University of Maryland samples (P < 0.005), and above the threshold for the LOD. To determine the effect of each matrix on the MAAB response, different concentrations of S. enterica serovar Typhimurium were spiked into each matrix and dose responses were determined (panel B). Whereas the presence of excreta from university-housed chickens significantly enhanced the sensor response to added S. enterica serovar Typhimurium (top curve; P < 0.005), the sensor response was significantly inhibited in the presence of excreta from the leisure farm chickens (bottom-most curve; P < 0.005). Furthermore, S. enterica serovar Typhimurium added to the excretal samples from the university chickens was detectable at 4 × 103 CFU/ml (8 × 103 CFU/g), well below the LOD for spiked buffer. On the other hand, given the lower slope of the leisure farm dose-response curve, a significantly higher concentration of S. enterica serovar Typhimurium was needed to boost the signal to levels that were significantly higher than those from unspiked excreta (4 × 106 CFU/ml = 8 × 106 CFU/g excreta; P < 0.05).
FIG. 4.
Detection of S. enterica serovar Typhimurium in chicken excreta. In both panels, values shown on the ordinate are mean normalized fluorescence values (± SEM; n = 10) from duplicate slides. (A) Unspiked excreta. Black bar, positive control (107 CFU/ml in buffer); white bars, samples from university facilities; striped bar, pooled sample from the leisure farm. The horizontal line (dashed) indicates the threshold value for determining the LOD. (B) Dose-response curves for spiked excreta. Open diamonds, excreta from university-raised chickens; circles, excreta from leisure farm-raised chickens. The dose-response curve for S. enterica serovar Typhimurium in buffer (closed diamonds) is shown for comparison. The inset shows the sensor responses for concentrations of spiked S. enterica serovar Typhimurium of <5 × 105 CFU/ml.
DISCUSSION
The MAAB is an optical biosensor designed for rapid and sensitive detection of multiple analytes in a single assay. The MAAB has been used for the detection of toxins, physiological markers of health, small organic molecules, a virus, and both gram-negative and gram-positive bacteria (48, 49, 51, 53). LODs are generally in the low-to-mid nanogram per milliliter range for proteins and small molecules and in the range of 103 to 105 CFU/ml and 107 PFU/ml for bacteria and viruses, respectively.
While bacteriological culture remains the most sensitive method and the “gold standard” for detecting the presence of Salmonella and other bacterial species in foods (3, 47, 66), the MAAB offers the advantage of detecting multiple analytes in a single test without significant sample preparation or aseptic technique. For both multianalyte and single-analyte assays, the two-dimensional nature of the MAAB's sensing surface allows for positive and negative controls, as well as standards, to be analyzed in parallel with noncharacterized or unknown samples, facilitating direct comparisons.
Rapid, single-analyte immunoassays were developed for the detection of S. enterica serovar Typhimurium, the type of Salmonella responsible for the largest percentage of salmonellosis cases in the United States in the past 12 years (42). Dose-response curves were created for spiked buffer samples to assess the MAAB's overall performance and to provide a basis for comparison with other so-called rapid methods of detection. The LODs determined here for both 15-min and 1-h protocols (∼104 CFU/ml) compare favorably with those for several other antibody-based systems (LODs in the range of mid-103 to 105 CFU/ml), including piezoelectric sensors (4, 33, 43, 45, 59), fiber optic sensors (44, 71, 72), an interferometer (57), surface plasmon resonance-based sensors (9, 56), and an electrochemical detector (22). ORIGEN, a system based on electrochemiluminescence, has proven to be the most sensitive rapid detection system, with a detection limit of 1,000 CFU/ml (70). Although several additional papers have reported lower LODs (33, 43, 69), data from appropriate blanks, controls, or replicates were not presented.
