Abstract
Chaperonins are absolutely required for the folding of a subset of proteins in the cell. An earlier proteome-wide analysis of Escherichia coli chaperonin GroEL/GroES (GroE) interactors predicted obligate chaperonin substrates, which were termed Class III substrates. However, the requirement of chaperonins for in vivo folding has not been fully examined. Here, we comprehensively assessed the chaperonin requirement using a conditional GroE expression strain, and concluded that only ∼60% of Class III substrates are bona fide obligate GroE substrates in vivo. The in vivo obligate substrates, combined with the newly identified obligate substrates, were termed Class IV substrates. Class IV substrates are restricted to proteins with molecular weights that could be encapsulated in the chaperonin cavity, are enriched in alanine/glycine residues, and have a strong structural preference for aggregation-prone folds. Notably, ∼70% of the Class IV substrates appear to be metabolic enzymes, supporting a hypothetical role of GroE in enzyme evolution.
Keywords: chaperonin, GroEL, metabolomics, protein folding, proteomics
Introduction
A protein must fold into the correct tertiary structure after emerging from the ribosome (Anfinsen, 1973). In the cell, protein folding is assisted by a variety of chaperones, which are also involved in other multiple cellular processes associated with the conformational changes of proteins, such as stress responses (Hartl and Hayer-Hartl, 2009). In addition to the ‘classical' functions, chaperones are proposed to promote protein evolution by buffering the destabilization of proteins caused by harmful genetic mutations (Jenkins et al, 1986; Van Dyk et al, 1989; Rutherford and Lindquist, 1998; Fares et al, 2002; Tokuriki et al, 2009). The absolute requirement of chaperones for cellular functions and for protein evolution might account for the fact that an organism lacking chaperones has not been identified.
The bacterial chaperonin GroEL and its co-factor GroES are the only indispensable chaperones for the viability of Escherichia coli (Fayet et al, 1989; Horwich et al, 1993). GroEL consists of two heptameric rings of 57 kDa subunits, and provides binding sites for non-native substrate proteins. GroEL can bind about half of the soluble E. coli proteins in their denatured states in vitro (Viitanen et al, 1992). The co-chaperonin GroES, a dome-shaped heptameric ring of ∼10 kDa subunits, caps GroEL in the presence of adenine nucleotides, forming a central cavity that can accommodate substrate proteins up to ∼60 kDa in size (Xu et al, 1997; Sakikawa et al, 1999; Hartl and Hayer-Hartl, 2002; Fenton and Horwich, 2003). GroEL mutations that affect either the cavity environments or the encapsulation of substrates within the cavity are lethal in E. coli, indicating the in vivo indispensability of the GroEL–GroES cavity (Koike-Takeshita et al, 2006; Tang et al, 2008).
An important goal in chaperonin biology is to identify a subset of obligate GroEL/GroES (hereafter, GroE) substrates that absolutely require GroE for folding in cells. Precise identification of the obligate GroE substrates should contribute to the identification of a distinctive role for GroE among chaperones, reveal the structural features of the obligate substrates, and shed light on the role of GroE in protein evolution. (Ewalt et al, 1997; Houry et al, 1999; Kerner et al, 2005).
One approach to identifying obligate GroE substrates is a detailed analysis of the phenotypes of GroE-depleted cells. Such analyses identified DapA and FtsE as obligate GroE substrates in the cell lysis and filamentous morphology phenotypes, respectively (McLennan and Masters, 1998; Fujiwara and Taguchi, 2007). Although a detailed phenotypic analysis can precisely identify obligate GroE substrates, this approach is limited, in that the substrates can only be identified one by one, and only in the cells with experimentally tractable phenotypes.
Another approach to substrate identification is a proteome-wide analysis. Hundreds of GroEL substrates have been identified using mass spectrometry (MS) (Kerner et al, 2005; Chapman et al, 2006). In particular, Kerner et al (2005) have identified ∼250 substrates that interact with GroE in E. coli, and categorized them into three classes depending on their enrichment in the GroE complex: Class I substrates as spontaneous folders, Class II as partial GroEL-dependent substrates, and Class III as the potential obligate GroE substrates. Notably, ∼84 Class III substrates are estimated to occupy ∼80% of the available GroEL capacity in the cell. This classification was primarily based on the proteomics of the GroE interactors. However, except for DapA, GatY, MetK, ADD, and YajO, which were verified as requiring GroE for folding (Kerner et al, 2005), the in vivo GroE dependency of the Class III substrates has not been tested. In fact, our earlier phenotype analysis revealed that one of the Class III proteins, ParC, was functional even under GroE-depleted conditions (Fujiwara and Taguchi, 2007), raising the possibility that the predicted Class III proteins are not necessarily obligate substrates of GroE.
Here, we comprehensively assessed the GroE dependency of the Class III substrates in cells by proteomics, metabolomics, and individual characterization of the GroE requirement (GR), and found that around 40% of the Class III substrates lack GroE dependence. Thus, only ∼60% of the Class III proteins, combined with at least eight substrates not previously categorized as Class III substrates, were GroE dependent in E. coli. As the in vivo GroE obligate substrates were not limited to the Class III substrates assigned previously, we have named the verified in vivo obligate GroE substrates as Class IV substrates. We found that Class IV substrates have a limited range of molecular weights and isoelectric points, are aggregation prone, and are structurally distinct. The features of Class IV substrates are consistent with a possible role of GroE in facilitating the evolution of enzymes involved in metabolic reactions and pathways.
Results
Proteomics of the soluble fraction in GroE-depleted E. coli
In GroE-depleted cells, the known obligate GroE substrates either aggregate (e.g. MetK) or are degraded (e.g. DapA, FtsE, and GatY) (McLennan and Masters, 1998; Kerner et al, 2005; Fujiwara and Taguchi, 2007). Thus, we expected that the abundance of other potential GroE obligate substrates would also be reduced in the soluble fraction of GroE-depleted cells. A proteome-wide analysis of the soluble fraction of GroE-depleted cells was therefore conducted to find candidate in vivo obligate GroE substrates. We used a conditional GroE expression strain, MGM100, in which the GroE promoter had been replaced by the arabinose-inducible (and glucose-inhibitable) BAD promoter (Figure 1A; McLennan and Masters, 1998). For proteomics, cells were subjected to a 2-h depletion of GroE in LB medium, during which the level of GroEL was reduced to <10% of that in undepleted cells (Supplementary Figure S1) (McLennan and Masters, 1998), and, as a control, cells with a normal level of GroE were also prepared. Note that diaminopimelic acid (DAP) was added to the medium to prevent cell lysis, a known consequence of DapA (a Class III substrate) deficiency in GroE-depleted cells (McLennan and Masters, 1998). The abundance of each protein was quantified by the exponentially modified Protein Abundance Index (emPAI), which provides an estimate of protein abundance by quantitating non-redundant peptides identified by MS (Ishihama et al, 2005, 2008). As a control for the sugar-associated changes in the proteome, MG1655, the wild-type parent strain of MGM100, was also examined by the same procedures.
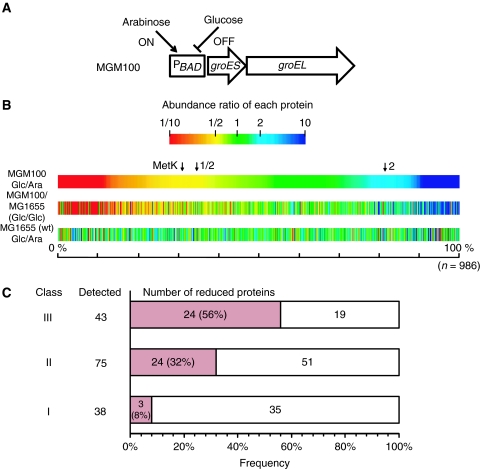
Figure 1.
