Abstract
Lactobacilli represent components of the commensal mammalian gastrointestinal microbiota and are useful as probiotics, functional foods, and dairy products. This study includes systematic polyphasic analyses of murine intestinal Lactobacillus isolates and correlation of taxonomic findings with data from cytokine production assays. Lactobacilli were recovered from mice with microbiota-dependent colitis (interleukin-10 [IL-10]-deficient C57BL/6 mice) and from mice without colitis (Swiss Webster and inducible nitric oxide synthetase-deficient C57BL/6 mice). Polyphasic analyses were performed to elucidate taxonomic relationships among 88 reference and murine gastrointestinal lactobacilli. Genotypic tests included single-locus analyses (16S ribosomal DNA sequencing and 16S-23S rRNA intergenic spacer region PCR) and genomic DNA profiling (repetitive DNA element-based PCR), and phenotypic analyses encompassed more than 50 tests for carbohydrate utilization, enzyme production, and antimicrobial resistance. From 20 mice without colitis, six Lactobacillus species were recovered; the majority of the mice were colonized with L. reuteri or L. murinus (72% of isolates). In contrast, only, L. johnsonii was isolated from 14 IL-10-deficient mice. Using an in vitro assay, we screened murine isolates for their ability to inhibit tumor necrosis factor alpha (TNF-α) secretion by lipopolysaccharide-activated macrophages. Interestingly, a subpopulation of lactobacilli recovered from mice without colitis displayed TNF-α inhibitory properties, whereas none of the L. johnsonii isolates from IL-10-deficient mice exhibited this effect. We propose that differences among intestinal Lactobacillus populations in mammals, combined with host genetic susceptibilities, may account partly for variations in host mucosal responses.
Lactobacillus species represent indigenous organisms of the mammalian gastrointestinal (GI) tract (19, 29) and have been used as probiotic agents for the treatment of GI infections and inflammatory bowel disease (IBD) (15, 16). Lactobacillus species have been isolated from the intestines of various mammals, including rodents (e.g., mice and rats), dogs, cats, ruminants, horses, nonhuman primates, and humans (19, 29, 41). These organisms are present in relatively high numbers in the GI tracts of mice and presumably play a beneficial role in healthy animals. Lactobacillus species colonize the murine stomach and intestine immediately after birth, adhere to epithelial cells, and are part of the stable intestinal microbiota of the animals during development and adulthood (30, 33).
Previous studies with Lactobacillus-deficient mice indicated that intestinal Lactobacillus species provide important biochemical functions for the murine intestine, including bile salt hydrolase (34) and azoreductase (18) activities. In addition to biochemical activities, Lactobacillus species may modulate host immune responses. Lactobacillus species differentially regulate cytokine production by dendritic cells (4) and cells derived from the intestinal mucosa (25). Probiotic Lactobacillus and Bifidobacterium strains stably colonize the intestinal lumen of laboratory mice (10, 39). Lactobacillus reuteri diminished inflammation in interleukin-10 (IL-10)-deficient mice predisposed to colitis (17). The functional importance of Lactobacillus species in the mammalian intestine highlights the need for detailed studies of enteric clones from laboratory mice, including studies of knockout mouse models of colitis.
Polyphasic approaches combining biochemical, molecular, and morphological data are important for the accurate classification of lactic acid bacteria (13). Lactobacillus species may be difficult to identify by conventional biochemical methods, although simplified approaches are useful for presumptively assigning organisms to this genus. Lactobacillus organisms are generally catalase negative, oxidase negative, vancomycin resistant (Vanr), and anaerobic and appear as gram-positive bacilli by Gram stain. Biochemical profiling has been useful for identifying species and groups of species. However, biochemical tests are limited with respect to species identification within species complexes, including the L. acidophilus and L. casei groups. For example, members of the L. casei complex have undergone several taxonomic changes (21), as have those of the L. acidophilus complex (5). DNA sequencing of informative target regions, such as the 16S rRNA gene and the 16S-23S ribosomal DNA intergenic spacer region (ISR), has resulted in useful strategies for definitive species identification within Lactobacillus species complexes (14, 32). Alternative approaches, such as plasmid profiling (30) and protein profiling (6), also have been used.
In this article, we present a polyphasic phenotypic and genotypic (phenogenetic) study of Lactobacillus isolates obtained from the intestines of laboratory mice. Lactobacillus isolates obtained from the GI tracts of mice were compared with reference isolates (e.g., American Type Culture Collection [ATCC] strains). In order to evaluate the Lactobacillus microbiota colonizing various regions of the GI tracts of mice in a mouse model of bacterium-dependent colitis (IL-10-deficient mice) and those of laboratory mice that are not models of bacterium-dependent colitis (Swiss Webster and inducible nitric oxide synthetase [iNOS]-deficient C57BL/6 mice; hereafter referred to as mice without colitis), intestinal lactobacilli were isolated from various regions of the GI tracts and feces. Candidate murine intestinal lactobacilli were cultivated on selective media and screened by Gram stain morphology and selected biochemical tests. Lactobacillus isolates were characterized by detailed biochemical studies, 16S rDNA sequencing, and genomic fingerprinting with repetitive DNA element-based PCR (rep-PCR). Biochemical profiling included 53 tests for carbohydrate utilization, enzyme production, and antimicrobial resistance. Substantial differences were observed in the nature of enteric Lactobacillus species and strains colonizing IL-10-deficient mice and mice without colitis.
MATERIALS AND METHODS
Animals.
Sentinel Swiss Webster mice, iNOS-deficient C57BL/6 mice, and IL-10-deficient C57BL/6 mice, ages 6 weeks to 10 months, were housed in an Association for Assessment and Accreditation of Laboratory Animal Care-approved facility (Division of Comparative Medicine, Massachusetts Institute of Technology) under specific-pathogen-free conditions in microisolator cages. All Swiss Webster mice were individually housed, while gene-deficient C57BL/6 (iNOS- and IL-10-deficient) mice were cohoused by gender and filial generation (except during breeding). Mice were kept free of known murine viruses, Salmonella spp., Citrobacter rodentium, ecto- and endoparasites, and known murine Helicobacter spp.
Bacterial isolation and culture.