MAAB assays were used to test spiked samples of foodstuffs that are often implicated in cases or outbreaks of salmonellosis (11, 39, 42, 63). While none of the unspiked foods gave rise to false positive results, some matrix effects were observed with sausage and sprout samples. Detection limits were higher for the sausage and sprout rinse samples, but a dampening of the sensor response was not noted. On the other hand, both the LOD and sensor response were significantly affected by the presence of sprout homogenate. This overall decrease in performance may be due to the high levels of polyphenols found in sprouts, which were previously shown to interfere with immunoassays (C. Fajardo-Lira, S. M. Henning, H. W. Lee, V. L. W. Go, and D. Heber, Abstr. 2002 Annu. Meet. Inst. Food Technologists, abstr. 46C-20, 2002); it has been speculated that polyphenols may adsorb to antibodies, preventing antibody-antigen binding (41). While the sensor performed poorly in the presence of homogenate, the LODs observed with sprout rinse samples, an accurate indicator of sprout contamination (60), were below the levels observed in pathogen growth studies using inoculated or naturally contaminated seeds (107 to 108 CFU/g) (21, 32).
Although the MAAB's LODs for spiked carcass rinse samples compared favorably with those of the interferometric sensor (57), the MAAB assays were less sensitive than ORIGEN (70) and an enzyme-catalyzed electrochemical system (12) in analogous assays. As with the MAAB, matrix effects were also observed with the ORIGEN system, most significantly in the presence of milk products and fish (70). The authors of the ORIGEN study conjectured that the presence of heavy metals and redox components in the fish samples interfered with the ORIGEN assays, giving high backgrounds (potential false positives); clumping of the magnetic particles was at least partially responsible for the false negative results obtained with milk. While the present study did not include dairy products, we recently observed a decrease in MAAB performance during analysis of milk samples for staphylococcal enterotoxin B (58); although the LOD was unchanged for the staphylococcal enterotoxin B assays, the dose response was significantly dampened in the presence of milk.
PCR is an exquisitely sensitive technique and is able to detect 1 to 100 salmonellae in pure culture (10, 35, 38). However, nucleic acid-based analysis of foods and fecal matter has proven challenging due to the presence of inhibitors of PCR amplification, such as proteases and phenolics (18). Thus, a pre-enrichment step is typically required to increase the number of viable cells while effectively diluting inhibitory substances present in the sample. A large number of papers describing PCR-based detection of Salmonella from spiked or naturally contaminated foods have claimed LODs ranging from 0.1 to 100 CFU/g (6, 10, 31, 37, 40). However, many of these LODs represent the concentrations of inoculum used rather than actual bacterial counts after enrichment; postenrichment concentrations can reach 106 to 109 CFU/ml (6, 7, 17, 23). Detection limits determined in studies reporting LODs based on postenrichment cell densities are in the range of 103 to 106 CFU/ml (5, 6, 17), comparable to the LOD determined for this study. A PCR system able to detect Salmonella in a carcass rinse sample without pre-enrichment has recently been described (30); the LOD reported (102 to 104 CFU/cm2) is comparable to that obtained for the MAAB (2.7 × 103 CFU/cm2).
The issue of cross-reactivity was addressed by performing assays in the presence of two other pathogens often found in the same types of foods. Whereas samples containing C. jejuni or E. coli alone were not cross-reactive in the Salmonella assays, some decrease in sensor response was observed for combination studies. These results were not surprising; cross-reactive capture of these other bacteria (present in large excess) would effectively reduce the concentration of sites available for binding Salmonella. The use of a monoclonal antibody as a tracer conferred selectivity to these assays; thus, signals from bound C. jejuni or E. coli were not significant. We have also recently tested Listeria monocytogenes and Shigella dysenteriae in assays for Salmonella and again observed only minimal levels of nonspecific binding (K. Sapsford and N. Kulagina, personal communications). Other detection systems have shown a similar overall lack of interference by the presence of nonrelevant bacteria (4, 12, 57, 59). Although we did not attempt to assess the specificity of the system using other Salmonella species, this method should prove amenable to detection of other salmonellae, provided that a more broadly reactive tracer antibody of similar affinity is used.
Fecal samples have been demonstrated to be good indicators of systemic infection (28, 34). Furthermore, chicken flocks with high levels of fecal contamination of Salmonella are up to 10 times more likely to produce contaminated eggs as those with low levels of fecal Salmonella (27). Thus, a rapid and sensitive method for measuring fecal load may prove a useful method for monitoring infected flocks and preventing contamination of eggs (and potentially meat) for human consumption.