Proteomes of GroE-normal and -depleted cells. (A) A schematic representation of the groE region of the E. coli strain (MGM100) used in this study. The chromosomal groE promoter was replaced by the BAD promoter, which is induced by arabinose and inhibited by glucose (McLennan and Masters, 1998). (B) Overview of the comparative proteomics of soluble lysates from cells grown with arabinose or with glucose. The abundance ratios were estimated by the emPAI values, which correlate well with the protein concentration (Ishihama et al, 2005, 2008). A total of 986 proteins were detected in MGM100 grown in arabinose (Ara) or glucose (Glc). Proteins were aligned by sorting the abundance ratios of the proteins (emPAIGlc/emPAIAra) in MGM100 cells in all colour maps. Each protein was coloured according to the abundance ratios of proteins in (MGM100 Glc/Ara, top), (MGM100 Glc/MGM1655 Glc, middle) and (MGM1655 Glc/Ara, bottom). Blue, proteins for which the abundance in the soluble fraction was higher with glucose; Green, proteins for which the abundance in the soluble fraction was not affected by the sugar; Red, proteins for which the abundance in the soluble fraction was higher with arabinose. Both 1/2 and 2 indicate emPAIGlc/emPAIAra. For reference, the position of a known in vivo obligate GroE substrate, MetK, is indicated by the arrow. (C) Numbers of proteins significantly reduced in GroE-depleted cells (red bars) in each GroEL substrate class. The proteins for which both emPAIGlc of MG1655 and emPAIAra of MGM100 were two-fold higher than emPAIGlc of MGM100, were defined as significantly reduced proteins. 56% of Class III substrates, 32% of Class II substrates, and 8% of Class I substrates were significantly reduced in GroE-depleted cells.
The proteomics quantified a total of 986 proteins in MGM100 cells under glucose and arabinose conditions. We then calculated the relative abundances of proteins defined as emPAIglucose/emPAIarabinose, and summarized the results in a colour map, in which the colours represent the reduction (red) or increase (blue) of protein production during glucose growth (Figure 1B). The map shows that ∼33% of proteins were reduced by >50% in MGM100 cells (Figure 1B, top). The tendency for the reduction was also obvious when we compared the emPAI values in MGM100 with those in MG1655 under glucose conditions (Figure 1B, middle). In addition, this reduction was not observed during glucose growth in MG1655 cells (Figure 1B, bottom). Collectively, the drastic reduction of many proteins during glucose growth in MGM100 cells was caused by the GroE depletion. We note that expression levels of a significant number of proteins were increased in the GroE-depleted cells (coloured blue), including methionine biosynthetic enzymes, such as MetE (Horwich et al, 1993; Chapman et al, 2006; Masters et al, 2009), and certain chaperones, such as DnaK and SecB (Supplementary Figure S2), possibly reflecting a stress response induced by GroE depletion. As the levels of chaperones other than GroE were not reduced, the changes we observed can be attributed to the reduced amount of GroE with confidence.
The proteome data were used to roughly choose candidate GroE substrates by the following criteria. First, the proteins with soluble abundance that was reduced during depletion to <50% of that found during arabinose growth in MGM100 (as a genetic control) were chosen. The 347 proteins chosen by the first criterion contained many false positives due to a sugar-associated reduction in their levels, and thus were filtered by a second criterion, in which the proteins with expression in MGM100 during glucose growth reduced to <50% of that found during glucose growth in MG1655 were chosen. The cutoff value of 50% was set to minimize false negatives, as the highest solubility of known in vivo obligate GroE substrates was 46%, as found with MetK in both cases. Using the genetic and sugar controls, 252 proteins among the detected 986 proteins met both criteria for rough candidate GroE substrates. The candidates included all of the in vivo tested obligate GroE substrates (MetK, GatY, and DapA), except for FtsE, which was not quantified in the proteomics, confirming the reasonableness of the selected threshold for the growth conditions used here. Then, the percentages of the candidates showing protein reductions in each of the GroEL substrate classes defined by Kerner et al (2005) were calculated. As shown in Figure 1C, 8% of Class I, 32% of Class II, and 56% of Class III substrates were reduced in the GroE-depleted cells. The fraction of class members showing reduced protein amounts increased with the degree of GroEL dependence. It is also noteworthy that about 44% of the Class III substrates (24 out of the 43 quantified proteins) did not meet our criteria for GroE obligate substrates. This again suggests that a significant fraction of Class III members, in addition to ParC, are not obligate substrates in vivo.
About 40% of Class III substrates do not require GroE for solubility
To assess whether ∼40% of the Class III substrates are not actually obligate GroE substrates, we developed methods, independent of proteomics, to verify their GR for solubility. The methods also aimed to comprehensively cover all of the Class III proteins suggested by Kerner et al (2005), as our proteomics detected only about half of the Class III substrates (43 of 84). We induced the expression of individual target proteins from a tac promoter in MGM100 cells after a 2-h depletion of GroE, and measured their total amounts and the proportion in the soluble fraction. The obligate GroE substrates would be expected to become insoluble or be degraded. To validate the strategy, we examined proteins for which the status of GroE dependence had already been verified: Enolase (spontaneously folding in vitro, Class I), GatD (partial GroE-dependent folding in vitro, Class II), MetK, FtsE, DapA (the in vivo obligate GroE substrates, Class III), and ParC (assigned as Class III, but functional in the GroE-depleted cells) (McLennan and Masters, 1998; Kerner et al, 2005; Fujiwara and Taguchi, 2007). Enolase, GatD, and ParC were soluble irrespective of the GroE level, whereas MetF, MetK, FtsE, and DapA aggregated in the GroE-depleted cells (Figure 2A). The disappearance or persistence of the bands under GroE-depleted conditions was almost complete, enabling easy and clear discrimination. Note that DapA was degraded in the earlier report (Kerner et al, 2005), probably reflecting a difference in the expression levels. Except for the difference of whether DapA was degraded or aggregated, the present results are consistent with the earlier data on the GroE dependency, suggesting that the overexpression strategy reflects the in vivo GroE dependency.
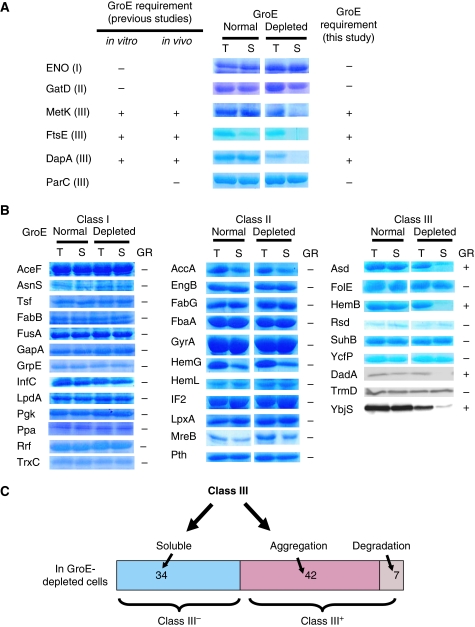
Figure 2.
Solubility of E. coli GroE substrates overexpressed in GroE-normal and -depleted cells. (A) Verification of the method. Solubilities of the proteins, for which the GR had been shown in earlier studies, after overexpression in GroE-normal and -depleted cells. Only the corresponding bands are shown. T, total lysates; S, soluble fractions; GroE-normal, cells cultivated with 0.2% arabinose; GroE-depleted, cells cultivated with 0.2% glucose. I, II, and III in the parentheses indicate the GroE substrate classes. The GroE dependency of ENO, GatD, MetK, and DapA in vitro and in vivo was shown earlier (McLennan and Masters, 1998; Kerner et al, 2005). FtsE in vitro and in vivo, and ParC in vivo were shown in our earlier studies (Fujiwara and Taguchi, 2007). (B) Solubility of substrates essential for the viability of E. coli after overexpression in GroE-normal and -depleted cells. All of the proteins, except for DadA, TrmD, and YbjS, were stained and visualized by Coomassie Brilliant Blue. DadA, TrmD, and YbjS were each fused to an HA tag at the C-terminus and were detected by immunoblotting. GR, GroE requirement. (C) Class III substrates were divided depending on the GroE dependency. Class III substrates with and without GroE dependency were termed Class III+ and Class III–, respectively.
