Abstract
Since its first administration in the 1940s, the influenza vaccine has provided tremendous relief against influenza infections. However, time has revealed the vaccine’s ultimate limit and the call for its reinvention has now come, just as we are beginning to appreciate the antibody immune responses vital in preventing infections. New strategies to design the influenza vaccine rely on selectively inducing broadly neutralizing antibodies that are specific for highly conserved viral epitopes. Such approaches take us away from the limited range of protection provided by current seasonal influenza vaccines and towards a future with a pan-influenza vaccine capable of providing universal strain coverage.
Reality check: the influenza vaccine dilemma
Seasonal influenza vaccinations have dramatically reduced influenza infections, however, significant morbidity and mortality still occur. Over 41,000 deaths in the U.S. every year result from influenza [1], and periodically, pandemics, such as the recent 2009 H1N1 pandemic, occur and cause this number to rise. Rare cases of zoological influenza strains being transmitted to humans (i.e., avian H5N1 strain) have led to disturbingly high mortality rates with 50% of infections leading to death [2]. Unfortunately, treatment of influenza infections with antiviral drugs is not a reliable option as it is often ineffective and leads to resistant viruses [3]. As such, despite its current limitations, prevention through vaccination is still the main protective measure against influenza for the general population. However, going forward, the vaccine needs to be improved to overcome the limited breadth of protection it offers in the face of a rapidly evolving pathogen. At the same time, novel prophylactic and therapeutic options need to be developed for immunocompromised populations, including the very young and very old, in whom vaccines are inevitably less effective. Emerging studies on reinventing the influenza vaccine and improving therapeutics rely heavily on isolating and characterizing neutralizing antibodies and the viral epitopes they bind. This review will highlight how such information on the immune responses against influenza has shaped the development of novel vaccination strategies that involve tactical selection of the prime/boost combination and carefully design of the immunogen. The strategies discussed here represent the beginnings of a new wave of research that will ultimately lead to the development of a viable universal influenza vaccine.
Tracking a moving target: the annual influenza vaccine
The current seasonal influenza vaccine is produced both as an inactivated virus vaccine and a live attenuated virus vaccine. Both types of vaccine reduce virus infectivity and restrict viral replication by inducing antibodies and cell-mediated immune memory against the virus. Antibodies are the primary means to prevent infections whereas cell-mediated immunity is important in clearing ongoing infections. The inactivated influenza vaccine is administered intramuscularly and induces primarily systemic antibodies while the live vaccine induces both systemic and mucosal antibodies due to its intranasal route of administration. Both forms of the vaccine are trivalent and are composed of three different virus strains: an influenza A H1N1 strain, influenza A H3N2 strain and influenza B strain. The components of the vaccine are determined by epidemiological evidence and virus surveillance. Genetic analysis on viral isolates from specimens collected around the world each year enables scientists to monitor the prevalent strains in the human population and the rate of spread [4]. Hence the components of the vaccine are tailored annually to match the strains that would most likely be dominant in the population for the upcoming influenza season.
A never-ending battle: why the current influenza vaccine is suboptimal
Monitoring the virus strains circulating in the human population every year and updating the vaccine accordingly, usually every 1–3 years, serves to keep up with only the antigenic drift of the most common influenza strains from the previous year. Antigenic drift refers to a continuous process in which mutations in the virus genome produce changes in the antibody-binding regions and give rise to new strains [5]. As first identified in HIV evolution [6], this drift is exacerbated by selecting for minimal changes in amino acid sequences that are targeted by post-translational modifications of the influenza virus [7]. Given that manufacturing the vaccine takes a few months, during which the influenza virus can rapidly mutate and evolve, there is an arms race between the virus and vaccine development. Not surprisingly, several occasions of mismatch between the vaccine components and the prevailing dominant strains, as seen in 1997/98 and 2003/04, have occurred [5]. In such scenarios, the vaccine does not confer the desired level of protection and the number of infections increases. Antigenic shift, another immune evasion mechanism of influenza, is not factored into the design of the current vaccine because it is simply too difficult to predict when and how a shift would occur. Antigenic shift refers to the reassortment of viral gene segments between various influenza viruses of human or zoological origin. This leads to the emergence of new strains that have caused most influenza pandemics [8]. In the most recent pandemic, the 2009 H1N1 pandemic, the seasonal vaccine did not contain the pandemic strain and a substantial number of infections and deaths occurred [9]. Consequently, a monovalent vaccine containing only the H1N1 pandemic strain was rapidly produced for administration along with the 2009/10 trivalent seasonal vaccine [10].
The prevalence of the virus in the human population is not the only factor taken into consideration when selecting vaccine strains. The virulence of the virus and ease of the manufacturing process are also evaluated and in doing so, compromises are made. Even if the vaccine components are a match to the circulating strains, there is limited vaccination in the general global population and on the individual level; many of the antibodies raised by the vaccine are unable to inhibit infections [11, 12] (discussed later). Further, the populations at greatest risk of mortality from influenza are the immunocompromised, the elderly, the very young, and pregnant women [13] and the ability of these populations to induce new responses to yearly vaccines is reduced. This limited penetrance of protection leads to a persistent number of infections each year. These infections then fuel the emergence of new influenza mutational variants and the reciprocal need for another new vaccine in a never-ending cycle. Yearly vaccinations are required since each seasonal vaccine elicits neutralizing antibodies that are specific only for the vaccine strains and closely related isolates, but rarely for divergent strains.