Mice were sacrificed by CO2 asphyxiation, and the entire GI tract was aseptically removed. Sections of the stomach, jejunum, cecum, and colon were cleared of luminal content by longitudinal incision of tissue followed by agitation in sterile buffered saline. Tissue specimens were homogenized in tryptic soy broth. Homogenates were streaked for isolation on DeMan-Rogosa-Sharpe (MRS) agar (Becton Dickinson, Sparks, Md.) and incubated anaerobically for 24 to 48 h with AnaeroGen sachets (Oxoid, Hampshire, England). Colonies resembling lactobacilli were subcultured and grown on MRS agar under microaerobic conditions (10% CO2, 10% H2, and 80% N2). The following 30 reference strains were used: L. acidophilus (ATCC 4356 and ATCC 4796), L. animalis (ATCC 35046), L. brevis subsp. gravesensis (ATCC 27305), L. brevis subsp. otakiensis (ATCC 27306), L. buchneri (ATCC 11577), L. casei (ATCC 334), L. delbrueckii subsp. bulgaricus (ATCC 11842), L. fermentum (ATCC 14931), L. gasseri (ATCC 33323), L. hilgardii (ATCC 8290), L. johnsonii (ATCC 33200), L. murinus (ATCC 35020), L. paracasei (ATCC 25302 and strain Shirota), L. plantarum (ATCC 11581, ATCC 14917, ATCC 49445, and ATCC 4008), L. reuteri (ATCC 23272, ATCC 53608, ATCC 53609, ATCC 55148, and SD2112), L. rhamnosus GG (ATCC 53103), L. ruminis (ATCC 25644), L. salivarius (ATCC 11471), L. vaginalis (ATCC 49540), and Lactobacillus strains ASF 360 and ASF 361. All lactobacilli were grown on MRS agar under anaerobic conditions at 37°C.
Lactobacilli isolated and characterized in this study represent aerotolerant populations colonizing the murine alimentary system. Our bacterial isolation methods included a combination of anaerobic and microaerobic cultivation approaches, and strictly anaerobic Lactobacillus species may not have been recovered from the intestines of the laboratory mice that we used.
ISR PCR.
Lactobacilli were assigned membership into four main taxonomic groups (I, II, III, and IV) by PCR-based approaches developed by Song et al. (32) and based on phylogenies derived from the 16S-23S rRNA ISR. Multiplex PCR was carried out with four forward primers (LU 1F, 5′-ATT GTA GAG CGA CCG AGA AG-3′; LU 3F, 5′-AAA CCG AGA ACA CCG CGT T-3′; LU 5F, 5′-CTA GCG GGT GCG ACT TTG TT-3′; and Ldel 7F, 5′-ACA GAT GGA TGG AGA GCA GA-3′) and one reverse primer (Lac 2R, 5′-CCT CTT CGC TCG CCG CTA CT-3′). Bacterial DNA was extracted by using an UltraClean microbial genomic DNA isolation kit (Mo Bio Laboratories, Inc., Solano Beach, Calif.). Genomic DNA was quantitated by absorbance spectrophotometry, and integrity was assessed by agarose gel electrophoresis followed by ethidium bromide staining. PCR was carried out with an ABI 2700 instrument (Applied Biosystems, Foster City, Calif.) under the following conditions: 95°C for 5 min; 35 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and 74°C for 5 min. Reaction mixtures (final volume, 50 μl) contained 1 μl of genomic DNA (at least 10 ng/μl), 50 pmol of each primer, 1.25 U of Amplitaq DNA polymerase (Applied Biosystems), 2.5 mM each deoxynucleoside triphosphate, 5 μl of 10× reaction buffer (supplied with enzymes), and 75 mM MgCl2. The expected sizes of the ISR amplicons were as follows: 450 bp (group I), 300 bp (group II), 400 bp (group III), and 350 bp (group IV).
16S rRNA gene sequencing.
Approximately 1,500 bp of the 16S rRNA gene was amplified with primers 16S-8F (5′-AGA GTT TGA TCY TGG YTY AG-3′) and 16S-1541R (5′-AAG GAG GTG WTC CAR CC-3′) under the following PCR conditions: 95°C for 5 min; 35 cycles of 95°C for 30 s, 57°C for 1 min, and 72°C for 1 min; and 72°C for 5 min. Each 50-μl PCR was carried out as described above. 16S rDNA amplicons were gel purified by using GFX PCR DNA and a gel band purification kit (Amersham Biosciences, Inc., Piscataway, N.J.) and a QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.). The 5′ terminus of the 16S rRNA gene was sequenced with primers 16S-8F and 16S-344F (5′-ACG GGA GGC AGC AGY-3′) by using an ABI Prism 3100 (Applied Biosystems) sequencing system and an ABI Prism BigDye Terminator cycle sequencing ready reaction kit, version 2.0 (Applied Biosystems), at the Baylor College of Medicine Core Sequencing Facility. All 16S rDNA amplicons were sequenced with two sets of primers, both oriented to amplify the sense strand, effectively resulting in a two-pass sequencing reaction. Additionally, electrophoretograms were inspected visually for appropriate signal peak intensity and spacing. Sequencing traces of amplicons containing ambiguous signals were resubmitted for sequencing. rDNA sequences were analyzed by using Lasergene, version 5.0 (DNAStar, Madison, Wis.). Contigs were generated by using SeqMan. Phylogenetic trees were constructed by aligning nucleotide positions 22 to 1004 (consensus positions; Escherichia coli ATCC 25922 cognates 30 to 885). Approximately 900 nucleotides were analyzed by using the MegAlign ClustalV algorithm. Isolates were identified by using the nucleotide-nucleotide Basic Local Alignment Search Tool (BLASTn) (www.ncbi.nlm.nih.gov/BLAST).
rep-PCR.
rep-PCR was performed as previously described (37, 38) with U-Prime Dt and E primer sets. Amplicons were resolved in 1.5% agarose gels and quantitatively analyzed by using GelComparII software, version 2 (Applied Maths, Kortrijk, Belgium). Similarity coefficients were calculated by using Pearson correlation and DNA profiles clustered by the unweighted pair-group method with arithmetic means.
Biochemical profiling.
Lactobacilli were grown on MRS agar and incubated under anaerobic conditions at 37°C for 24 to 48 h. All isolates were visualized by Gram staining. Biochemical testing was performed with API 50CH strips (BioMerieux, Hazelwood, Mo.) according to the manufacturer's instructions. Catalase and oxidase spot tests were performed according to the supplier's recommendations (Becton Dickinson). Urease was detected by culturing of lactobacilli on Christensen's urea slants (Remel, Lenexa, Kans.) and incubation under anaerobic conditions for up to 5 days. Vancomycin susceptibility was assessed by a modified Kirby-Bauer disk diffusion test. Briefly, lactobacilli were inoculated into buffered saline to a 0.5 McFarland standard and swabbed onto MRS agar. Vancomycin-impregnated disks (5 μg; Becton Dickinson) were applied to bacterial cultures, which then were grown anaerobically for 48 h. Isolates displaying zones of clearance of greater than 15 mm were considered susceptible.