Next, we extended the method to all of the essential genes in the three GroEL substrate classes, to test the GR for solubility in cells. The solubility of the essential Class I and Class II proteins (proteins with low expression levels were not measured) was independent of the GroE levels (Figure 2B), confirming that Classes I and II were not dependent on GroE for folding. The GroE-independence of Ppa (Class I), GatD, LpxA, HemL, and FabG (Class II), which were candidates of GroE substrates identified by our proteomics, indicated that not all of the candidates predicted by the proteomics are in vivo obligate GroE substrates. For the Class III substrates, the essential proteins with low expression (DadA, TrmD, and YbjS) were fused with an HA tag sequence at the C-terminus to facilitate western blotting. Seven proteins (MetK, FtsE, and DapA (Figure 2A), Asd, HemB, DadA, and YbjS (Figure 2B)) were found to be aggregated in the GroE-depleted cells. In contrast, six other essential Class III proteins (ParC, FolE, Rsd, SuhB, YcfP, and TrmD) were soluble even after GroE depletion (Figure 2A and B). Taken together, the results showed that the in vivo obligate GroE substrates were enriched in Class III, but not in Class I and II proteins. More importantly, the results also indicated that approximately half of the Class III proteins did not require GroE for solubility, as already suggested by the above proteomics data. Depending on the in vivo GR for solubility, we divided the Kerner's Class III substrates into Class III+ (plus; GroE dependent for solubility in vivo) and Class III− (minus; not GroE dependent for solubility in vivo). Finally, we tested all of the remaining Class III substrates, including the proteins not detected in our proteomics survey, except for the plasmid origin protein Ypt1, for an in vivo GR for solubility. A total of 59 Class III proteins were tested by an overexpression-Coomassie staining method (Supplementary Figure S3) or the HA tag fusion expression-immunoblotting method (Supplementary Figure S4A). In some cases, the proteins that were expressed in the GroE-normal cells had entirely disappeared from the GroE-depleted cells. As the disappearance after GroE depletion of obligate substrates, such as DapA, GatY, and FtsE, had previously been reported, we regarded the seven Class III proteins that disappeared in the depleted cells as Class III+ substrates. As the overexpression of some proteins resulted in aggregate formation even in the GroE-normal cells, we reduced their expression to that which ‘leaked' from the tac promoter in the absence of inducer (Supplementary Figure S4B and C). After confirmation of the method using the several substrates validated above (Supplementary Figure S4B), 11 HA tag-fused Class III proteins (Supplementary Figure S4C) were tested. When all of the solubility assays for the Class III substrates were combined (Figure 2A and B; Supplementary Figures S3 and S4), Class III was divided into 49 Class III+ (7 in Figure 2; 20 in Supplementary Figure S3; 22 in Supplementary Figure S4) and 34 Class III− substrates (Figure 2C).
When measured by proteomics data, we found that all of the Class III+ substrates (except for AraA and Nfo) were specifically reduced in the GroE-depleted cells (Supplementary Figure S5). As the proteomics method did not involve overexpression, we conclude that the aggregation found in the GroE-depleted cells in which overexpression was used was not merely the consequence of the overexpression of the individual candidate substrates.
Regarding the Class III− proteins, ∼80% of the Class III− members quantified in the proteomics were not reduced. However, the amounts of the remaining proteins, such as SuhB, TrmA, and YcfP, were specifically reduced, prompting us to test whether the Class III− members are physiologically functional when synthesized in the absence of GroE.
Activities of Class III− proteins in GroE-depleted cells
In contrast to the Class III+ proteins, the Class III− proteins not only retained their solubility in the absence of GroE, but the amounts of 2/3 of them remained unchanged, as assessed by proteomic measurements. As the ultimate test of GroE dependence is whether it is required for functionality, we tested whether the Class III− enzymes that retained their solubility were also functional.
Our first test used metabolomics, using a capillary electrophoresis (CE)-MS system (Ishii et al, 2007). We quantified the concentrations of 187 metabolites, and found that 9 metabolites were significantly increased and 20 were reduced when the GroE was depleted (Supplementary Table S1). Dysfunction of an enzyme should cause an accumulation of its precursor and/or a reduction in the product of the catalysed reaction (Supplementary Figure S6A). Indeed, 5-aminolevulinate and N-acetylornithine, the precursors of the reactions catalysed by the Class III+ proteins HemB and ArgE, respectively, accumulated in GroE-depleted cells (Supplementary Figure S6B; Supplementary Table S1). Likewise, S-adenosylmethionine and N-acetylglucosamine, the products of the Class III+ enzymes MetK and NagZ, respectively, were decreased. Thus, the metabolomics data support the dysfunction of several Class III+ substrates (Supplementary Figure S6C; Supplementary Table S1).
For Class III−, we found that the intracellular thymidine concentration was not decreased in the GroE-depleted cells (Figure 3A), implying that FolE, one of the essential Class III− proteins, is functional in the cells, as FolE-defective cells only grow in thymidine-supplemented rich medium (El Yacoubi et al, 2006).
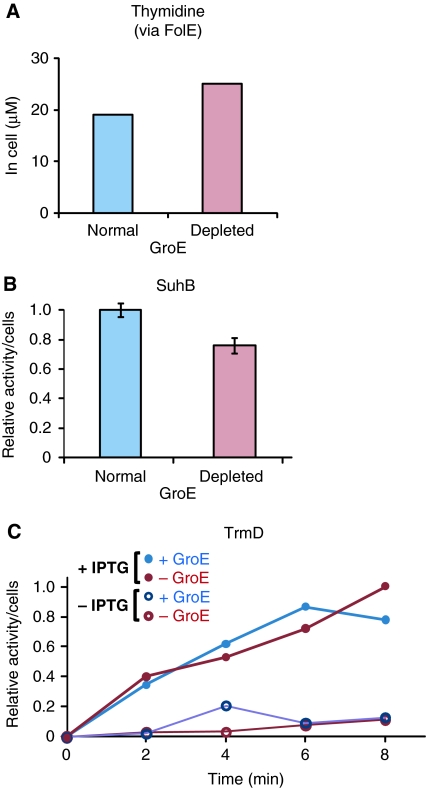
Figure 3.
Functions of essential Class III− proteins in GroE-depleted cells. (A) The concentration of thymidine in GroE-normal and -depleted cells. Thymidine is known to be reduced in FolE-depleted cells. (B) Inositol monophosphatase activity of SuhB in cell lysates of the SuhB-overexpressing GroE-normal and -depleted cells. Relative activities were determined in the cell lysates under each GroE condition, in the presence or absence of 1 mM IPTG. (C) Time course of tRNA(Leu) methylase activity of TrmD in GroE-normal (+GroE) and -depleted (−GroE) cell lysates. + IPTG: TrmD was induced by 1 mM IPTG; -IPTG: cells without IPTG treatment.
Second, the enzymatic activities of the Class III− proteins were directly assayed in the E. coli lysates. The activities of two essential Class III− proteins, an inositol monophosphatase, SuhB, and a tRNA methylase, TrmD, were measured in the lysates after the overexpression of Class III− proteins. The enzymes were active in both the GroE-depleted and -normal cells (Figure 3B and C), indicating that the enzymes are both soluble and functional in the GroE-depleted cells.
Among the Class III− substrates, only four proteins (ParC, FolE, SuhB, and TrmD) are essential. Although the functionality of the remaining Class III− proteins was not tested, we could show that at least all of the essential Class III− proteins were physiologically functional even in the GroE-depleted cells, further supporting the validity of the Class III+ and III− grouping.
Identification of other in vivo obligate GroE substrates that were not previously assigned as Class III substrates
After the complete survey of the Class III substrates, we searched for other novel GroE obligate substrates besides the identified Class III proteins. The metabolomics data showed that the level of O-phosphoserine, the product of a Class II substrate, SerC, was reduced in the GroE-depleted cells, suggesting that SerC was not active in the cells (Supplementary Figure S6C; Supplementary Table S1). In addition, the proteomics data also suggested that SerC was reduced in the supernatant of the GroE-depleted cells (Figure 4A). We then tested the solubility of SerC by the overexpression method, and found that it was aggregated in the GroE-depleted cells (Figure 4B), strongly suggesting that the in vivo obligate GroE substrates are not confined to the identified Class III substrates.