Taking careful aim: the ideal vaccination outcome
To combat these problems and limitations and to stop the spread of influenza indefinitely, the influenza vaccine must be reinvented into a single broadly protective vaccine or what has been termed a universal pan-influenza vaccine. Such a vaccine would trigger a B cell response that provides long-term protection against a wide variety of strains and could be administered progressively to the entire population. The characteristics of an ideal anti-influenza antibody response are two-fold: 1) induction of broadly neutralizing antibodies that bind epitopes critical for viral function and can incapacitate large swathes of influenza phylogeny, and 2) induction of antibodies that are able to aide in clearance of influenza through interactions with other immune cells. To design a vaccine that elicits antibodies with these characteristics it is first necessary to understand how these antibodies are induced during the course of a natural immune response.
Engaging the enemy: direct antibody-mediated neutralization of influenza
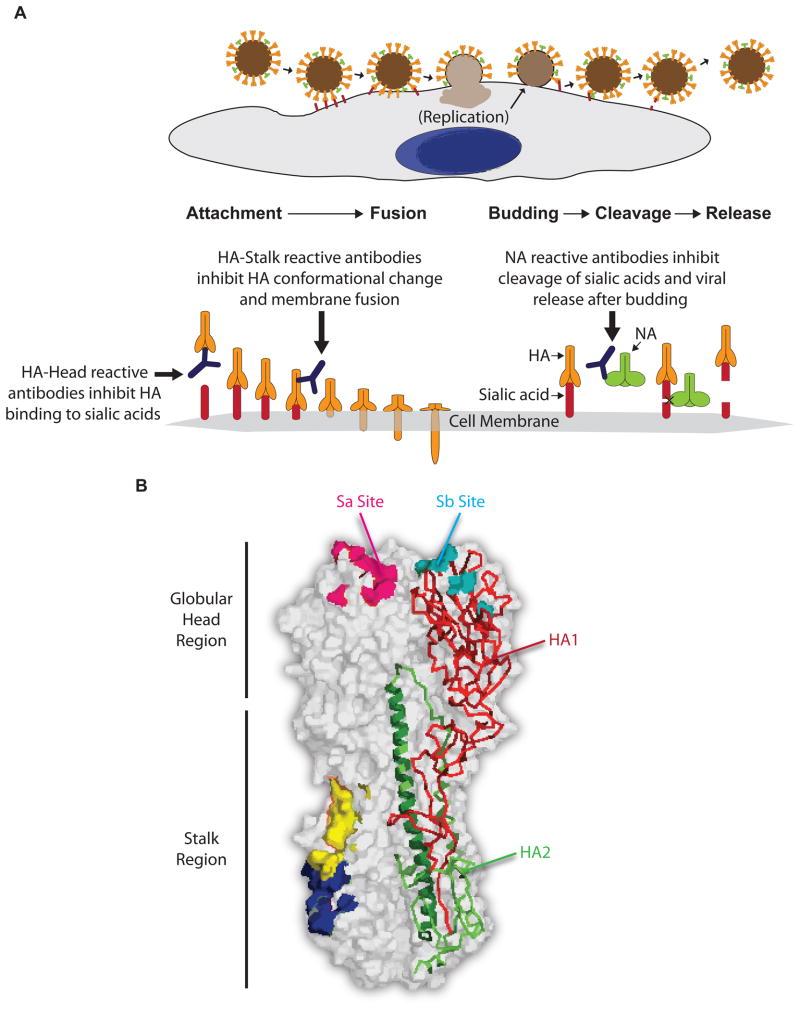
Certain antibodies, without the aid of other cell types, are able to directly neutralize influenza by binding to and inhibiting the function of viral proteins most commonly necessary in two separate stages of the influenza life cycle [14]. Such antibodies are specific for epitopes in the hemagglutinin (HA) and neuraminidase (NA) proteins and can prevent host cell entry or budding of virions, respectively (Figure 1A).