Excreta from the different sources differed widely in their responses in the MAAB assays. While none of the university samples gave false positive signals, the sensor response for dose-response curves was enhanced for these samples. On the other hand, signals from unspiked leisure farm excreta were significantly higher than those from negative controls, but the presence of this matrix resulted in an overall dampening of the sensor response with increasing concentrations of added S. enterica serovar Typhimurium. While it is likely that the leisure farm excreta contained significant quantities of endogenous Salmonella, institutional regulations limiting the growth and use of live S. enterica serovar Typhimurium prevented the determination of actual bacterial counts from these samples; thus, it is not known whether the high signals in unspiked leisure farm excreta were due to endogenous Salmonella or should be considered false positives. In spite of these limitations, detection limits for all spiked fecal samples (from the university and the leisure farm) were below or in the same range as concentrations observed for excreta from experimentally infected birds (29).
Various biosensors have been developed for their potential application in rapid testing of foodstuffs for Salmonella. To date, however, none of these sensors has found widespread use for this purpose and, with the exception of ORIGEN (70), none have been tested with more than one or two different sample types. The results described here indicate that the MAAB not only has similar or better detection limits than other sensors, but also requires only 15 min for analysis of prepared samples. Sample processing is simple, requiring only homogenization of the foodstuff or excreta, followed by coarse filtration or centrifugation; this ease of sample preparation contrasts greatly with the large numbers of manipulations typically required for nucleic acid-based techniques (6, 10, 31), even with immunomagnetic separation.
One obvious limitation of the MAAB, as well as other described biosensors, is its sensitivity. The LODs obtained for this study are not sufficient for on-site testing of foodstuffs, even with the 1-h procedure. While the LOD determined here is below the commonly quoted infective dose of 105 CFU (8), infective doses as low as 1 to 6 CFU have been reported when Salmonella was ingested with a food source (8, 14, 26). Inclusion of a short pre-enrichment step in the MAAB procedure may improve sensitivity, but the additional steps increase both the complexity of the system and the time until results are known. We are currently exploring methods to increase the signal-to-noise ratio in hopes of attaining the appropriate level of sensitivity required for direct testing of foods in the field. However, the successful detection of S. enterica serovar Typhimurium in spiked chicken excreta (and possibly in unspiked excreta from leisure farm chickens) demonstrates the potential of this instrument for preharvest testing of poultry and other livestock; chickens, turkeys, swine, cattle, and other livestock could be screened for asymptomatic infection, allowing the producer and processor to treat or remove suspect animals and their associated foodstuffs from production.
This paper describes the first step in developing a portable immunosensor capable of rapidly analyzing a wide variety of different foodstuffs for the presence of multiple disease-causing analytes. Here we demonstrated the ability to detect S. enterica serovar Typhimurium in five different food preparations; testing of chicken excreta demonstrated the potential for use of the MAAB as a screening tool for identification of infected chickens. Future efforts will focus on improving detection limits, decreasing the effect of certain sample matrices, and integrating additional assays for food-borne pathogens into a single multianalyte assay.
Acknowledgments
Funding for this work was provided by the National Aeronautics and Space Administration (NASA) and the Office of Naval Research (ONR).
We express our gratitude to Ellen Goldman for her assistance in collecting the leisure farm excretal samples and to Lisa Shriver-Lake for her helpful suggestions and input.
The views expressed here are those of the authors and do not reflect the views of the U.S. Navy, the U.S. Department of Defense, the University of Maryland, or the U.S. government.
REFERENCES
- 1.Anderson, G. P., and N. L. Nerurkar. 2002. Improved fluoroimmunoassays using the dye Alexa Fluor 647 with the RAPTOR, a fiber optic biosensor. J. Immunol. Methods 271:17-24. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, C. D., L. Dillard, M. Pratt, and J. Rivera. 2001. Chapter 4A Addendum. FSIS procedure for the use of Salmonella rapid screening immunoassay kits, p. 4A-1-4A-4. In B. P. Dey and C. P. Lattuada (ed.), USDA/FSIS microbiology laboratory guidebook, 3rd ed. U.S. Department of Agriculture, Washington, D.C.
- 3.Andrews, W. H., and T. S. Hammack. 2001. Salmonella. In Bacteriological analytical manual, 8th ed. U.S. Department of Agriculture, Washington, D.C.