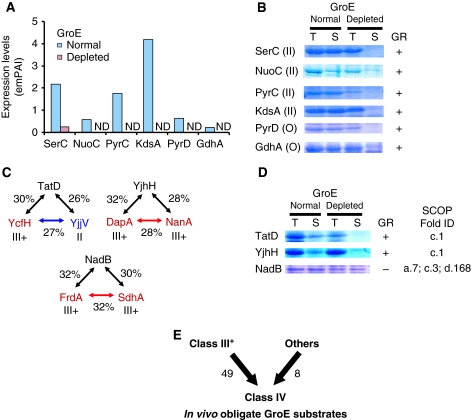
Figure 4.
Obligate GroE substrates that were not derived from Class III substrates. (A) Expression levels of Class II substrates and proteins that were not identified as GroEL interactors, but were significantly reduced in GroE-depleted cells. Expression levels were determined by emPAI (Ishihama et al, 2005). ND, not detected. (B) Solubility of the proteins referred to in (A) in GroE-normal and -depleted cells. The number in parentheses indicates the GroE substrate class of the protein. ‘O' indicates that the protein had not appeared among the GroEL interactors. GR, GroE requirement. (C) Homologs of Class III and II proteins. Percents indicate the amino-acid sequence identities between the indicated proteins pairs. Class IV (red), III− (blue), and all E. coli cytosolic proteins (green). (D) Solubility of the homologs of Class III+ substrates in GroE-normal and -depleted cells. (E) Class IV substrates defined as the in vivo obligate GroE substrates. In total, 49 Class III+ substrates, combined with eight newly identified obligate substrates, were classified as Class IV substrates.
Other putative GroE substrates were also identified. First, using the proteomics data, we verified the GR for a dozen drastically reduced proteins in the GroE-depleted cells. A severe cutoff value, ∼12% of solubility, was introduced to choose the candidates, as the solubility of SerC was 12%. These included three Class II proteins (KdsA, PyrC, and NuoC) and six proteins that had not appeared among the GroEL interactors (GuaC, ThiL, SdaB, PyrD, NemA, and GdhA). The solubility assays of these overexpressed candidate proteins revealed that all of the Class II candidates and two of the other six candidates (PyrD and GdhA) behaved as in vivo GroE obligate substrates (Figure 4B; Supplementary Figure S7).
We also identified the homologs of Class III+ substrates on a database, and evaluated the GR for their solubility in GroE-depleted cells. Several Class III+ proteins share homology (Figure 4C). These proteins include two pairs, DapA and NanA (28% identity), and FrdA and SdhA (32% identity). YjhH (a homolog of DapA and NanA), NadB (a homolog of FrdA and SdhA), and TatD (a homolog of YcfH and a Class II protein YjjV) were examined by the solubility assay. Among these proteins, TatD and YjhH required GroE for solubility, whereas NadB did not (Figure 4D).
Taken together, we found eight new in vivo GroE obligate substrates not previously identified as Class III proteins. We have combined these eight in vivo substrates with the 49 Class III+ members, and now suggest their classification together to form a new group, the Class IV substrates, for which folding is obligatorily dependent on GroE in vivo (Figure 4E; Table 1).
Table 1. Class IV substrates.
| Gene | b num | ESa | Solb | MWc | A/Gd | pI | Foldse | Function |
|---|---|---|---|---|---|---|---|---|
| Metabolic reactions | ||||||||
| yqaB | b2690 | 0 | 13% | 20 757 | 20.2% | 5.5 | c.108 | Fructose-1-phosphatase |
| rfbC | b2038 | 0 | ND | 21 246 | 11.4% | 5.5 | b.82 | dTDP-4-deoxyrhamnose-3,5-epimerase |
| acpH | b0404 | 0 | 14% | 22 938 | 9.8% | 5.9 | Acyl carrier protein phosphodiesterase | |
| serCf | b0907 | 0 | 17% | 28 177 | 19.3% | 5.4 | c.67 | 3-Phosphoserine/phosphohydroxythreonine aminotransferase |
| gatY | b2096 | 0 | 11% | 30 782 | 19.0% | 5.9 | c.1 | D-tagatose 1,6-bisphosphate aldolase 2, catalytic subunit |
| dapA | b2478 | 1 | ND | 31 238 | 18.5% | 6.0 | c.1 | Dihydrodipicolinate synthase |
| nanA | b3225 | 0 | 16% | 32 556 | 17.8% | 5.6 | c.1 | N-acetylneuraminate lyase |
| metF | b3941 | 0 | 26% | 33 068 | 15.5% | 6.0 | c.1 | 5,10-Methylenetetrahydrofolate reductase |
| dusC | b2140 | 0 | 10% | 35 162 | 16.2% | 6.1 | c.1 | tRNA-dihydrouridine synthase C |
| hemB | b0369 | 1 | 7% | 35 580 | 20.1% | 5.3 | c.1 | Porphobilinogen synthase |
| dusB | b3260 | 0 | 16% | 35 830 | 17.8% | 6.3 | c.1 | tRNA-dihydrouridine synthase B |
| lipA | b0628 | 0 | 9% | 36 043 | 15.3% | 8.1 | Lipoate synthase | |
| add | b1623 | 0 | 15% | 36 355 | 20.4% | 5.4 | c.1 | Adenosine deaminase |
| yajO | b0419 | 0 | 5% | 36 374 | 15.7% | 5.2 | c.1 | 2-Carboxybenzaldehyde reductase |
| ltaE | b0870 | 0 | 10% | 36 455 | 21.6% | 5.8 | c.67 | L-allo-threonine aldolase, PLP dependent |
| pyrDg | b0945 | 0 | 13% | 36 775 | 17.0% | 7.7 | c.1 | Dihydro-orotate oxidase, FMN linked |
| nagZ | b1107 | 0 | 12% | 37 556 | 19.6% | 5.9 | c.1 | β-N-acetyl-glucosaminidase |
| fbaB | b2097 | 0 | 5% | 38 071 | 20.3% | 6.2 | c.1 | Fructose-bisphosphate aldolase Class I |
| kdsAf | b1215 | 1 | 30% | 38 808 | 18.0% | 6.3 | c.1 | 3-Deoxy-D-manno-octulosonate 8-phosphate synthase |
| pyrCf | b1062 | 0 | 4% | 38 817 | 14.9% | 5.8 | c.1 | Dihydro-orotase |
| dadX | b1190 | 0 | 3% | 38 842 | 21.1% | 6.6 | c.1; b.49 | Alanine racemase 2, PLP binding |
| asd | b3433 | 1 | 19% | 39 970 | 17.7% | 5.4 | c.2; d.81 | Aspartate-semialdehyde dehydrogenase, NAD(P) binding |
| fadA | b3845 | 0 | 10% | 40 872 | 24.3% | 6.3 | c.95 | 3-Ketoacyl-CoA thiolase (thiolase I) |
| bioF | b0776 | 0 | 3% | 41 557 | 22.1% | 6.6 | c.67 | 8-Amino-7-oxononanoate synthase |
| metK | b2942 | 1 | 48% | 41 898 | 18.8% | 5.1 | d.130 | Methionine adenosyltransferase 1 |
| argE | b3957 | 0 | 42% | 42 301 | 15.9% | 5.5 | d.58; c.56 | Acetylornithine deacetylase |
| lldD | b3605 | 0 | 4% | 42 683 | 22.7% | 6.3 | c.1 | L-lactate dehydrogenase, FMN linked |
| fabF | b1095 | 0 | 8% | 42 999 | 25.2% | 5.7 | c.95 | 3-Oxoacyl-[acyl-carrier-protein] synthase II |
| thiH | b3990 | 0 | ND | 43 279 | 14.9% | 6.6 | Thiamine biosynthesis ThiGH complex subunit | |