Figure 1.
a) Mode of action of neutralizing antibodies against influenza virus. The influenza viral life cycle is depicted with magnification of critical steps in infection: HA mediated cell entry and NA mediated release. Neutralizing antibodies against influenza virus prevent infections by inhibiting either viral entry into host cells or release of virions from infected cells. HA-specific antibodies are responsible for preventing viral entry into host cells by binding two regions on HA: the head region and the stalk region. HA-head reactive antibodies block binding of the HA head region to sialic acids on host cells thus preventing attachment of virus to host cells while HA-stalk reactive antibodies inhibit conformational changes required in HA for fusion of the virus with host cell membrane. Generally, HA-head reactive antibodies are strain specific since the head region is mostly variable while some HA-stalk reactive antibodies can be broadly neutralizing if they bind a highly conserved and vital region of HA. NA-reactive antibodies block the later stages of the virus replication cycle by inhibiting the cleavage of sialic acids by NA and the release of new virions. This then prevents further infection of other cells. b) Structure of Hemagglutinin. A space-filling model of a trimer of HA is shown (PDB ID: 3LZG) [35]. One of the HA monomers is illustrated in Ribbons representation with HA1 colored in red and HA2 colored in green. The region of HA2, which is colored in darker green and shown as an alpha helix, is representative of the LAH construct used in designing a minimal immunogen for vaccination [40]. Antibodies that are hemagglutinin inhibiting (HAI) most commonly bind to the antigenic Sa and Sb sites on the receptor-binding domain of the globular head region. Broadly neutralizing antibodies typically bind critical conserved epitopes in the stalk region of HA and two examples of such epitopes are shown in blue [25] and in yellow [26].
As HA executes cell entry in a two-step manner (Box 1 and Figure 1A), antibodies are presented with two opportunities to neutralize the protein, either by preventing 1) attachment to sialic acid or 2) the conformational changes necessary for membrane fusion. Antibodies can prevent sialic acid:HA ligation by binding at or near the receptor-binding site on HA thus sterically-inhibiting the interaction [14, 15]. An example of this mode of neutralization was described in recent crystallographic studies on a human monoclonal antibody (mAb) that binds the receptor-binding domain of HA, with its epitope including the antigenic site designated Sb [15] (Figure 1B). The study revealed that the mAb inserts a loop into the receptor-binding pocket of HA and in doing so, mimics interactions otherwise provided by sialic acid. This binding mechanism enabled the mAb to neutralize a broad range of H1 influenza strains. Unfortunately, there are multiple non-neutralizing epitopes on the head region of HA that do not affect sialic acid:HA ligation and have no impact on the viral life cycle. In fact, most natural human antibodies to influenza vaccination [11, 12] or infection [16] are unable to inhibit infection. These epitopes, however, may contribute to viral clearance by other cell-mediated means (discussed later). Notably, the epitopes in the head region that have a functional role in receptor binding are less likely to mutate, while those in the rest of the globular head are very mutable and effectively mask the proximal active sites. Therefore, antibodies that bind one strain of influenza at these highly mutable sites are often useless against other strains.
Box 1. Description of influenza viral surface proteins: hemagglutinin (HA) and neuraminidase (NA).
There are 16 different influenza subtypes of HA and they are clustered into two groups: group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16) and group 2 (H3, H4, H7, H10, H14, and H15). A transmembrane trimer expressed on the surface of influenza virions, it is first produced as the homotrimer HA0, which is cleaved to give rise to a final product consisting of two subunits, HA1 and HA2 [47–49]. During infection, HA1 - corresponding to the variable head region of hemagglutinin - binds sialic acid on the surface of host cells via its receptor-binding site [50]. Following internalization into the host cell’s acidic endosome, HA2, the conserved stalk region, undergoes a pH dependent change, allowing the virion to fuse with the host cell membrane and transfer its genetic material into the cytoplasm [51–53]. Antibodies that prevent influenza infections neutralize the virus by blocking these two vital regions on HA – the receptor-binding site and the conserved stalk region. Neutralization capacity of antibodies can be observed in the laboratory using two main assays. The hemagglutination inhibition (HAI) assay, only indicates if an antibody has successfully blocked the sialic acid:HA interaction, and does not measure neutralization via the HA stalk or other viral epitopes. The microneutralization assay accounts for both mechanisms of neutralization as it relies on detecting antibodies that have prevented viral infection of cells in vitro. Besides HA, a second viral epitope important in direct antibody neutralization of influenza is neuraminidase (NA), a mushroom shaped homotetramer expressed on the surface of the virion [54, 55]. Following viral replication, viral particles must bud from host cells. Neuraminidase facilitates this by removing sialic acid from the glycoproteins of budding virions, preventing them from aggregating [56]. Like HA, NA is necessary for the viral life cycle, appears as several distinct subtypes, and accumulates mutations that contribute to antigenic drift [57].
Similar to the receptor-binding site in the head region, the membrane fusion activity of HA, which is dependent on protein conformational changes, can be inhibited by antibodies specific for epitopes in the stalk region (Box 1 and Figure 1) [17–19]. It has been demonstrated that neutralizing antibodies to the stalk region arise during influenza infection [16] and vaccination [11, 20–23]. Importantly, since the stalk region is fairly conserved over influenza strains, these antibodies bind and neutralize a diverse panel of strains including various H1, H2, H5, H6, H8, and H9 (group 1) strains of influenza [11, 16, 18, 19, 21, 24] and more recently, H3 and H7 strains (group 2) [25]. The latest and most exciting discovery was of a neutralizing antibody that recognized a stalk epitope conserved by virtually all influenza A strains that are infectious to humans [26] (Figure 1B). Hence, to elicit these potently broadly neutralizing antibodies with a universal vaccine, the conserved HA stalk epitopes need to be targeted. Further, because these antibodies have demonstrable efficacy against influenza in protecting [11, 16, 19, 25, 26] or even treating [16] mice and ferrets in vivo, there is also a substantial interest in developing HA-stalk reactive mAbs as the next class of broad-spectrum anti-influenza therapeutics.