Data for biochemical tests were transformed into binomial values (0, negative or sensitive; 1, positive or resistant). Lactobacilli were clustered by using the unweighted nearest-neighbor method by calculating the squared Euclidean distances of binary measures or measures of similarity (Jaccard, Sokal, and Sneath matching coefficients and simple matching coefficient). Discriminant and factorial analyses were used to determine the most useful biochemical tests for presumptive identification. Lactobacillus phenograms were generated by using statistical software (SPSS for Windows, version 11.0.1; SPSS Inc., Chicago, Ill.).
Bioassays.
In vitro bioassays were carried out as previously described (24). Briefly, media conditioned by lactobacilli were tested for the ability to inhibit tumor necrosis factor alpha (TNF-α) production by lipopolysaccharide (LPS)-activated macrophages. Naive RAW 264.7 (ATCC CRL-2278) macrophages were exposed to purified E. coli (serotype O127:B8) LPS (Sigma, St. Louis, Mo.) and Lactobacillus-conditioned media (L-cm). Culture supernatants were collected 5 h postactivation, and TNF-α levels were measured by using a quantitative enzyme-linked immunosorbent assay (Biosource, Camarillo, Calif.).
RESULTS
16S rDNA sequence-based identification of lactobacilli.
Detailed biochemical and molecular studies of murine intestinal Lactobacillus isolates highlighted the presence of distinct Lactobacillus populations. A total of 58 murine isolates, representing oral (2 of 58), jejunal (13 of 58), colonic (14 of 58), and fecal (29 of 58) lactobacilli from 31 mice (12 Swiss Webster, 5 iNOS-deficient C57BL/6, and 14 IL-10-deficient C57BL/6), were studied along with 30 reference strains. To verify the identities of the reference strains used in this study, 16S rRNA genes were amplified and sequenced. With BLASTn, sequence analyses of the 16S rRNA gene yielded identification at the species level. With the exception of L. reuteri ATCC 53609 (found by BLASTn to be most similar to L. fermentum) and L. plantarum ATCC 49445 (found by BLASTn to be most similar to L. sakei), all reference strains were found to be most similar to their species designations. L. reuteri ATCC 53609 and L. plantarum ATCC 49445 were subjected to all of the genetic and phenotypic tests used here but were excluded from any of the analyses correlating sequence, biochemical, and functional (bioassay) data. A second analysis of 16S rDNA sequences with Ribosomal Database Project II (http://rdp.cme.msu.edu/html/) yielded similar results, providing identification. The same queries were performed for all murine isolates (Table 1).
TABLE 1.
16S rDNA sequence-based identification of murine GI lactobacilli isolated in this studya
| Mouse without colitis | BLASTn identification |
|---|---|
| pupjm-1b | L. reuteri |
| 6801 cm-1b | L. reuteri |
| 6801 jm-1b | L. reuteri |
| 6799 jm-1b | L. reuteri |
| 6800 cmb | L. reuteri |
| 6800 jm-1b | L. reuteri |
| 6798-1b | L. reuteri |
| 6798 cm-1b | L. reuteri |
| 6798 jm-1b | L. reuteri |
| 6799b | L. reuteri |
| 1662 | L. reuteri |
| 1650 | L. reuteri |
| 1604-1 | L. reuteri |
| 1603-1 | L. vaginalis |
| 1598-1 | L. vaginalis |
| 1600-1 | L. reuteri |
| 1583 | L. reuteri |
| 562N-1 | L. murinus |
| 562N-2 | L. murinus |
| 987 col-1 | L. murinus |
| 988 cm | L. murinus |
| 988 col | L. murinus |
| 1604-2 | L. murinus |
| 4901 | L. johnsonii |
| 4903 | L. johnsonii |
| 4931 | L. johnsonii |
| 4938 | L. johnsonii |
| 1598-2 | L. intestinalis |
| 1602 | L. paracasei |
| IL-10 2 cm-1 | L. johnsonii |
| IL-10 2 jm-2 | L. johnsonii |
| IL-10 2 col-1 | L. johnsonii |
| IL-10 2 col-2 | L. johnsonii |
| IL-10 4 jm-1 | L. johnsonii |
| IL-10 4 jm-2 | L. johnsonii |
| IL-10 4 col-1 | L. johnsonii |
| IL-10 4 col-2 | L. johnsonii |
| IL-10 5 jm-1 | L. johnsonii |
| IL-10 5 jm-2 | L. johnsonii |
| IL-10 5 col-1 | L. johnsonii |
| IL-10 5 col-2 | L. johnsonii |
| IL-10 6 jm-1 | L. johnsonii |
| IL-10 6 jm-2 | L. johnsonii |
| IL-10 6 col-1 | L. johnsonii |
| IL-10 6 col-2 | L. johnsonii |
| IL-10 2-1 | L. johnsonii |
| IL-10 3-1 | L. johnsonii |
| IL-10 6-1 | L. johnsonii |
| IL-10 7-1 | L. johnsonii |
| IL-10 7-2 | L. johnsonii |
| IL-10 11-1 | L. johnsonii |
| IL-10 12-1 | L. johnsonii |
| IL-10 13-2 | L. johnsonii |
| IL-10 14-2 | L. johnsonii |
| IL-10 16-1 | L. johnsonii |
| IL-10 18-1 | L. johnsonii |
| IL-10 20-1 | L. johnsonii |
| IL-10 21-1 | L. johnsonii |
There were 29 strains in each group.
Isolates recovered from iNOS-deficient (C57BL/6) mice. All other isolates from mice without colitis were recovered from Swiss Webster sentinel mice.
Phenotypic analyses of lactobacilli.
Distinct microscopic morphologies were observed after Gram staining of broth-grown lactobacilli. At least four microscopic rod-shaped morphologies could be distinguished and were randomly assigned as types I, II, III, and IV (Fig. 1 and Table 2). All Lactobacillus strains tested were unable to utilize glycerol, erythritol, l-xylose, inositol, glycogen, xylitol, d-arabitol, l-arabitol, and 2-keto-gluconate. All strains were found to be catalase negative (at 3% [vol/vol] H2O2) and oxidase negative. Differences among species were noted for the following biochemical tests and used to construct phenograms: d-arabinose, l-arabinose, ribose, d-xylose, adonitol, β-methyl-xyloside, galactose, d-glucose, d-fructose, d-mannose, l-sorbose, rhamnose, dulcitol, mannitol, sorbitol, α-methyl-d-mannoside, α-methyl-d-glucoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, cellobiose, maltose, lactose, melibiose, saccharose, trehalose, inulin, melezitose, d-raffinose, β-gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, gluconate, and 5-keto-gluconate.