| csdB | b1680 | 0 | 11% | 44 390 | 19.0% | 5.9 | c.67 | Selenocysteine lyase, PLP dependent |
| rspA | b1581 | 0 | 4% | 45 919 | 16.3% | 5.7 | d.54; c.1 | Mannonate/altronate dehydratase |
| deoA | b4382 | 0 | 13% | 47 148 | 21.6% | 5.2 | d.41; a.46; c.27 | Thymidine phosphorylase |
| dadA | b1189 | 0 | 12% | 47 558 | 19.2% | 6.2 | c.5; d.16; c.3; c.4; c.2 | D-amino-acid dehydrogenase |
| gdhAg | b1761 | 0 | 22% | 48 530 | 21.5% | 6.0 | c.2; c.58 | Glutamate dehydrogenase |
| eutB | b2441 | 0 | 13% | 49 334 | 18.3% | 4.8 | a.105; c.1 | Ethanolamine ammonia-lyase, large subunit, heavy chain |
| xylA | b3565 | 0 | ND | 49 691 | 18.6% | 5.8 | c.1 | D-xylose isomerase |
| uxaC | b3092 | 0 | 8% | 53 925 | 14.7% | 5.4 | c.1 | Uronate isomerase |
| araA | b0062 | — | 8% | 56 021 | 16.4% | 6.1 | c.118; b.71 | L-arabinose isomerase |
| aldB | b3588 | 0 | ND | 56 306 | 19.1% | 5.4 | c.82 | Aldehyde dehydrogenase |
| sdhA | b0723 | 0 | 10% | 64 355 | 19.6% | 5.9 | a.7; c.3; d.168 | Succinate dehydrogenase, flavoprotein subunit |
| frdA | b4154 | 0 | 51% | 65 904 | 21.1% | 5.9 | a.7; c.3; d.168 | Fumarate reductase (anaerobic) catalytic and NAD/ flavoprotein subunit |
| nuoCf | b2286 | 0 | 18% | 68 683 | 13.5% | 6.0 | e.18 | NADH: ubiquinone oxidoreductase, chain C, D |
| Other processes | ||||||||
| ftsE | b3463 | 1 | 12% | 24425 | 18.5% | 9.4 | c.37 | Predicted transporter subunit: ATP-binding component of ABC superfamily |
| fucR | b2805 | 0 | 36% | 27342 | 13.6% | 7.8 | a.4; c.35 | DNA-binding transcriptional activator |
| tatDg | b4483 | 0 | ND | 28961 | 17.7% | 5.2 | c.1 | DNase, magnesium dependent |
| nfo | b2159 | 0 | 14% | 31444 | 20.4% | 5.4 | c.1 | Endonuclease IV with intrinsic 3′-5′ exonuclease activity |
| argP | b2916 | 0 | 17% | 33438 | 13.1% | 6.4 | c.94; a.4 | DNA-binding transcriptional activator, replication initiation inhibitor |
| tldE | b4235 | 0 | 16% | 48313 | 20.0% | 5.4 | Protease involved in Microcin B17 maturation and in sensitivity to the DNA gyrase inhibitor LetD | |
| pepQ | b3847 | 0 | 15% | 50122 | 15.3% | 5.6 | d.127 | Proline dipeptidase |
| tldD | b3244 | 0 | 91% | 51295 | 20.6% | 4.9 | Protease involved in Microcin B17 maturation and in sensitivity to the DNA gyrase inhibitor LetD | |
| Unknown | ||||||||
| ycfH | b1100 | 0 | ND | 29 772 | 14.7% | 5.2 | c.1 | Predicted metallodependent hydrolase |
| yafD | b0209 | 0 | 10% | 29 972 | 14.7% | 9.6 | d.151 | Conserved protein |
| ybjS | b0868 | 0 | 7% | 38 089 | 15.1% | 8.8 | c.2 | Predicted NAD(P)H-binding oxidoreductase |
| yneB | b1517 | 0 | 34% | 31 859 | 18.6% | 6.1 | c.1 | Predicted aldolase |
| yjhHg | b4298 | 0 | 8% | 32 714 | 16.9% | 5.3 | c.1 | Predicted lyase/synthase |
| yjjU | b4377 | 0 | 7% | 39 794 | 15.7% | 8.7 | Predicted esterase | |
| yfbQ | b2290 | 0 | 21% | 45 468 | 14.6% | 5.9 | c.67 | Predicted aminotransferase |
| ND, not determined as in Niwa et al (2009). | ||||||||
| aES, essentiality of the gene (Baba et al, 2006). 1 and 0 indicate essential and nonessential, respectively. | ||||||||
| bSol, solubility in translation without any chaperone (Niwa et al, 2009). | ||||||||
| cMW, molecular weight (Da). | ||||||||
| dA/G, content of alanine and glycine. | ||||||||
| eSCOP-fold ID of the proteins. | ||||||||
| fPreviously classified as Class II substrates in Kerner et al (2005). | ||||||||
| gNot identified as GroE interactors as in Kerner et al (2005). | ||||||||
GroE dependency of in vitro translated Class III− and newly identified Class IV proteins
To elucidate the in vitro GroE dependency of the newly identified substrates, a Class III− protein (FolE) and several Class IV proteins (DapA as a known obligate substrate, and SerC and KdsA as newly identified Class IV substrates) were translated by a reconstituted in vitro cell-free translational system, PURE system (Shimizu et al, 2001; Ying et al, 2005; Niwa et al, 2009), which does not contain any chaperones. The requirements of the DnaK (DnaK, DnaJ, and GrpE) and GroE (GroEL and GroES) chaperone systems on the folding were monitored by the solubility and the appearance of folded structures, defined as a sharp band in native PAGE. As shown in Supplementary Figure S8, FolE (Class III−) was soluble and formed a folded structure even in the absence of chaperones. In contrast, all of the Class IV proteins tested (DapA, SerC, and KdsA) were aggregation prone without chaperones. The addition of the DnaK system increased the solubilities of the Class IV proteins to a greater or lesser extent, but the folded structures were not detected in native PAGE, implying that the soluble but unfolded structures in the presence of DnaK might be easily degraded in vivo. The Class IV proteins were soluble and formed folded structures only in the presence of GroE. The in vitro folding assay further confirmed our conclusion, in which the Class IV substrates, including the substrates that were not originally assigned as Class III (SerC and KdsA), stringently require GroE for correct folding.
Amino-acid sequence features of Class IV and III− proteins
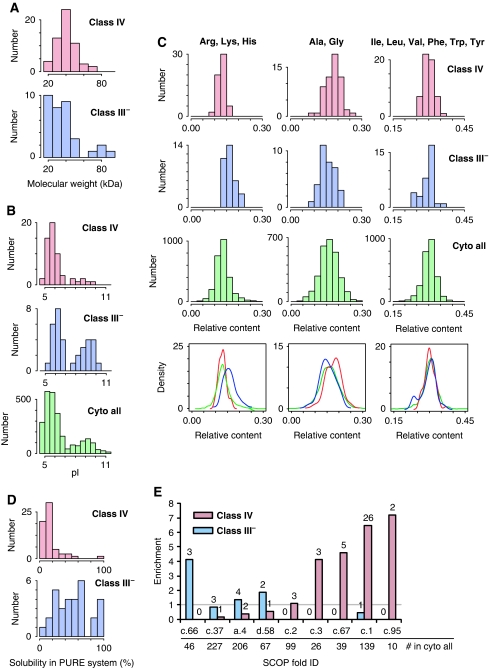
To define the features that are correlated with in vivo GroE dependency, we compared the physicochemical properties of the Class IV and III− proteins. First, the molecular weights of the Class IV substrates were distributed normally, with a peak around 40 kDa, and ranging from 21 to 68 kDa (Figure 5A), whereas the molecular weights of the 34 Class III− proteins ranged broadly, including five proteins smaller than 20 kDa and four proteins larger than 70 kDa, including ParC (84 kDa).
Figure 5.