Neutralizing antibodies directed against NA are also important in inhibiting the viral life cycle. Specifically, antibodies that block the active site of NA and inhibit it from cleaving sialic acid, prevent release of new virions from the infected host cell (Box 1 and Figure 1A). This then leads to large aggregates of influenza virions that are unable to infect other host cells [27, 28].
Diplomacy and public relations: indirect antibody clearance of virions
Antibodies binding non-neutralizing epitopes facilitate viral clearance through a number of mechanisms. While neutralization takes place exclusively between the viral epitope and the variable arms of the antibody, indirect antibody clearance also relies on the heavy chain’s constant (Fc) domain. After a viral particle has been opsonized by IgG antibodies, the Fc regions bind to Fc-receptors (FcRs) expressed on phagocytic cells, leading to ingestion and destruction of the pathogen [28]. This antibody-mediated uptake of pathogens, termed FcR-mediated phagocytosis, is also vital in enabling presentation of influenza peptides on MHC class II molecules by phagocytic cells for the induction of T cell mediated immune responses. FcRs are also transmembrane proteins that can transmit stimulatory or inhibitory signals to most immune cells, thus allowing antibodies to indirectly orchestrate the type and strength of the innate and cellular immune responses (reviewed in [29]).
In another mechanism called antibody dependent cell-mediated cytotoxicity (ADCC), antigen bound IgG1 and IgG3 antibodies bind FCRyIII on natural killer (NK) cells, which are then able to kill virions and virally infected cells. The role of NK cells in clearing flu particles in the absence of neutralizing antibodies was demonstrated in mice immunized with the non-neutralizing flu antigen M2 [30]. The third antibody dependent mechanism important in the clearance of influenza is complement dependent cytotoxicity (CDC) [30, 31]. In this mechanism, IgG antibodies bound to the target surface activate complement proteins in the host serum, which leads to inflammation and a stepwise recruitment of proteins that ultimately puncture the lipid membrane of the pathogen or infected host cell. One study showed that mice lacking C3, an important intermediate in the complement cascade, were unable to successfully clear influenza after being vaccinated with M2 [32], suggesting that clearance of influenza virions is at least partially reliant on CDC.
Taken together, these factors establish that antibodies to both neutralizing and non-neutralizing epitopes of influenza are important for successful disease outcome. Despite this, the vaccine community focuses primarily on the direct and immediate prophylactic protection provided by neutralizing antibodies. While such antibodies are valuable, the protection they provide is tempered by the fact that they also drive viral evolution due to selective pressure, making the neutralizing epitopes a moving target. It is possible that a balanced response, including non-neutralizing antibodies specific for conserved epitopes, would provide broader protection against influenza escape mutants.
History is the key: what can humoral immunity teach us about successful vaccine design?
Rational vaccine design relies on synthesizing what we know about the humoral responses to influenza into novel strategies that can specifically elicit the ideal responses. By examining the outcome of past vaccination strategies and infections, tactics to enhance our vaccines to universal anti-influenza potential are becoming apparent. For now, two strategies – prime/boost and minimalistic immunogen design - are showing great promise in laying the foundations for the design of a universal influenza vaccine (Table 1).
Table 1.
Current and potential strategies for influenza vaccination
| Strategy | Description | References |
|---|---|---|
| Seasonal/Annual Vaccination | Trivalent vaccine consisting of H1N1, H3N2 and B strains chosen based on strain prevalence in the past year | [4] |
| Prime/boost with viruses | Two highly divergent virus strains used in a sequential vaccination program | [16, 37, 38] |
| Prime/boost with HA proteins and whole virus | Priming with one HA protein or several antigenically distinct HA proteins in a sequential manner and boosting with whole virus | [20, 23] |
| Minimalistic immunogen | Vaccinating with a specially designed immunogen that consists of only the highly conserved HA stalk regions | [21, 40, 41] |
Cutting them off at the pass: Prime/boost strategies
The humoral response to influenza is comprised of both newly activated naive cells and recalled memory B cells. As many influenza epitopes shift each year, newly activated cells will be specific for influenza antigens unique to the latest variant, whereas the recalled memory cells would be specific for epitopes that have not changed. In 2008, it was demonstrated that individuals born in 1915 or earlier still had circulating memory B cells to the 1918 influenza pandemic strain [33], indicating that the B cell memory compartment is extremely long lived. In addition, despite little cross-reactivity with any recent vaccine strains, the very same antibodies isolated against the 1918 pandemic strain cross-reacted to a conserved epitope on the 2009 pandemic H1N1 strain [34, 35].