- 4.Babacan, S., P. Pivarnik, S. Letcher, and A. Rand. 2002. Piezoelectric flow injection analysis biosensor for the detection of Salmonella typhimurium. J. Food Sci. 67:314-320. [Google Scholar]
- 5.Bailey, J. S. 1998. Detection of Salmonella cells within 24 to 26 hours in poultry samples with the polymerase chain reaction BAX system. J. Food Prot. 61:792-795. [DOI] [PubMed] [Google Scholar]
- 6.Bhaduri, S., and B. Cottrell. 2001. Sample preparation methods for PCR detection of Escherichia coli O157:H7, Salmonella typhimurium, and Listeria monocytogenes on beef chuck shoulder using a single enrichment medium. Mol. Cell. Probes 15:267-274. [DOI] [PubMed] [Google Scholar]
- 7.Bhagwat, A. A. 2003. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 84:217-224. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, M. J., and L. S. Newman. 1982. A review of human salmonellosis. I. Infective dose. Rev. Infect. Dis. 4:1096-1106. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. W., Y. Li, J. A. Seelenbinder, P. Pivarnik, A. G. Rand, S. V. Letcher, O. J. Gregory, and M. J. Platek. 1998. Immunoassays based on surface-enhanced infrared absorption spectroscopy. Anal. Chem. 70:2991-2996. [DOI] [PubMed] [Google Scholar]
- 10.Carli, K. T., C. B. Unal, V. Caner, and A. Eyigor. 2001. Detection of Salmonellae in chicken feces by a combination of tetrathionate broth enrichment, capillary PCR and capillary gel electrophoresis. J. Clin. Microbiol. 39:1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC. 2002. Salmonella surveillance: annual summary, 2001. U.S. Department of Health and Human Services, CDC, Atlanta, Ga.
- 12.Che, Y. H., Y. Li, M. Slavik, and D. Paul. 2000. Rapid detection of Salmonella typhimurium in chicken carcass wash water using an immunoelectrochemical method. J. Food Prot. 63:1043-1048. [DOI] [PubMed] [Google Scholar]
- 13.Cras, J. J., C. A. Rowe-Taitt, D. Nivens, and F. S. Ligler. 1999. Comparison of chemical cleaning methods of glass in preparation for silanization. Biosens. Bioelectron. 14:683-688. [Google Scholar]
- 14.D'Aoust, J. Y. 1985. Infective dose of Salmonella typhimurium in cheddar cheese. Am. J. Epidemiol. 122:717-719. [DOI] [PubMed] [Google Scholar]
- 15.Delehanty, J. B., and F. S. Ligler. 2002. A microarray immunoassay for simultaneous detection of proteins and bacteria. Anal. Chem. 74:5681-5687. [DOI] [PubMed] [Google Scholar]
- 16.Delehanty, J. B., and F. S. Ligler. 2003. A method for printing functional microarrays. BioTechniques 34:380-385. [DOI] [PubMed] [Google Scholar]
- 17.De Medici, D., L. Croci, E. Delibato, S. Di Pasquale, E. Filetici, and L. Toti. 2003. Evaluation of DNA extraction methods for use in combination with SYBR Green I real-time PCR to detect Salmonella enterica serotype Enteritidis in poultry. Appl. Environ. Microbiol. 69:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickinson, J. H., R. G. Kroll, and K. A. Grant. 1995. The direct application of the polymerase chain reaction to DNA extracted from foods. Lett. Appl. Microbiol. 20:212-216. [DOI] [PubMed] [Google Scholar]
- 19.El-Gazzar, R. E., and E. H. Marth. 1992. Foodborne disease: investigative procedures and economic assessment. J. Environ. Health 55:24-27. [Google Scholar]
- 20.Feldstein, M. J., J. P. Golden, C. A. Rowe, B. D. MacCraith, and F. S. Ligler. 1999. Array biosensor: optical and fluidics systems. J. Biomed. Microdevices 1:139-153. [DOI] [PubMed] [Google Scholar]
- 21.Fett, W. 2000. Naturally occurring biofilms on alfalfa and other types of sprouts. J. Food Prot. 63:625-632. [DOI] [PubMed] [Google Scholar]
- 22.Gehring, A. G., C. G. Crawford, R. S. Mazenko, L. J. Van Houten, and J. D. Brewster. 1996. Enzyme-linked immunomagnetic electrochemical detection of Salmonella typhimurium. J. Immunol. Methods 195:15-25. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, C., D. Winters, A. O'Leary, and M. Slavik. 2003. Development of a triplex PCR assay for the specific detection of Campylobacter jejuni, Salmonella spp. and Escherichia coli O157:H7. Mol. Cell. Probes 17:135-138. [DOI] [PubMed] [Google Scholar]
- 24.Golden, J. P. October1998. Chemical sensor using two-dimensional lens array. U.S. patent 5,827,748.