Physicochemical features of Class IV and III− proteins. (A) Histograms of molecular weights of Class IV and III− proteins. (B) Histograms of isoelectric points (pI) of Class III–, IV, and all cytosolic E. coli proteins. (C) Histograms (above) and Kernel-type density maps (bottom) of amino-acid compositions in Class IV, III−, and all cytosolic E. coli proteins. In the Kernel-type density maps, red, blue, and green lines indicate Class IV, III−, and all cytosolic proteins in E. coli, respectively. (D) Aggregation propensities. Histograms of inherent protein solubility for Class IV and III− proteins, determined by a global aggregation analysis after chaperone-free translation (Niwa et al, 2009). (E) Enrichment of structural folds (SCOP classification) in Classes III− and IV. Enrichment factors are defined as the relative frequency of the fold in Class IV or III− per that in all E. coli cytosolic proteins. Numbers indicate the frequency of the fold in Class III− or Class IV substrates. The grey line indicates an enrichment factor of 1. c.66, S-adenosyl-L-methionine-dependent methyltransferases; c.37, P-loop containing nucleoside triphosphate hydrolases; a.4, DNA/RNA-binding three-helical bundle; d.58, Ferredoxin like; c.2, NAD(P)-binding Rossmann-fold domains; c.3, FAD/NAD(P)-binding domain; c.67, PLP-dependent transferase like; c.1, TIM β/α-barrel; c.95, thiolase like.
Second, the pI distribution and the hydrophobicity (Kyte and Doolittle, 1982) of Classes IV and III− were compared. The pI values of the Class IV substrates were distributed with a single peak around pI 5.8 (Figure 5B), whereas the pI distribution of the Class III− members was bimodal and similar to that of all cytosolic proteins of E. coli, although the acidic peak of Class III− was slightly higher (Figure 5B). The hydrophobicity distribution of the Class IV substrates was similar to that of all cytosolic proteins, whereas the Class III− proteins had lower hydrophobicity than either Class IV or all cytosolic proteins (Supplementary Figure S9A).
Third, we analysed the amino-acid compositions of the Class IV and III− proteins. As expected from the differences in the pI distributions and hydrophobicity, the amino-acid compositions also differed between Classes IV and III−. Specifically, positively charged amino acids (arginine, lysine, and histidine) were enriched among the Class III− members (Figure 5C). Among the Class IV substrates, alanine and glycine (Ala/Gly) were weakly but significantly enriched, whereas the enrichment of positively charged amino acids was not observed (Figure 5C). On average, the Ala/Gly enrichment corresponded to six additional alanine or glycine residues in a protein with 300 amino acids. Neither hydrophobic amino acids (phenylalanine, tyrosine, tryptophan, isoleucine, leucine, and valine) nor other amino acids (negative, polar, neutral, and sulphur-containing amino acids) were enriched in either Class IV or III− (Figure 5C; Supplementary Figure S10).
Finally, we analysed the sequences of the GroE substrates with FoldIndex, a tool to predict intrinsically unfolded proteins (Prilusky et al, 2005), as earlier studies suggested that this index provides a folding propensity that significantly differentiates GroEL-interacting proteins from the E. coli proteome (Noivirt-Brik et al, 2007). However, the FoldIndex distribution of the Class IV substrates was almost the same as that of the E. coli cytosolic proteins (Supplementary Figure S9B), suggesting that FoldIndex is not correlated with the in vivo GroEL/ES dependency. Nevertheless, we note that the FoldIndex distribution of Class III− was lower than that of the E. coli cytosolic proteins (Supplementary Figure S9B), suggesting that FoldIndex might predict the preferential binding of proteins to GroE in cells.
Class IV substrates are inherently aggregation prone
We recently conducted a comprehensive aggregation analysis of all E. coli proteins, using an in vitro reconstituted translation system (Niwa et al, 2009). As the reconstituted translation system is chaperone free, the inherent aggregation propensities of thousands of proteins were elucidated. The histograms of the inherent solubilities of Classes IV and III− indicated a striking difference (Figure 5D). The Class IV substrates were inherently highly aggregation prone, whereas the Class III− proteins were broadly distributed, from soluble to aggregation prone.
Structural features of Class IV substrates
Earlier work revealed that TIM-barrel folds were substantially enriched in Class III proteins (Kerner et al, 2005). Astonishingly, all of the TIM-barrel folds identified in the Class III proteins, except one (GatZ), were within the Class IV substrates (Table 1). As a result, the TIM-barrel folds were further enriched in the Class IV substrates, with 25 out of 57 Class IV substrates identified in this study possessing one (Table 1). This enrichment further supports the notion that the TIM-barrel fold is correlated with GroE dependency (Kerner et al, 2005; Masters et al, 2009). Not only TIM-barrel folds (c.1 in SCOP database terminology; Murzin et al, 1995), but also FAD/NAD(P)-binding domains (c.3), PLP-dependent transferase-like folds (c.67), and thiolase folds (c.95) were highly enriched in the Class IV substrates, as compared with the frequency of their appearance in all cytosolic proteins (Figure 5E).
Recently, Masters et al (2009) pointed out that the aldolase superfamily (c.1.10) subclass of TIM-barrel folds is preferred among Class III proteins. Our own analysis of the superfamilies of TIM folds concurs with this; we found the aldolase and metallo-dependent hydrolase (c.1.9) superfamily folds at higher frequencies than the others, within the Class IV members (Supplementary Figure S11).
Class IV homologs in a GroE-lacking organism did not exhibit GroE dependency in E. coli
The genome of Ureaplasma urealyticum lacks the groELS gene (Glass et al, 2000). Therefore, we tested whether the Class IV homologs in Ureaplasma are GroE dependent. Five homologs of Class IV members (UuDeoA, UuCsdB, UuGatY, UuMetK, and UuYcfH) were found in Ureaplasma, by BLAST searching at a threshold E value of 1 × e−35. The FoldIndex (Prilusky et al, 2005) did not detect significant differences between the five homologs in Ureaplasma and the corresponding Class IV proteins. The genes encoding UuDeoA, UuMetK, and UuYcfH were cloned and overexpressed in MG1655, and their solubilities were assessed. Strikingly, all of the Class IV homologs tested were soluble, even in the GroE-depleted cells (Supplementary Figure S12A), indicating that the GroE dependency was not conserved among the homologs. In addition, the S-adenosylmethionine synthase activity of UuMetK in the lysate of the GroE-depleted cells was comparable to that of the GroE-normal cells (Supplementary Figure S12B). Moreover, we noticed that the leaky UuMetK expression suppressed the overexpression of MetE (Supplementary Figure S12C), which is one of the hallmarks of GroE-depleted cells (Chapman et al, 2006; Masters et al, 2009). Collectively, the UuMetK data show that UuMetK is active in the GroE-depleted cells. These are the first demonstrations, as far as we know, that the orthologs of the GroE obligate substrates in a chaperonin-defective organism do not require GroE for correct folding in E. coli.
Intriguingly, the amino-acid compositions of the Ureaplasma Class IV homologs revealed that the Ala/Gly fractions in all of the homologs were lower than those in their E. coli counterparts (Table 2), whereas the contents of other amino-acid groups, including aromatic, hydrophobic, and positive amino acids, were indistinguishable from those in the E. coli counterparts (data not shown). The amino-acid content analysis again raises the possibility that a high Ala/Gly content might be involved in GroE dependency.
Table 2. Ureaplasma urealyticum orthologs of Class IV substrates.
| Protein | Organism | IDa | MWb | pI | A/Gc | Foldsd | GRe |
|---|---|---|---|---|---|---|---|
| DeoA | E. coli | 36% | 47 148 | 5.21 | 21.6% | d.46; a.46; c.27 | + |
| U. urealyticum | 48 678 | 7.57 | 12.8% | − | |||
| MetK | E. coli | 43% | 41 898 | 5.10 | 18.6% | d.130 | + |
| U. urealyticum | 41 988 | 6.41 | 13.8% | − | |||
| YcfH | E. coli | 31% | 29 772 | 5.19 | 14.7% | c.1 | + |
| U. urealyticum | 30 929 | 6.15 | 9.1% | − | |||
| aID, identity between the two sequences. | |||||||
| bMW, molecular weight (Da). | |||||||
| cA/G, alanine/glycine contents in the sequences. | |||||||
| dSCOP-fold ID of the Class IV proteins in E. coli. | |||||||
| eGR, E. coli GroE requirement. Plus (+) indicates that the protein has in vivo GroE requirement. GroE requirement for the Ureaplasma orthologs was assessed by the solubility in GroE-normal and -depleted cells, as shown in Supplementary Figure S12A. | |||||||
Discussion
Enriched GroE interactors (Class III substrates) are not necessarily obligate GroE substrates in vivo
On the basis of our earlier study, in which we found one of the Class III proteins, ParC, to be physiologically functional in GroE-depleted cells, we have systematically evaluated the in vivo GroE dependence (for solubility) of all Class III substrates. The results clearly showed that ParC was not unique; the in vivo solubility of ∼40% (34 out of 84) of the Class III substrates was independent of GroE (Figures 2A and B and 3; Supplementary Figures S3 and S4). On this basis, we divided the Class III substrates into two classes: GroE-independent Class III− and GroE obligate Class III+ substrates (Figure 2C).