To this end, prime/boost strategies, in which an individual is primed with one strain and later boosted with another strain, are being developed to enrich the humoral response with antibodies to conserved influenza epitopes. Unfortunately, as previously encountered in the design of seasonal vaccines, many non-neutralizing epitopes do not mutate drastically between similar variants of influenza and most antibodies induced are not neutralizing. On the other hand, a prime/boost strategy using highly divergent strains of influenza in which only functional epitopes are conserved would recall only the memory B cells that are specific for these vulnerable epitopes. In this way, the immune response can be enriched for broadly neutralizing antibodies.
Evidence supporting the prime/boost approach
A recent study of human antibody immune responses to seasonal influenza vaccination identified 20 mAbs which cross-reacted to H1, H2, H5, H6, and H9 strains, including the 2009 H1N1pandemic strain [11]. Interestingly, the mAbs used different V genes and had high frequencies of somatic hypermutation, suggesting that they arose from memory cells. While one antibody bound the head region of HA, the remaining 19 bound the stalk region, consistent with the functional importance of this region. Importantly, four of the 20 mAbs were able to protect mice from lethal challenge with other influenza subtypes. While these antibodies were rare, these findings, along with complementary studies [34, 36], firmly establish that routine vaccination can produce powerful, heterosubtypic antibodies that provide protection against new and novel strains of influenza. The key to a universal vaccine is to induce these antibodies in a more dominant fashion.
Several recent reports on experiments performed in mice, ferrets, and nonhuman primates [20, 37, 38], suggest that not only do broadly neutralizing, heterosubtypic antibodies arise during the natural course of infection and vaccination, they are especially prevalent when a prime/boost strategy involving divergent influenza strains is used. The 2009 H1N1 pandemic presented a unique opportunity to investigate the effect of a novel influenza strain on the antibody repertoire. In a study of individuals diagnosed with the highly unique 2009 H1N1 pandemic strain, it was found that their antibody repertoire was indeed characterized by frequent highly cross-reactive and neutralizing mAbs [16]. Similar to the findings by Corti et al., these antibodies had high levels of somatic hypermutation, suggesting they arose from memory B cells specific for conserved, functional domains on multiple influenza strains. Furthermore, the mAbs were found to be specific for conserved epitopes in the head and stalk regions of various H1N1 and H5N1 strains. Representative antibodies with broad-reactivity were shown to rescue mice lethally infected with the 2009 H1N1 pandemic strain even when administered more than 2 days after infection. This study demonstrated that in people with a diverse immunological history, by using the appropriate influenza immunogen, an immune response in which majority of neutralizing antibodies are broadly protective could be induced.
Demonstrated successes
To enhance the production and breadth of broadly neutralizing antibodies, a vaccination regimen incorporating two immunizations in a prime/boost combination was recently developed. In this protocol, mice were vaccinated with plasmid DNA encoding H1N1 HA or H3N2 HA from the 2006/07 vaccine strains and boosted with the trivalent 2006/07 seasonal vaccine [20]. Sera from mice given the prime/boost vaccination showed increased neutralization of diverse H1N1 strains compared to sera from mice given only DNA or seasonal vaccine. The prime/boost combination also provided mice and ferrets protection against divergent H1N1 viruses and resulted in increased survival rates and lower weight loss than vaccination with only DNA or the seasonal vaccine. Using a competition neutralization assay, the authors showed that the antibodies in the sera bound to the conserved HA stem region.
In another study, a slightly different protocol was established to promote the production of cross-reactive antibodies. Mice were sequentially immunized with DNA coding for the HA of different influenza A H3 virus strains arising approximately 10 years apart: A/Hong Kong/1/1968, A/Alabama/1/1981, A/Beijing/47/1992 and boosted with another H3 virus, A/Wyoming/3/2003 (see Glossary for an explanation of the strain naming convention) [23]. The aim of this step-wise approach was to trigger the repeated expansion of B cells specific for conserved epitopes on antigenically distinct HAs. Indeed, this strategy successful elicited neutralizing antibodies with broad reactivity against various H3 viruses and with capacity to protect mice against challenge with diverse H3 viruses.
The above studies highlight how the vaccination regimen can impact the production of broadly neutralizing antibodies and hence shape the design of the universal influenza vaccine. However, while the prime/boost strategy demonstrates increased production of broadly neutralizing antibodies, it is not the complete answer to reinventing the influenza vaccine. Other innovative strategies must be considered to truly maximize the broadly neutralizing antibody response. How can we trigger immune responses to induce sufficient concentrations of broadly neutralizing mAbs in the plasma and at mucosal surfaces to truly provide protection against novel strains? On the flip side, how can we design the vaccine such that antibody responses to non-conserved or variable regions of the virus are minimized and are instead focused on the conserved regions of the virus? One potential approach is to expand the immunogenicity of the influenza HA protein to include more diverse head and stalk region epitopes by using immunostimulatory adjuvants such as MF59 [39]. Another approach, as described below, takes the opposite tack and aims to simplify the immunogen.