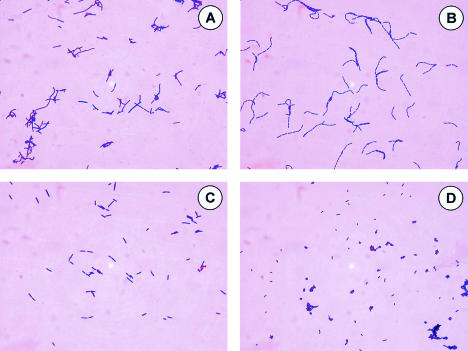
FIG. 1.
Gram stain morphologies of lactobacilli grown in MRS broth. Representatives of the four distinct morphologies are depicted. (A) Type I morphology (L. johnsonii ATCC 33200). (B) Type II morphology (L. rhamnosus GG). (C) Type III morphology (L. murinus 35020). (D) Type IV morphology (L. reuteri 53608) (see Table 2).
TABLE 2.
Microscopic morphologies of lactobacillia
| Microscopic morphology (type) | Width/length ratio | Description | Species exhibiting the morphology |
|---|---|---|---|
| I | 1:5-1:8 | Large irregular cells; sausage- or coryneform-like; chains of 2-4 cells | L. acidophilus group |
| II | 1:3-1:5 | Small, thin rectangular cells; box-like with square edges; chains of 3-8 cells | L. casei group |
| III | 1:2-1:5 | Medium-sized cells; single cells or pairs of cells | L. murinus |
| IV | |||
| A | 1:1-1:1.5 | Short, pleomorphic cells with round edges; chains of 2 or 3 cells | L. reuteri |
| B | 1:1.5-1:3 | Similar to IV-A but with longer chains of cells | L. vaginalis |
Gram-stained smears were prepared from MRS broth-grown lactobacilli.
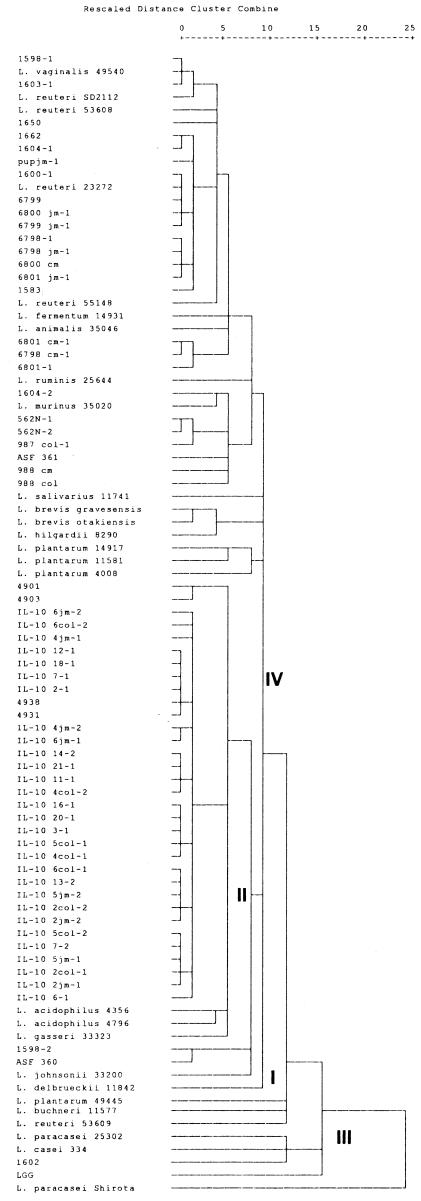
Cluster analyses of biochemical data (Fig. 2) can be used to classify lactobacilli into taxa that resemble Lactobacillus grouping schemes based on 16S-23S rRNA ISR sequences. With discriminant and factorial analyses of data from all biochemical tests, a presumptive identification scheme for lactobacilli was formulated based on biochemical properties. In conjunction with Vanr and Gram stain morphology, biochemical profiling can be used to differentiate major groups of murine GI lactobacilli (up to a 95% confidence interval). Overall, biochemical data-based cluster analyses were consistent with sequence-based cluster identification. Although no approved values for zones of clearance have been published by NCCLS for the genus Lactobacillus, the 15-mm cutoff value was empirically determined from reference strains (ATCC and other well-characterized strains) used in this study. This value appears to concur with published disk diffusion standard data for other gram-positive bacteria (e.g., enterococci and Staphylococcus aureus) (22).
FIG. 2.
Biochemical profile-based clustering of lactobacilli. Phenograms were generated by using the nearest-neighbor clustering algorithm and the simple matching coefficient. Lactobacilli clustered into four groups that match 16S-23S rRNA-based taxonomy (32). Group I, L. delbrueckii; group II, L. acidophilus complex (including L. acidophilus, L. intestinalis [ASF 360], L. gasseri, and L. johnsonii); group III, L. casei complex (including L. casei, L. paracasei, and L. rhamnosus); group IV, L. animalis, L. murinus, L. brevis, L. buchneri, L. hilgardii, L. fermentum, L. plantarum, L. reuteri, L. ruminis, L. salivarius,and L. vaginalis.
In our abbreviated identification scheme, lactobacilli can be presumptively grouped into four taxa (groups I through IV) when biochemical tests are combined with microscopic morphologies. Since only a single strain of group I, L. delbrueckii subsp. bulgaricus, was used in the analyses, no identification scheme could be generated for this group. Lactobacilli can be divided into vancomycin-resistant (groups III and IV) and vancomycin-susceptible (groups I and II) groups. Following vancomycin susceptibility testing, lactobacilli can be grouped further by utilization of d-raffinose or saccharose. Vanr isolates are tested for d-raffinose utilization. Members of group III generally fail to ferment d-raffinose, while group IV lactobacilli (except for L. fermentum and L. brevis) are positive for d-raffinose. On the other hand, Vans isolates generally are group II lactobacilli.
Genotypic analyses of lactobacilli.