One intriguing possibility is that the classification of Classes III+ and III−; that is, the in vivo GroEL dependence, is correlated with an enrichment of the substrates in Class III+ among the GroE interactors. An analysis of the raw data on the enrichment (U Hartl, personal communication), however, did not reveal a statistically significant difference between Classes III+ and III− in terms of the GroEL interaction. This analysis is consistent with the notion that the in vivo GroE dependency is not determined by the enrichment of the GroE interaction.
The Class III− proteins: in vivo preferential GroEL interactors are not dependent on GroE for solubility
One of the novel concepts in this paper is that of the Class III− proteins, defined as Class III proteins that are soluble in GroE-depleted cells. We also showed that four Class III− proteins that are essential for viability were functional (Figure 3), indicating that these Class III− proteins can fold correctly in vivo without GroE.
However, several caveats should be stated regarding the activity of the Class III− proteins. Observing enzyme activity in GroE-depleted cells does not necessarily mean that the folding of these enzymes is GroE independent, as a small amount of folded enzyme may suffice, depending on the substrate concentration and the enzyme's kinetic parameters. In addition, we cannot rule out the possibility that other Class III− proteins are soluble but inactive in the absence of GroE, as we have not measured the activities of all of the Class III− proteins in the GroE-depleted cells. We also note that a situation may occur when upregulation of some enzymes and downregulation of others will result in smaller than one would expect changes in the amounts of precursor and/or product. Assuming that the soluble but inactive states result from insufficient folding, we might expect that the insufficiently folded proteins would be easily degraded by cellular proteases, as observed for some of the GroE obligate substrates (Class III+ or IV), such as TldE (Supplementary Figure S3). However, we found that most of the Class III− proteins are resistant to proteases, even when overexpressed (Figure 2; Supplementary Figures S3 and S4), suggesting that the Class III− proteins can at least fold into protease-resistant conformational states. In addition, we found several striking differences between Classes III− and III+ (IV) in terms of multiple physicochemical properties (Figure 5), reinforcing the validity of their separation into two groups.
The occupancy of GroE by the Class III− substrates was not provided from our experiments. How much of the GroE capacity Class III+ (IV) versus Class III− substrates occupy must depend at least on chaperone abundance, protein abundance and their binding affinity. Nevertheless, the Class III− proteins, representing ∼40% of Class III, should engage a substantial amount of the GroE in the cell, as the Class III substrates were originally defined as highly enriched GroE interactors (Kerner et al, 2005). What is the role of these preferential interactions, despite the apparent absence of GroE dependency?
One plausible explanation is that the Class III− substrates might interact with GroEL for relatively longer periods after translation, irrespective of the GR for folding. The prolonged interaction with GroEL, probably due to a strong affinity, would result in an enrichment of the substrates in the GroE complex, leading to the assignment of these proteins as Class III substrates, which were originally defined as being enriched with GroEL (Kerner et al, 2005). In this context, the earlier observation that some proteins >60 kDa exhibited very slow release from GroEL (Houry et al, 1999) seems to be correlated with our assignment of all Class III proteins >70 kDa as Class III−.
An alterative, though not mutually exclusive, hypothesis is that the preferred function of the Class III− proteins is nucleotide binding, as half of the Class III− proteins were RNA or DNA-binding proteins (i.e. transcriptional regulators or proteins using RNA as substrates) (Supplementary Table S2). In support of this hypothesis, our bioinformatics analysis revealed that the Class III− proteins were enriched in positively charged amino acids (Figure 5C). To accommodate the negative charges of polynucleotides, the protein surfaces of RNA or DNA-binding proteins typically contain positive charges. Therefore, the Class III− proteins might naturally associate with the very acidic inside surface of the GroE cavity for a purpose other than assistance with folding (Shimamura et al, 2004; Tang et al, 2006). In fact, the contribution of the electrostatic interaction in the GroEL–substrate interactions has been reported in a thermodynamic analysis (Aoki et al, 1997).
The Class IV substrates: in vivo obligate GroE substrates
Our comprehensive analysis revealed that 60% (49 out of 84) of the Class III substrates were Class III+, partly verifying the original proposal that the preferential GroE interactors are in vivo obligate GroE substrates. The further identification of eight in vivo obligate substrates that were not previously included in Class III prompted us to define a new GroE substrate class, Class IV, defined as in vivo obligate GroE substrates (Figure 4E; Table 1).
Intriguingly, the Class IV substrates had a slight but significant positive bias in alanine and glycine (Ala/Gly) content. Although we lack a plausible interpretation, the short side chains of Ala/Gly might confer more flexibility to the proteins in their denatured states, possibly leading to aggregation-prone properties. In this context, we note that the Ala/Gly content was lower in the Ureaplasma Class IV homologs, which did not require GroE for folding, than in the counterpart E. coli proteins (Supplementary Figure S12).
Our in vivo evaluation of GroE dependence showed that the tested Class I and II substrates contained few in vivo obligate GroE substrates (Figure 2B), suggesting that among the previously identified GroE interactors, the Class IV members would be drawn entirely from Class III. Therefore, almost all of the Class IV substrates of known GroEL interactors were identified in this study. However, we identified some Class IV proteins among those not identified as GroE interactors, raising the question of the number of unidentified Class IV substrates still remaining to be found in E. coli. Over a thousand soluble proteins remain to be examined for GroE dependency, as the number of cytosolic proteins in E. coli has been predicted to be ∼2200 (Niwa et al, 2009), as compared with the 986 proteins quantified by our proteomics. In this regard, our survey of GroE substrates is not comprehensive of the whole E. coli proteome. Further considerations of approaches, such as metabolomics, homology searching, or more accurate comparative proteomics with the transcriptome, would be required to search for the unidentified GroE substrates.
We compared the Class IV substrates with the list of GroE substrates reported by Chapman et al (2006). Chapman et al observed global aggregation in an E. coli strain with a temperature-sensitive GroEL mutation, and then identified the insoluble proteins as GroE substrates. Among the ∼300 substrates identified by Chapman et al, only 17 of the 57 Class IV substrates overlapped. The poor overlapping of the substrates might be attributed to the different E. coli strains. Indeed, Masters et al (2009) observed that insoluble proteins did not accumulate in GroE-depleted MGM100 cells, suggesting that the intracellular milieu in the temperature-sensitive strain and that in the GroE-depleted cells are quite different.
Essential genes in Class IV substrates
Our verified obligate GroE substrates included only six genes (DapA, ASD, MetK, FtsE, HemB, and KdsA) essential for the viability of E. coli in rich medium. Although we anticipate that unidentified essential Class IV substrates exist among the proteins that were not tested, we can predict the possible phenotypic defects caused by their inactivation in the GroE-depleted cells (Supplementary Figure S13A). If there are no further essential GroE-dependent proteins, then the complementation of these six essential genes by some means should generate an E. coli strain that can grow without groEL/ES, as also discussed by Masters et al (2009) (Supplementary Figure S13B). Such viable groEL/ES-knockout E. coli would provide the answer to the long-standing question of why GroE is essential for cell viability. Alternatively, the complementation of the six essential genes in E. coli-lacking groEL/ES could be still lethal, due to the presence of unidentified essential Class IV substrates. In such a case, we can extend the phenotypic analysis to find the unidentified Class IV members, using the engineered E. coli.