Sharp shooting: a minimalistic approach
To reduce skewing the antibody response towards the variable domains of the virus, a minimalistic approach in designing the components of the vaccine has been proposed [21, 40, 41]. The current influenza vaccine is made of whole influenza viruses and consequently induces an immune response that is dominated by non-neutralizing antibodies and homosubtypic neutralizing antibodies [11, 12]. Using whole virus or large viral proteins in the vaccine provides B cells with a wide variety of binding sites. This dilutes the desired broadly protective antibody response with non-neutralizing specificities as well as specificities that are confined to strains within the same subtype. Conversely, using a minimum but highly conserved viral epitope as the immunogen in the vaccine would direct and focus the antibody response on the conserved epitope. Taking such an approach would bring us closer to a universal vaccine capable of eliciting broadly neutralizing antibodies and providing protection against the wide varieties of newly emerging influenza viruses.
Evidence supporting the minimalistic approach
The rationale for this approach is the evidence provided by several studies that broadly neutralizing mAbs bind to the conserved HA stem region and can confer protection against various strains [19, 25, 26, 42, 43]. In one such study, broadly neutralizing mAbs were recovered from combinatorial display libraries constructed from human IgM+ memory B cells of patients immunized with the seasonal vaccine [43]. These broadly neutralizing mAbs were heterosubtypic in that they had neutralizing activity against antigenically diverse H1, H2, H5, H6, H8 and H9 influenza subtypes (the group 1 subtypes) and one of the mAbs (CR6261) was able to confer prophylactic protection and therapeutic efficacy in mice challenged with H5N1 or H1N1 virus. Importantly, it was discovered that these broadly neutralizing mAbs bind to a conserved region of the HA stem domain. In fact, crystallography on CR6261 with H1 and H5 HAs revealed that the mAb recognizes a highly conserved helical region in the membrane-proximal stem domain of HA1 and HA2 which enables the mAb to block conformational rearrangements of HA thus inhibiting membrane fusion [18].
On the same note, a human antibody phage display library selected on the H5 HA ectodomain was used to recover ten broadly neutralizing mAbs. Crystal structure of one of these neutralizing mAbs bound to H5N1 HA revealed that its heavy chain inserts into a highly conserved pocket in the HA stem, inhibiting the conformational changes needed for membrane fusion [19]. Another mAb, C179, which recognizes a common epitope among H1, H2, H5, and H6 HAs, was shown to protect mice from a lethal challenge with various H5N1 viruses and the 2009 pandemic H1N1 virus [42, 44]. We now know that a similar epitope on the HA stalk can be targeted for broad protection against H3 and H7 (group 2) influenza strains [25] or even to all influenza A strains that infect humans [26] (Figure 1B). By designing a multivalent immunogen that comprises the conserved HA stalk epitopes of both the group 1 and group 2 viruses, a vaccine protective against all influenza A strains could be developed.
Besides the conserved HA stalk region, other studies have emerged to show that the conserved extracellular domain of the viral matrix 2 protein (M2e) is able to elicit cross-reactive antibodies that can confer protection in mice challenged with H5N1 or H1N1 viruses [45, 46].
Demonstrated successes
The minimalistic approach was recently taken a step closer to reality by a demonstration of its success in mice. Mice were vaccinated with a novel immunogen constructed with the complete HA2 polypeptide and regions of HA1 contributing to the stalk region but lacking the globular head [21]. This construct, termed as headless HA, retains the structural integrity of the conserved stalk domain. By immunizing with the headless HA construct it was hypothesized that broadly cross-reactive antibodies would be specifically elicited. Indeed, sera isolated from mice vaccinated with the headless HA showed broader reactivity against heterologous strains than sera from mice vaccinated with full-length HA. In addition, mice vaccinated with the headless HA vaccine were protected against lethal virus challenge. Protection against homologous virus challenge was also seen in another study in which mice were vaccinated with an immunogen (HA6) consisting mostly of the HA2 subunit with only proximal regions of HA1 to support its stable conformation at neutral pH [41].
In perhaps the most profound example of the minimalistic approach one group engineered a broadly reactive immunogen based on only a 60 amino acid peptide. For this, a broadly neutralizing mouse mAb (12D1) was utilized to identify a short but highly conserved region on the HA2 subunit that could act as the immunogen in the vaccine [40]. The region identified consists of amino acids 76–130 and is referred to as the long α-helix (LAH) of HA2 (Figure 1B). In constructing the vaccine, the LAH was coupled to a carrier protein (KLH) to increase its antigenicity. Stunningly, vaccination with this 60 amino acid construct protected mice against challenge with divergent subtypes of influenza viruses, namely H3N2, H5N1 and H1N1. Sera from mice immunized with the LAH-KLH conjugate also showed broad reactivity against various other group 1 and group 2 HAs. This work provides direct evidence that through careful design of the immunogen used in the vaccine, it is possible to target B cell responses to only the most conserved portions of HA.
Conclusion: an armistice in sight?