- 25.Golden, J. P., and F. S. Ligler. 2002. A comparison of imaging methods for use in an array biosensor. Biosens. Bioelectron. 17:719-725. [DOI] [PubMed] [Google Scholar]
- 26.Greenwood, M. H., and W. L. Hooper. 1983. Chocolate bars contaminated with Salmonella napoli: an infectivity study. Br. Med. J. 286:1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanzler, D. J., D. C. Kradel, and W. M. Sischo. 1998. Management and environmental risk factors for Salmonella enteritidis contamination of eggs. Am. J. Vet. Res. 59:824-829. [PubMed] [Google Scholar]
- 28.Hinton, M. 1988. Salmonella infection in chicks following the consumption of artificially contaminated feed. Epidemiol. Infect. 100:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt, P. S., R. J. Buhr, D. L. Cunningham, and R. E. Porter, Jr. 1994. Effect of two different molting procedures on Salmonella enteritidis infection. Poult. Sci. 73:1267-1275. [DOI] [PubMed] [Google Scholar]
- 30.Hong, Y., M. E. Berrang, T. Liu, C. L. Hofacre, S. Sanchez, L. Wang, and J. J. Maurer. 2003. Rapid detection of Campylobacter coli, Campylobacter jejuni, and Salmonella enterica on poultry carcasses by using PCR-enzyme-linked immunosorbent assay. Appl. Environ. Microbiol. 69:3492-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsih, H.-Y., and H.-Y. Tsen. 2001. Combination of immunomagnetic separation and polymerase chain reaction for the simultaneous detection of Listeria monocytogenes and Salmonella spp. in food samples. J. Food Prot. 64:1744-1750. [DOI] [PubMed] [Google Scholar]
- 32.Jaquette, C. B., L. R. Beuchat, and B. E. Mahon. 1996. Efficacy of chlorine and heat treament in killing Salmonella stanley inoculated onto alfalfa seeds and growth and survival of the pathogen during sprouting and storage. Appl. Environ. Microbiol. 62:2212-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, G.-H., A. G. Rand, and S. V. Letcher. 2003. Impedance characterization of a piezoelectric immunosensor, part II: Salmonella typhimurium detection using magnetic enhancement. Biosens. Bioelectron. 18:91-99. [DOI] [PubMed] [Google Scholar]
- 34.Leach, S. A., A. Williams, A. C. Davies, J. Wilson, P. D. Marsh, and T. J. Humphrey. 1999. Aerosol route enhances the contamination of intact eggs and muscle of experimentally infected laying hens by Salmonella typhimurium DT104. FEMS Microbiol. Lett. 171:203-207. [DOI] [PubMed] [Google Scholar]
- 35.Liao, C.-H., and L. M. Schollenberger. 2003. Detection of Salmonella by indicator agar media and PCR as affected by alfalfa seed homogenates and native bacteria. Lett. Appl. Microbiol. 36:152-156. [DOI] [PubMed] [Google Scholar]
- 36.Ligler, F. S., and C. A. Rowe Taitt, ed. 2002. Optical biosensors: present and future. Elsevier, Amsterdam, The Netherlands.