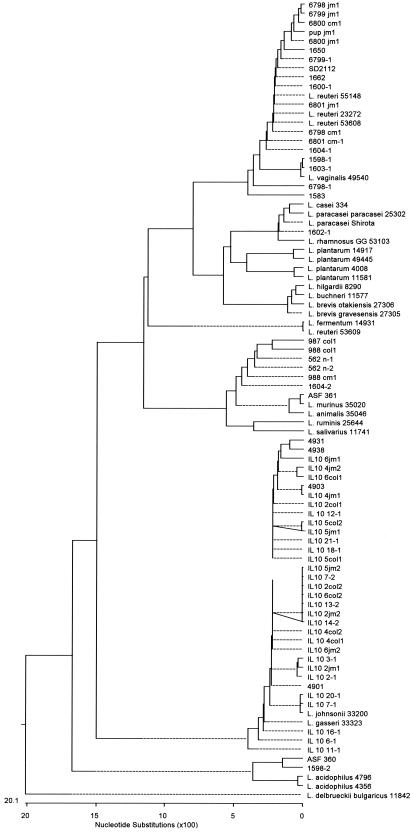
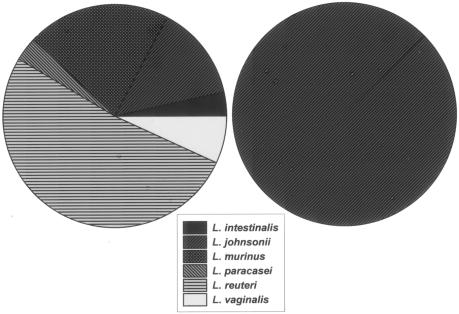
Intestinal Lactobacillus isolates obtained from the same animals and cultured from different regions of the GI tract were clustered by using 16S rDNA sequence analyses (Fig. 3). Qualitatively, no differences were observed in species isolated from different regions of the GI tract in any single animal. With respect to mice without colitis, 15 individually housed, nonlittermate Swiss Webster mice as well as 4 pair-housed iNOS-deficient C57BL/6 mice and 1 progeny iNOS-deficient C57BL/6 mouse were surveyed. Isolates from animals without colitis were identified by 16S rDNA sequencing with BLASTn as L. reuteri (15 of 29), L. murinus (6 of 29), L. johnsonii (4 of 29), L. vaginalis (2 of 29), L. intestinalis (1 of 29), and L. paracasei (1 of 29) (Fig. 4A). In contrast, all 29 isolates from IL-10-deficient mice were identified as L. johnsonii (Fig. 4B). Since the need to identify lactobacilli derived from the IL-10-deficient mice was paramount, care was taken to scrutinize all phenogenetic information and assign isolates to a species. Biochemical profiling and ISR PCR successfully identified the isolates as members of group II (L. acidophilus complex). When biochemical characteristics were used to cluster these isolates with ATCC reference strains, these IL-10-deficient mouse-derived isolates clustered more closely with group II-A (L. acidophilus).
FIG. 3.
16S rDNA sequence-based cluster analysis. Dendrograms based on ∼900 nucleotides (lactobacilli consensus positions 22 to 1004) were generated by using the ClustalV algorithm and Lasergene.
FIG. 4.
Lactobacillus species isolated from the murine GI tract. (A) Twenty-nine isolates recovered from mice without colitis (Swiss Webster and iNOS-deficient C57BL/6 mice). (B) Twenty-nine isolates obtained from a mouse model of colitis (IL-10-deficient C57BL/6 mice). Note the homogeneity of species recovered from IL-10-deficient mice compared to mice without colitis.
Variable regions within 16S rRNA genes have been used to discriminate among closely related species of lactobacilli, specifically within the L. acidophilus complex (14). With the first variable region (V1 region), representing a 26-nucleotide segment (consensus sequence positions 80 to 105), the isolates from IL-10-deficient mice were identified unambiguously as L. johnsonii. Species belonging to group II-A (including reference strains L. acidophilus and ASF 360 [L. intestinalis]) harbor nucleotide sequence gaps (consensus positions 83 to 89 and 99 to 101) with a single insertion (position 97), permitting differentiation from members of group II-B (L. johnsonii and L. gasseri). Four notable substitutions at positions T81A, A85G, T98C, and A100G in the V1 region distinguish these mouse isolates from L. gasseri, while these same positions are identical to those found in L. johnsonii (data not shown). In order to determine phylogenetic relationships among mouse isolates and reference strains, the same 16S rDNA sequences were used to cluster lactobacilli into the “true” phylogenetic tree. With this analysis, we found that reference lactobacilli cluster into clades that coincide with the expected topology.
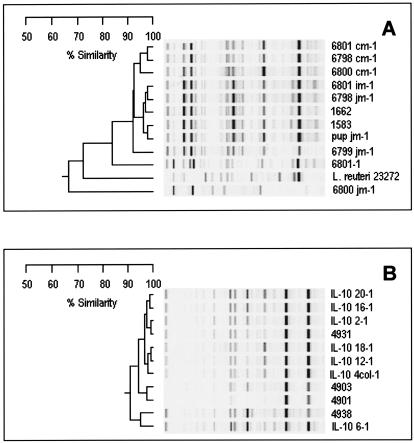
Consistent with these data, isolates from the IL-10-deficient mice and mice without colitis clearly were distinguishable by cluster analyses based on rep-PCR fingerprinting (with U-prime E primers). L. reuteri represents the majority of isolates isolated from mice without colitis (52%, or 15 of 29), as determined by 16S rDNA sequencing. With DNA fingerprint analysis of a subset of L. reuteri isolates, 11 of 15 L. reuteri isolates clustered into one heterogeneous clade (similarity ranging from 65 to 99%) with a single outlier (Fig. 5A). With a second DNA fingerprint analysis, the same L. reuteri isolates clustered into one major clade (R > 0.95); a second, minor clade (range of R, 0.43 to 0.7) and an outlier were evident (data not shown). L. reuteri recovered from the small and large intestines of the same mice displayed DNA fingerprint patterns that indicate subspecies variations (e.g., 6801-1 and 6801 cm-1 are both L. reuteri from one animal but display only ∼85% DNA profile similarity, as shown in Fig. 5A).
FIG. 5.
rep-PCR fingerprint analyses. (A) Selected murine L. reuteri strains were analyzed together with L. reuteri ATCC 23272. (B) Selected murine L. johnsonii strains from IL-10-deficient mice and mice without colitis were analyzed. Note the relative homogeneity in the DNA fingerprint profiles of L. johnsonii strains recovered from mice with colitis. In contrast, L. johnsonii strains recovered from mice without colitis (Swiss Webster mouse isolates 4901, 4903, and 4938) appear to be different strains.