Adopting a minimalistic approach in designing the components of the influenza vaccine seems promising and may certainly pave the way towards a universal influenza vaccine. Keeping in mind that manipulating the vaccination regimen or prime/boost combination can further enhance the quantity and quality of the antibody response, a yearly influenza vaccine may well become a thing of the past. While it seems certain that the influenza vaccine will be reinvented in the near future, one should remain cautionary of having too high of an expectation. To this point, influenza has always won the war: could selective pressure on a new class of broadly conserved epitopes drive the evolution of even more virulent and resistant escape variants? Only time and the continued creative efforts of the scientific community will tell if we can ever win the war on influenza through vaccination.
Acknowledgments
This work was supported by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) grant numbers: 2U54AI057158-06, 2U19AI057266-06, 5U19AI062629-07, 1U19AI082724-02, and U19AI09023-01.
Glossary
- Antigenic drift
a continuous process in which mutations in the virus genome produce changes in the antibody-binding regions and give rise to new strains
- Antigenic shift
the reassortment of viral gene segments between influenza viruses of human or zoological origin which leads to the emergence of new strains
- Antigenic
having the ability to induce an immune response
- Broadly neutralizing antibody
an antibody that binds to a conserved epitope, that is vital in the viral life cycle, and is able to therefore bind to and neutralize various virus strains
- Epitope
the site on the surface of an antigen that is recognized by an antibody
- Fc domain
the constant region of an antibody of which there are five different isotypes (eg. IgG, IgA)
- Heterosubtypic antibodies
antibodies that bind multiple subtypes of influenza viruses
- Heterologous strains
strains of different influenza subtypes
- Homologous strains
strains within the same influenza subtype
- Immunogen
an antigen that is able to elicit an immune response
- Homosubtypic antibodies
antibodies that bind influenza virus strains within the same subtype
- Influenza virus strain naming convention
“Strain type A or B/geographical origin of strain/numbered isolate from that year and origin/year (hemagglutinin and neuraminidase type)”, i.e., “A/Hong Kong/1/1968 (H3N2)”
- Neutralizing antibody
an antibody that binds a vulnerable region of a pathogen and directly prevents infection by the pathogen
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beigel JH. Influenza. Critical Care Medicine. 2008;36:2660–2666. doi: 10.1097/CCM.0b013e318180b039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol. 2007;7:267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland GA, et al. Influenza Virus Resistance to Antiviral Agents: A Plea for Rational Use. Clinical Infectious Diseases. 2009;48:1254–1256. doi: 10.1086/598989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr IG, et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: Basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 Northern Hemisphere season. Vaccine. 2010;28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 5.Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25:6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 6.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 7.Wei CJ, et al. Cross-Neutralization of 1918 and 2009 Influenza Viruses: Role of Glycans in Viral Evolution and Vaccine Design. Science Translational Medicine. 2010;2 doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilbourne ED. Influenza pandemics of the 20th century. Emerging Infectious Diseases. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swerdlow DL, et al. 2009 H1N1 Influenza Pandemic: Field and Epidemiologic Investigations in the United States at the Start of the First Pandemic of the 21(st) Century. Clinical Infectious Diseases. 2011;52:S1–S3. doi: 10.1093/cid/ciq005. [DOI] [PubMed] [Google Scholar]
- 10.Kuehn BM. H1N1 Vaccine. Jama-Journal of the American Medical Association. 2009;302:375–375. [Google Scholar]
- 11.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. Journal of Clinical Investigation. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrammert J, et al. Rapid cloning of high affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WH, et al. Vaccination in the elderly: an immunological perspective. Trends in Immunology. 2009;30:351–359. doi: 10.1016/j.it.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annual Review of Biochemistry. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 15.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. The Journal of experimental medicine. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai M, et al. Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Research. 1998;53:129–139. doi: 10.1016/s0168-1702(97)00143-3. [DOI] [PubMed] [Google Scholar]
- 18.Ekiert DC, et al. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui JH, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Structural & Molecular Biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei CJ, et al. Induction of Broadly Neutralizing H1N1 Influenza Antibodies by Vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 21.Steel J, et al. Influenza Virus Vaccine Based on the Conserved Hemagglutinin Stalk Domain. Mbio. 2010;1 doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui J, et al. Wide Prevalence of Heterosubtypic Broadly Neutralizing Human Anti-Influenza A Antibodies. Clinical Infectious Diseases. 2011;52:1003–1009. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TT, et al. Broadly Protective Monoclonal Antibodies against H3 Influenza Viruses following Sequential Immunization with Different Hemagglutinins. Plos Pathogens. 2010;6 doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashem AM, et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochemical and Biophysical Research Communications. 2010;403:247–251. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 25.Ekiert DC, et al. A Highly Conserved Neutralizing Epitope on Group 2 Influenza A Viruses. Science. 2011 doi: 10.1126/science.1204839. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corti D, et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science. 2011 doi: 10.1126/science.1205669. Published online. [DOI] [PubMed] [Google Scholar]