- 37.Lin, J. S., and H. Y. Tsen. 1999. Development and use of polymerase chain reaction for the specific detection of Salmonella typhimurium in stool and food samples. J. Food Prot. 62:1103-1110. [DOI] [PubMed] [Google Scholar]
- 38.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: toward an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nastasi, A., C. Mammina, and R. Mioni. 1999. Detection of Salmonella spp. in food by a rapid PCR-hybridization procedure. Microbiologica 22:195-202. [PubMed] [Google Scholar]
- 41.Ogunjimi, A. A., and P. V. Choudary. 1999. Adsorption of endogenous polyphenols relieves the inhibition by fruit juices and fresh produce of immuno-PCR detection of Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 23:213-220. [DOI] [PubMed] [Google Scholar]
- 42.Olsen, S. J., L. C. MacKinon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne disease outbreaks, United States, 1993-1997. Morb. Mortal. Wkly. Rep. 49:1-51. [PubMed] [Google Scholar]
- 43.Pathirana, S. T., J. Barbaree, B. A. Chin, M. G. Hartell, W. C. Neely, and V. Vodyanoy. 2000. Rapid and sensitive biosensor for Salmonella. Biosens. Bioelectron. 15:135-141. [DOI] [PubMed] [Google Scholar]
- 44.Pivarnik, P., H. Cao, S. Letcher, A. Pierson, and A. G. Rand. 1999. Magnetic focusing immunosensor for the detection of Salmonella typhimurium in foods. Proc. SPIE 2544:41-49. [Google Scholar]
- 45.Prusak-Sochaczewski, E., J. H. T. Luong, and G. G. Guibault. 1990. Development of a piezoelectric immunosensor for the detection of Salmonella typhimurium. Enzyme Microb. Technol. 12:173-177. [DOI] [PubMed] [Google Scholar]
- 46.Rogers, K. R., and A. Mulchandani. 1998. Affinity biosensors: techniques and protocols. Humana, Totowa, N.J.
- 47.Rose, B. E. 1998. Isolation and identification of Salmonella from meat, poultry, and egg products, p. 4-1-4-14. In B. P. Dey and C. P. Lattuada (ed.), USDA/FSIS microbiology laboratory guidebook, 3rd ed. U.S. Department of Agriculture, Washington, D.C.
- 48.Rowe, C. A., L. M. Tender, M. J. Feldstein, J. P. Golden, S. B. Scruggs, B. D. MacCraith, J. J. Cras, and F. S. Ligler. 1999. Array biosensor: simultaneous analysis for bacterial, viral, and toxic analytes in blind samples. Anal. Chem. 71:3846-3852. [DOI] [PubMed] [Google Scholar]
- 49.Rowe, C. A., S. D. Balderson, M. J. Feldstein, J. P. Golden, and F. S. Ligler. 1999. An array immunosensor for simultaneous detection of clinical analytes. Anal. Chem. 71:433-439. [DOI] [PubMed] [Google Scholar]
- 50.Rowe-Taitt, C. A., J. J. Cras, C. H. Patterson, J. P. Golden, and F. S. Ligler. 2000. A ganglioside-based assay for cholera toxin using an array biosensor. Anal. Biochem. 281:123-133. [DOI] [PubMed] [Google Scholar]
- 51.Rowe-Taitt, C. A., J. P. Golden, M. J. Feldstein, J. J. Cras, K. E. Hoffman, and F. S. Ligler. 2000. Array biosensor for detection of biohazards. Biosens. Bioelectron. 14:785-794. [DOI] [PubMed] [Google Scholar]
- 52.Rowe-Taitt, C. A., J. W. Hazzard, K. E. Hoffman, J. J. Cras, J. P. Golden, and F. S. Ligler. 2000. Simultaneous detection of six biohazardous agents using a planar waveguide array biosensor. Biosens. Bioelectron. 15:579-589. [DOI] [PubMed] [Google Scholar]
- 53.Sapsford, K. E., P. T. Charles, C. H. Patterson, Jr., and F. S. Ligler. 2002. Demonstration of four immunoassay formats using the array biosensor. Anal. Chem. 74:1061-1068. [DOI] [PubMed] [Google Scholar]
- 54.Sapsford, K. E., Z. Liron, Y. S. Shubin, and F. S. Ligler. 2001. Kinetics of antigen binding to arrays of antibodies in different sized spots. Anal. Chem. 73:5518-5524. [DOI] [PubMed] [Google Scholar]