The population of L. johnsonii isolates from IL-10-deficient mice was more homogeneous (i.e., relatively clonal) than that of L. reuteri isolates from mice without colitis. A subset of L. johnsonii isolates (7 of 29) from IL-10-deficient mice clustered into a single clade (Fig. 5B). This clade appeared to be relatively homogeneous, with a correlation coefficient of approximately 0.90 (or 90% similarity). L. johnsonii isolates recovered from mice without colitis (isolates 4901, 4903, and 4938) also clustered with other L. johnsonii isolates but appeared to be different strains. DNA fingerprint analysis with a second set of primers (U-Prime Dt primers) also grouped these IL-10-deficient isolates into a single clade with 9 of 10 isolates having a correlation coefficient of >0.85 and with a single outlier (R = 0.79) (data not shown).
In vitro immunofunctional analyses of lactobacilli.
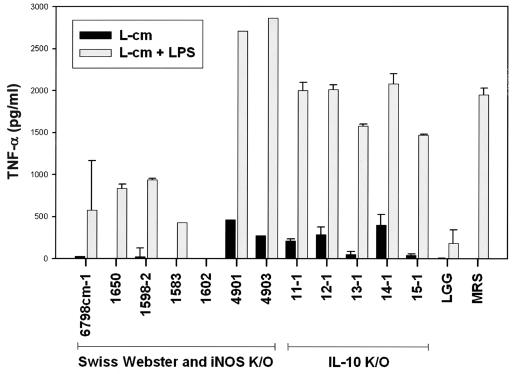
To correlate immunomodulatory activity with characterization of strains recovered from the mouse intestine, cell-free L-cm were tested for effects on proinflammatory cytokine output by LPS-stimulated murine macrophages. Of 29 lactobacilli isolated from mice without colitis, 6 (21%) displayed TNF-α inhibitory effects on LPS-stimulated macrophages when coincubated with cell-free L-cm. The magnitude of inhibition of TNF-α production varied among the isolates, indicating functional differences among these potential probiotic isolates. In contrast, none of the 29 lactobacilli recovered from IL-10-deficient mice demonstrated immunomodulatory activity (Fig. 6).
FIG. 6.
In vitro immunomodulatory activity of lactobacilli, determined by measuring TNF-α inhibition in LPS-activated murine macrophages. Selected murine Lactobacillus strains are shown. Note that all of the L. johnsonii strains, regardless of origin, failed to diminish TNF-α inhibition. A subset of lactobacilli recovered from mice without colitis displayed immunomodulatory activity. LGG, L. rhamnosus GG; K/O, knockout (deficient). LPS was from E. coli O127:B8. Error bars indicate standard deviations.
DISCUSSION
In our study, mice without colitis were colonized by several Lactobacillus species, with L. reuteri being the predominant species. Two indigenous Lactobacillus species, L. reuteri (27) and L. murinus (8), represented 72% of Lactobacillus species isolated from these animals, whereas IL-10-deficient animals were colonized with a single enteric Lactobacillus species, L. johnsonii. None of the L. johnsonii isolates from either IL-10-deficient mice or mice without colitis down-regulated murine TNF-α production. Interestingly, a subset of enteric Lactobacillus clones recovered from healthy mice inhibited TNF-α production in vitro, demonstrating anti-inflammatory activity. L. reuteri isolates from mice without colitis were genetically heterogeneous at the clonal level, with different strains being found in different areas of the intestine. In contrast, L. johnsonii isolates from IL-10-deficient animals were relatively homogeneous and represented the sole species colonizing different intestinal regions of IL-10-deficient mice.
Early detailed studies of intestinal microecology of rodents (29, 30) were performed prior to the development of molecular phylogenetic approaches. rDNA sequences of 62 Lactobacillus species are available in the current version of Ribosomal Database Project II. For 25 of these 62 species, several subspecies and/or strain sequences have been deposited. A four-group or complex classification for lactobacilli has been suggested based on sequence analyses of the 16S-23S rRNA ISR (32). That study described the identification in the mouse intestine of six Lactobacillus species belonging to three of the four groups. According to this group-based classification, lactobacilli cluster into group I (L. delbrueckii group), group II (L. acidophilus group), group III (L. casei group), and group IV (L. salivarius-L. reuteri-L. plantarum-L. animalis-L. murinus group). Within the L. acidophilus complex (group II), two subgroups were delineated based on DNA-DNA hybridization (12) and sequences in the V1 region of the 16S rRNA gene (14). Within the L. casei group (group III), several methods distinguished the group members, including assessment of sequence variations in the 16S rRNA gene (ribotyping) (28) and the 16S-23S rRNA ISR (35). Alternative single-locus targets for phylogenetic classification of lactobacilli include tuf, the gene encoding elongation factor Tu (3).
DNA sequencing of rRNA genes appear to be sufficient for the identification of most lactobacilli. As with other bacteria, single-locus sequencing approaches generally do not distinguish organisms at the subspecies level, nor do they identify particular species, such as L. vaginalis, the L. animalis-L. murinus complex, or other potentially significant intestinal lactobacilli. Our data obtained by 16S-23S rRNA ISR-based PCR grouping and 16S rDNA-based cluster analysis for Lactobacillus identification generally are consistent with data from other phylogenetic studies of this genus. While 16S rRNA analyses have provided robust phylogenetic positioning of gram-positive and gram-negative bacteria, taxonomic units may not be clearly distinguishable on the basis of rRNA gene sequences alone (7).
Multilocus molecular strategies, such as randomly amplified polymorphic DNA analysis (42) and rep-PCR DNA typing (37, 38), are required for clonal analyses of bacteria, including lactobacilli. Both methods have been useful and reliable for the identification and typing of various Lactobacillus species (2, 6, 31, 36). The present investigation included rep-PCR studies for clonal analyses of enteric Lactobacillus isolates and correlation of genomic profiling to biochemical and sequencing studies. Relative levels of genetic heterogeneity differed, depending on the species of lactobacilli and mouse population studied. The rep-PCR clustering by multiple approaches was consistent with sequencing-based species identification. L. johnsonii isolates from IL-10-deficient animals were genetically homogeneous and distinct from isolates obtained from mice without colitis.
Phenogenetic approaches result in accurate identification and characterization of Lactobacillus species, including probiotic strains (13). The use of biochemical information alone is less reliable than the use of genotypic methods (23). Initial presumptive tests for the identification of lactobacilli include modified Kirby-Bauer disk diffusion antimicrobial susceptibility testing, biochemical screening tests, and morphological examination. Definitive identification requires 16S rDNA sequence-based identification. If clonal or strain-level differentiation is required, the analysis should include whole genomic fingerprinting, such as rep-PCR. The need for such rigorous characterization, especially in lactobacilli, is due to the uncertainty surrounding the identities of lactobacilli that may be of biological consequence. For example, in a rodent study of Lactobacillus-mediated colitis attenuation, it was suggested that beneficial effects might be restricted to individual species or clones (9). In humans, no association has yet been established between groups of lactobacilli and disease in the intestine (20).