- 27.Webster RG, Laver WG. PREPARATION AND PROPERTIES OF ANTIBODY DIRECTED SPECIFICALLY AGAINST NEURAMINIDASE OF INFLUENZA VIRUS. Journal of Immunology. 1967;99:49. [PubMed] [Google Scholar]
- 28.Monto AS, Kendal AP. EFFECT OF NEURAMINIDASE ANTIBODY ON HONG-KONG INFLUENZA. Lancet. 1973;1:623–625. doi: 10.1016/s0140-6736(73)92196-x. [DOI] [PubMed] [Google Scholar]
- 29.Nimmerjahn F, Ravetch JV. Antibody-mediated modulation of immune responses. Immunological reviews. 2010;236:265–275. doi: 10.1111/j.1600-065X.2010.00910.x. [DOI] [PubMed] [Google Scholar]
- 30.Jegerlehner A, et al. Influenza a vaccine based on the extracellular domain of M2: Weak protection mediated via antibody-dependent NK cell activity. Journal of Immunology. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 31.Mozdzanowska K, et al. Treatment of influenza virus-infected SCID mice with nonneutralizing antibodies specific for the transmembrane proteins matrix 2 and neuraminidase reduces the pulmonary virus titer but fails to clear the infection. Virology. 1999;254:138–146. doi: 10.1006/viro.1998.9534. [DOI] [PubMed] [Google Scholar]
- 32.Wang RF, et al. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Research. 2008;80:168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Yu XC, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–U541. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause JC, et al. Naturally Occurring Human Monoclonal Antibodies Neutralize both 1918 and 2009 Pandemic Influenza A (H1N1) Viruses. Journal of Virology. 2010;84:3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hancock K, et al. Cross-Reactive Antibody Responses to the 2009 Pandemic H1N1 Influenza Virus. New England Journal of Medicine. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 37.Medina RA, et al. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nature Communications. 2010;1 doi: 10.1038/ncomms1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manicassamy B, et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 2010;6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khurana S, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bommakanti G, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13701–13706. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuno Y, et al. A COMMON NEUTRALIZING EPITOPE CONSERVED BETWEEN THE HEMAGGLUTININS OF INFLUENZA-A VIRUS H1 AND H2 STRAINS. Journal of Virology. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Throsby M, et al. Heterosubtypic Neutralizing Monoclonal Antibodies Cross-Protective against H5N1 and H1N1 Recovered from Human IgM(+) Memory B Cells. Plos One. 2008;3 doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakabe S, et al. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Research. 2010;88:249–255. doi: 10.1016/j.antiviral.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grandea AG, et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao GY, et al. An H5N1 M2e-based multiple antigenic peptide vaccine confers heterosubtypic protection from lethal infection with pandemic 2009 H1N1 virus. Virology Journal. 2010;7:6. doi: 10.1186/1743-422X-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klenk HD, et al. ACTIVATION OF INFLUENZA-A VIRUSES BY TRYPSIN TREATMENT. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 48.Skehel JJ, Waterfield MD. STUDIES ON PRIMARY STRUCTURE OF INFLUENZA-VIRUS HEMAGGLUTININ. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:93–97. doi: 10.1073/pnas.72.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarowitz SG, Choppin PW. ENHANCEMENT OF INFECTIVITY OF INFLUENZA-A AND INFLUENZA-B VIRUSES BY PROTEOLYTIC CLEAVAGE OF HEMAGGLUTININ POLYPEPTIDE. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 50.Weis W, et al. STRUCTURE OF THE INFLUENZA-VIRUS HEMAGGLUTININ COMPLEXED WITH ITS RECEPTOR, SIALIC-ACID. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 51.Cross KJ, et al. Composition and Functions of the Influenza Fusion Peptide. Protein and Peptide Letters. 2009;16:766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 52.Daniels RS, et al. FUSION MUTANTS OF THE INFLUENZA-VIRUS HEMAGGLUTININ GLYCOPROTEIN. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 53.Skehel JJ, et al. CHANGES IN THE CONFORMATION OF INFLUENZA-VIRUS HEMAGGLUTININ AT THE PH OPTIMUM OF VIRUS-MEDIATED MEMBRANE-FUSION. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varghese JN, et al. STRUCTURE OF THE INFLUENZA-VIRUS GLYCOPROTEIN ANTIGEN NEURAMINIDASE AT 2.9-A-RESOLUTION. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 55.Burmeister WP, et al. THE 2.2-A RESOLUTION CRYSTAL-STRUCTURE OF INFLUENZA-B NEURAMINIDASE AND ITS COMPLEX WITH SIALIC-ACID. Embo Journal. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang IC, et al. Influenza A virus neuraminidase limits viral superinfection. Journal of Virology. 2008;82:4834–4843. doi: 10.1128/JVI.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colman PM. STRUCTURAL BASIS OF ANTIGENIC VARIATION - STUDIES OF INFLUENZA-VIRUS NEURAMINIDASE. Immunology and Cell Biology. 1992;70:209–214. doi: 10.1038/icb.1992.26. [DOI] [PubMed] [Google Scholar]