- 55.Schuderer, J., A. Akkoyun, A. Brandenburg, U. Bilitewski, and E. Wagner. 2000. Development of a multichannel fluorescence affinity sensor system. Anal. Chem. 72:3942-3948. [DOI] [PubMed] [Google Scholar]
- 56.Seelenbinder, J. A., C. W. Brown, P. Pivarnik, and A. G. Rand. 1999. Colloidal gold filtrates as metal substrates for surface-enhanced infrared absorption spectroscopy. Anal. Chem. 71:1963-1966. [DOI] [PubMed] [Google Scholar]
- 57.Seo, K. H., R. E. Brackett, N. F. Hartman, and D. P. Campbell. 1999. Development of a rapid response biosensor for detection of Salmonella typhimurium. J. Food Prot. 62:431-437. [DOI] [PubMed] [Google Scholar]
- 58.Shriver-Lake, L. C., Y. S. Shubin, and F. S. Ligler. 2003. Detection of staphylococcal enterotoxin B in spiked food samples. J. Food Prot. 66:1851-1856. [DOI] [PubMed] [Google Scholar]
- 59.Si, S., F. Ren, W. Cheng, and S. Yao. 1997. Preparation of a piezoelectric immunosensor for the detection of Salmonella paratyphi A by immobilization of antibodies on electropolymerized films. Fresenius J. Anal. Chem. 357:1101-1105. [Google Scholar]
- 60.Stewart, D. S., K. F. Reineke, J. M. Ulaszek, and M. L. Tortorello. 2001. Growth of Salmonella during sprouting of alfalfa seeds associated with salmonellosis outbreaks. J. Food Prot. 64:618-622. [DOI] [PubMed] [Google Scholar]
- 61.Taitt, C. R., G. P. Anderson, B. D. Lingerfelt, and F. S. Ligler. 2002. Nine-analyte detection using an array sensor. Anal. Chem. 74:6114-6120. [DOI] [PubMed] [Google Scholar]
- 62.Taitt, C. R., J. P. Golden, Y. S. Shubin, L. C. Shriver-Lake, K. E. Sapsford, A. Rasooly, and F. S. Ligler. A portable array biosensor for detecting multiple analytes in complex samples. Microb. Ecol., in press. [DOI] [PubMed]
- 63.Taormina, P. J., L. R. Beuchat, and L. Slutsker. 1999. Infections associated with eating seed sprouts: an international concern. Emerg. Infect. Dis. 5:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas, N. L. 1978. Observations of the relationship between the surface area and weight of eviscerated carcasses of chickens, ducks, and turkeys. J. Food Technol. 13:81-86. [Google Scholar]
- 65.Todd, E. C. D. 1989. Preliminary estimates of costs of foodborne disease in the United States. J. Food Prot. 52:595-601. [DOI] [PubMed] [Google Scholar]
- 66.USDA. 1995. Food safety and inspection service: procedure for isolation of Salmonella from food. Laboratory communication no. 75. U.S. Department of Agriculture, Washington, D.C.
- 67.Wadkins, R. M., J. P. Golden, and F. S. Ligler. 1997. Patterned planar array immunosensor for multianalyte detection. J. Biomed. Optics 2:74-79. [DOI] [PubMed] [Google Scholar]
- 68.Wadkins, R. M., J. P. Golden, L. M. Pritsiolas, and F. S. Ligler. 1998. Detection of multiple toxic agents using a planar array immunosensor. Biosens. Bioelectron. 13:407-415. [DOI] [PubMed] [Google Scholar]
- 69.Weimer, B. C. June2002. Real time detection of antigens. U.S. patent 6,399,317.
- 70.Yu, H., and J. G. Bruno. 1996. Immunomagnetic-electrochemiluminescent detection of Escherichia coli O157 and Salmonella typhimurium in foods and environmental water samples. Appl. Environ. Microbiol. 62:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, C., P. Pivarnik, A. G. Rand, and S. V. Letcher. 1998. Fiber-optic biosensor based on ultrasonic concentration of particles and cells. Biosens. Bioelectron. 13:495-500. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, C., P. Pivarnik, S. Auger, A. Rand, and S. Letcher. 1997. A compact fiber-optic immunosensor based on evanescent wave excitation using a semiconductor laser. Sens. Actuat. B 42:169-175. [Google Scholar]