Differences in the relative heterogeneities of enteric lactobacilli from animals without colitis and IL-10-deficient animals may reflect differences in the housing of sentinel mice or the impact of host susceptibility on colonization patterns in the intestine. Genetically similar L. reuteri isolates were derived from iNOS-deficient C57BL/6 mice that were pair housed or cohoused. Accordingly, a somewhat clonal population of L. johnsonii was found in IL-10-deficient mice, possibly due to cohousing or derivation from parental littermates. Since all gene-deficient mice were raised and housed in the same barrier facility and given standard chow, we believe that random acquisition and enrichment of intestinal lactobacilli should be similar for all mice. Sentinel Swiss Webster mice were housed in the same specific-pathogen-free animal facility and were used regularly for routine murine pathogen surveillance. Host genetic differences undoubtedly influence the composition of the intestinal microbiota (i.e., microbiota of Swiss Webster mice likely will be different from that of C57BL/6 mice). It is interesting that similar Lactobacillus species colonize the intestines of mice without colitis (i.e., Swiss Webster and iNOS-deficient C57BL/6 mice). In contrast, iNOS-deficient and IL-10-deficient mice, both from a C57BL/6 background, have strikingly different intestinal lactobacillus populations despite being housed in the same facility and having a genetic difference in only a single locus.
Distinct intestinal Lactobacillus species predominate in mice without colitis and IL-10-deficient mice. These results are in concurrence with the findings of Madsen et al. (17), who described studies of enteric Lactobacillus populations in IL-10-deficient animals. In that study (17), however, isolates were described at the species level, and subspecies assessments of clonal population structures were not included. Nevertheless, in two geographically distinct colonies of IL-10-deficient mice (Alberta, Canada, and Massachusetts), L. johnsonii was recovered as the predominant species in mice that can spontaneously develop colitis due to the microbiota (17). L. johnsonii may represent an inert bystander organism in the IL-10-deficient mouse intestine, while Lactobacillus clones found in wild-type mice, such as L. reuteri, may interact with cells of the intestinal mucosa in a manner beneficial to the host (e.g., immunomodulatory properties). Another possibility is that L. johnsonii in the IL-10-deficient mouse model somehow promotes inflammation, similar to the effects of Enterococcus faecalis in gnotobiotic IL-10-deficient mice (1). Interestingly, monoxenic mice experimentally colonized with L. johnsonii displayed evidence of bacterial translocation into mucosal lymphoid organs and stimulation of Lactobacillus-specific humoral immune responses (11). Different strains of L. johnsonii were recovered from mice without colitis and IL-10-deficient mice, possibly indicating biologically relevant differences among various clones of L. johnsonii.
Peña and Versalovic previously described an in vitro assay demonstrating that particular lactobacilli were capable of decreasing TNF-α production in LPS-activated murine macrophages (24). In IBD, macrophages represent primary producers of the proinflammatory cytokine TNF-α, amplify the host immune response in the intestine, and represent a primary target of immunotherapy (26). Depletion of peritoneal macrophages in IL-10-deficient mice prevents IBD (40), indicating a primary role for macrophages and TNF-α in intestinal inflammation. Murine GI lactobacilli isolated in the present study were assayed for inhibition of TNF-α production. All lactobacilli isolated from IL-10-deficient mice failed to decrease TNF-α production, whereas six Lactobacillus isolates from mice without colitis, identified by 16S rDNA sequencing as being most similar to L. reuteri (four of six), ASF 360 (L. intestinalis) (one of six), or L. paracasei (one of six), significantly inhibited TNF-α production. Specific lactobacilli may be relevant to disease induction or progression in the IL-10-deficient mouse model of colitis. For the murine L. reuteri population in this study, genomic fingerprinting and biochemical profiling analyses indicated that multiple clones inhibit TNF-α production. These results indicate that multiple strains of L. reuteri may be capable of probiotic activity but, conversely, that not all strains of L. reuteri have in vitro immunomodulatory activity.
As the clone is the fundamental unit of pathogenesis, we propose that the fundamental probiotic unit is also the bacterial clone. The widespread use of lactobacilli in the food and dairy industry, their apparent role in GI health, and their use in probiotic therapy have stimulated a more thorough examination of the genus Lactobacillus. Detailed phenogenetic studies of this genus will be necessary to understand its biological role in the intestinal microbiota and the relevance of lactobacilli to animal and human health.
Acknowledgments
We thank members of the Division of Comparative Medicine at Massachusetts Institute of Technology (MIT; Cambridge, Mass.), especially James G. Fox, for the animals used in this study and access to the corresponding animal facilities. We acknowledge technical support kindly provided by C. Corcoran, E. Buckley, J. Cline, and M. Ihrig at MIT as well as R. A. Luna, S. C. Jones, and Y. P. Lin at Texas Children's Hospital and Baylor College of Medicine.
This work was supported by several awards to J.V., including the First Award of the Crohn's & Colitis Foundation of America (CCFA) and the National Institutes of Health (K08-DK02705). J.V. also was supported by U.S. Public Health Service grant DK56338, which funds the Texas Gulf Coast Digestive Diseases Center and The Moran Foundation.
REFERENCES
- 1.Balish, E., and T. Warner. 2002. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am. J. Pathol. 160:2253-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouton, Y., P. Guyot, E. Beuvier, P. Tailliez, and R. Grappin. 2002. Use of PCR-based methods and PFGE for typing and monitoring homofermentative lactobacilli during Comte cheese ripening. Int. J. Food Microbiol. 76:27-38. [DOI] [PubMed] [Google Scholar]
- 3.Chavagnat, F., M. Haueter, J. Jimeno, and M. G. Casey. 2002. Comparison of partial tuf gene sequences for the identification of lactobacilli. FEMS Microbiol. Lett. 217:177-183. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, H., H. Frokiaer, and J. Pestka. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168:171-178. [DOI] [PubMed] [Google Scholar]
- 5.Fujisawa, T., Y. Benno, T. Yaeshima, and T. Mitsuoka. 1992. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int. J. Syst. Bacteriol. 42:487-491. [DOI] [PubMed] [Google Scholar]
- 6.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 7.Gupta, R. S. 2002. Phylogeny of bacteria: are we now close to understanding it? ASM News 68:284-291. [Google Scholar]
- 8.Hemme, D., et al. 1980. Lactobacillus murinus n. sp., une nouvell a espece de la flore dominante autochthone du tube digestif du rat et de la souris. Ann. Microbiol. 131A:297-308. [PubMed] [Google Scholar]
- 9.Holma, R., P. Salmenpera, J. Lohi, H. Vapaatalo, and R. Korpela. 2001. Effects of Lactobacillus rhamnosus GG and Lactobacillus reuteri R2LC on acetic acid-induced colitis in rats. Scand. J. Gastroenterol. 36:630-635. [DOI] [PubMed] [Google Scholar]
- 10.Holzapfel, W. H., P. Haberer, R. Geisen, J. Bjorkroth, and U. Schillinger. 2001. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 73:365S-373S. [DOI] [PubMed] [Google Scholar]
- 11.Ibnou-Zekri, N., S. Blum, E. J. Schiffrin, and T. von der Weid. 2003. Divergent patterns of colonization and immune response elicited from two intestinal Lactobacillus strains that displayed similar properties in vitro. Infect. Immun. 71:428-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, M. C., B. Ray, and T. Bhowmik. 1987. Selection of Lactobacillus acidophilus strains for use in “acidophilus products.” Antonie Leeuwenhoek 53:215-231. [DOI] [PubMed] [Google Scholar]
- 13.Klein, G., A. Pack, C. Bonaparte, and G. Reuter. 1998. Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 41:103-125. [DOI] [PubMed] [Google Scholar]
- 14.Kullen, M. J., R. B. Sanozky-Dawes, D. C. Crowell, and T. R. Klaenhammer. 2000. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511-516. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlane, G., and J. Cummings. 2002. Probiotics, infection and immunity. Curr. Opin. Infect. Dis. 15:501-506. [DOI] [PubMed] [Google Scholar]
- 16.Madsen, K. 2001. The use of probiotics in gastrointestinal disease. Can. J. Gastroenterol. 15:817-822. [DOI] [PubMed] [Google Scholar]
- 17.Madsen, K. L., J. S. Doyle, L. D. Jewell, M. M. Tavernini, and R. N. Fedorak. 1999. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116:1107-1114. [DOI] [PubMed] [Google Scholar]
- 18.McConnell, M., and G. Tannock. 1991. Lactobacilli and azoreductase activity in the murine cecum. Appl. Environ. Microbiol. 57:3664-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsuoka, T. 1982. Recent trends in research on intestinal flora. Bifidobacteria Microbiota 1:3-24. [Google Scholar]
- 20.Molin, G., et al. 1993. Numerical taxonomy of Lactobacillus spp. associated with healthy and diseased mucosa of the human intestines. J. Appl. Bacteriol. 74:314-323. [DOI] [PubMed] [Google Scholar]
- 21.Mori, K., et al. 1997. Comparative sequence analyses of the genes coding for 16S rRNA of Lactobacillus casei-related taxa. Int. J. Syst. Bacteriol. 47:54-57. [DOI] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests, 8th ed., vol. 23, p. 37-50. Approved standard M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Nigatu, A. 2000. Evaluation of numerical analyses of RAPD and API 50 CH patterns to differentiate Lactobacillus plantarum, Lact. fermentum, Lact. rhamnosus, Lact. sake, Lact. parabuchneri, Lact. gallinarum, Lact. casei, Weissella minor and related taxa isolated from kocho and teff. J. Appl. Microbiol. 89:969-978. [DOI] [PubMed] [Google Scholar]
- 24.Peña, J. A., and J. Versalovic. 2003. Lactobacillus rhamnosus GG decreases TNF-alpha production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell. Microbiol. 5:277-285. [DOI] [PubMed] [Google Scholar]
- 25.Perdigon, G., C. Maldonado Galdeano, J. Valdez, and M. Medici. 2002. Interaction of lactic acid bacteria with the gut immune system. Eur. J. Clin. Nutr. 56(Suppl. 4):S21-S26. [DOI] [PubMed] [Google Scholar]
- 26.Podolsky, D. 2002. Inflammatory bowel disease. N. Engl. J. Med. 347:417-429. [DOI] [PubMed] [Google Scholar]
- 27.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:45-53. [PubMed] [Google Scholar]
- 28.Ryu, C. S., J. W. Czajka, M. Sakamoto, and Y. Benno. 2001. Characterization of the Lactobacillus casei group and the Lactobacillus acidophilus group by automated ribotyping. Microbiol. Immunol. 45:271-275. [DOI] [PubMed] [Google Scholar]
- 29.Savage, D., R. Dubos, and R. Schaedler. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J. Exp. Med. 127:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaedler, R., R. Dubos, and R. Costello. 1965. The development of the bacterial flora in the gastrotinestinal tract of mice. J. Exp. Med. 122:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sohier, D., J. Coulon, and A. Lonvaud-Funel. 1999. Molecular identification of Lactobacillus hilgardii and genetic relatedness with Lactobacillus brevis. Int. J. Syst. Bacteriol. 49:1075-1081. [DOI] [PubMed] [Google Scholar]
- 32.Song, Y., et al. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 33.Tannock, G. 1997. Normal microbiota of the gastrointestinal tract of rodents, p. 187-215. In R. Mackie et al. (ed.), Gastrointestinal microbiology, vol. 2. Chapman & Hall, New York, N.Y. [Google Scholar]
- 34.Tannock, G., M. Dashkevicz, and S. Feighner. 1989. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl. Environ. Microbiol. 55:1848-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tannock, G. W., A. Tilsala-Timisjarvi, S. Rodtong, J. Ng, K. Munro, and T. Alatossava. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl. Environ. Microbiol. 65:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ventura, M., and R. Zink. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 217:141-154. [DOI] [PubMed] [Google Scholar]
- 37.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Versalovic, J., M. Schneider, F. de Bruijn, and J. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR). Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 39.Wagner, R., T. Warner, L. Roberts, J. Farmer, and E. Balish. 1997. Colonization of congenitally immunodeficient mice with probiotic bacteria. Infect. Immun. 65:3345-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe, N., et al. 2003. Elimination of local macrophages in intestine prevents chronic colitis in interleukin-10-deficient mice. Dig. Dis. Sci. 48:408-414. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, T., M. Morotomi, N. Suegara, Y. Kawai, and M. Mutai. 1977. Distribution of indigenous lactobacilli in the digestive tract of conventional and gnotobiotic rats. Microbiol. Immunol. 2:183-191. [DOI] [PubMed] [Google Scholar]
- 42.Williams, J., A. Kubelik, K. Livak, J. Rafalski, and S. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]