Aggregation-prone folds are common in Class IV substrates
A striking feature of Class IV is the inherent tendency of its members to aggregate during chaperone-free translation in vitro (Niwa et al, 2009); the solubilities of ∼90% of the Class IV substrates are <30% (Figure 5D; Table 1). This tendency to aggregate may well be a prerequisite for in vivo GroE dependency. One exception is TldD (51 kDa), which was highly soluble in a cell-free translation reaction without chaperones (>90% solubility; Niwa et al, 2009). One intriguing possibility is that TldD would not require GroE for folding per se, but for accelerated folding (Brinker et al, 2001; Tang et al, 2006, 2008), which might be critical for its physiological function, as discussed earlier (Jewett and Shea, 2010).
A bioinformatic analysis revealed distinct structural preferences among the Class IV substrates. The TIM-barrel fold (c.1), which has been proposed as the preferred folding topology for GroE substrates (Houry et al, 1999; Kerner et al, 2005), was found in close to half (25/57) of the Class IV proteins. However, we also found that other fold classes, such as the FAD/NAD(P)-binding domain (c.3), the PLP-dependent transferase-like fold (c.67), and the thiolase fold (c.95) were also overrepresented in Class IV, although less so than the TIM-barrel fold proteins. Strikingly, all of the fold classes overrepresented among the Class IV members are aggregation-prone folds, as described in our earlier study (Niwa et al, 2009) (Note that the thiolase fold (c.95) is not included among the aggregation-prone folds due to the lack of statistical significance, but 70% of c.95-containing proteins were <50% soluble).
Evolutionary consideration of Class IV substrates
Finally, our analysis revealed that >70% (42 out of 57) of the Class IV substrates, including enzymes that are closely coupled, sequentially aligned, or redundant (Supplementary Figure S14), are likely to be involved in metabolic reactions (Table 1). Although the preferential role of GroE in the folding of metabolic enzymes has already been proposed (Houry et al, 1999; Kerner et al, 2005), this study, combined with other recent work, allows for the development of a plausible hypothesis for the evolutionary relationship between GroE and the obligate substrates, as follows.
Chaperones are known to provide a buffering system for genetic mutations, and thus promote genetic diversity (Rutherford and Lindquist, 1998; Queitsch et al, 2002). A recent quantitative assessment clearly showed that GroE promotes enzyme evolution by buffering the destabilizing mutations that confer improved enzymatic activities (Tokuriki et al, 2009). As the destabilization of proteins generally results in their intracellular aggregation, aggregation-prone proteins, such as the Class IV substrates, could survive mutations if their aggregation is prevented by GroE, leading to the acquisition of diversity and/or the potential improvement of the enzymes in the Class IV substrates.
Materials and methods
Plasmids
pMCS (Fujiwara and Taguchi, 2007), in which the T7 promoter region of the pET15b(+) (Novagen) was replaced with the tac promoter, was used for constructing all plasmids. A description of the detailed cloning strategies used in this study is provided in the Supplementary data. Essential genes in rich medium were deduced from earlier data (Baba et al, 2006). Because of the small number of Class III proteins essential in rich medium, 13 Class III proteins essential in minimal medium (Kerner et al, 2005) were identified. As TGA encodes tryptophan in Ureaplasma, the Ureaplasma genes were optimized to the codon use of E. coli, and were chemically synthesized (GenScript).
Analysis of GroEL/ES dependency in GroE-depleted cells
E. coli MGM100 (MG1655 groE∷araC-PBAD-groE (Kanr)) cells, harbouring one of the plasmids encoding a candidate GroEL/ES obligate substrate, were grown in LB medium with arabinose at 37 °C to log phase. After washing, the cells were diluted 1:100 into LB with 1 mM diaminopimelate (DAP, WAKO), containing either 0.2% arabinose or 0.2% glucose. After 2 h of cultivation, protein expression was induced with 1 mM isopropyl α-D-thiogalactopyranoside (IPTG) for 1 h. For leaky expression, this IPTG-inducing step was omitted, and the cells were cultivated for a total of 5 h after the sugar shift. The cells were then harvested, suspended in STE lysis buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, and 1 mM ethylenediaminetetraacetic acid) for adjustment to equivalent OD660 units, and sonicated (Branson sonifier). The soluble fraction was obtained by centrifugation (20 000 × g, 30 min). Total and soluble extracts were analysed by SDS–PAGE and were detected by Coomassie Brilliant Blue staining or by immunoblotting using an anti-HA monoclonal antibody (SIGMA).
Proteomics and metabolomics
MGM100 or MG1655 cells were grown in LB medium containing arabinose or glucose for 5 h. For proteomics, trypsin-digested lysates of 1.6 OD600 units were analysed by LC-MS. The protein abundance based on emPAI was calculated according to the previously reported methods (Ishihama et al, 2005). For metabolomics, an aliquot containing 20 OD600 units of cells was passed through a 0.45-μm pore size filter (Durapore HVLP09050, Millipore). Metabolites were extracted by 5 ml of methanol. After the addition of 4 ml of chloroform and 1.6 ml of Milli-Q water, the 4 ml water layer was isolated, lyophilized, and dissolved in 50 μl of Milli-Q water containing a migration time standard, before analysis by CE-MS. Detailed conditions of both analyses are available in earlier reports (Ishii et al, 2007; Iwasaki et al, 2009) and in the Supplementary data.
Enzyme assays
The plasmid-encoded SuhB, TrmD, and MetK proteins were overexpressed by IPTG induction in GroEL/ES-normal or -depleted cells. The relative activities of these enzymes were analysed without purification. Unless noted, the lysates from cells without IPTG-induction but harbouring the corresponding plasmid were used as a control. The relative activities were defined as the activity of cell lysates from GroEL/ES-sufficient cells with IPTG induction as 1.0. The details of the enzyme assays are described in the Supplementary data.
Supplementary Material
Acknowledgments
We thank Mr M Honma, Ms M Ohishi, Ms C Kimura, and Mr Y Igarashi for technical assistance with the metabolome and proteome analyses. We also thank Dr Ulrich Hartl for sharing the MS data from the earlier study and helpful discussions, Dr Millicent Masters for critical reading and generous editing of the manuscript, Professors Masaru Tomita and Takuya Ueda for their supports, and Dr Ayumi Koike-Takeshita for providing the GroES antibody. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (19037007, 19058002 to HT) from MEXT, and by research funds from Yamagata Prefecture and Tsuruoka City to Keio University. KF was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science. KF, YI, KN, TS, and HT designed the research; KF, YI, and KN performed the experiments; KF, YI, KN, and HT analysed the data; and KF, YI, KN, and HT wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181: 223–230 [DOI] [PubMed] [Google Scholar]
- Aoki K, Taguchi H, Shindo Y, Yoshida M, Ogasahara K, Yutani K, Tanaka N (1997) Calorimetric observation of a GroEL-protein binding reaction with little contribution of hydrophobic interaction. J Biol Chem 272: 32158–32162 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M (2001) Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107: 223–233 [DOI] [PubMed] [Google Scholar]
- Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, Horwich AL (2006) Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA 103: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, de Crecy-Lagard V (2006) Discovery of a new prokaryotic type I GTP cyclohydrolase family. J Biol Chem 281: 37586–37593 [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Hendrick JP, Houry WA, Hartl FU (1997) In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90: 491–500 [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E (2002) Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417: 398. [DOI] [PubMed] [Google Scholar]
- Fayet O, Ziegelhoffer T, Georgopoulos C (1989) The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol 171: 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton WA, Horwich AL (2003) Chaperonin-mediated protein folding: fate of substrate polypeptide. Q Rev Biophys 36: 229–256 [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Taguchi H (2007) Filamentous morphology in GroE-depleted Escherichia coli induced by impaired folding of FtsE. J Bacteriol 189: 5860–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY, Cassell GH (2000) The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407: 757–762 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K (1993) Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell 74: 909–917 [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402: 147–154 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y et al. (2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316: 593–597 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Masuda T, Tomita M, Ishihama Y (2009) Chemical cleavage-assisted tryptic digestion for membrane proteome analysis. J Proteome Res 8: 3169–3175 [DOI] [PubMed] [Google Scholar]
- Jenkins AJ, March JB, Oliver IR, Masters M (1986) A DNA fragment containing the groE genes can suppress mutations in the Escherichia coli dnaA gene. Mol Gen Genet 202: 446–454 [DOI] [PubMed] [Google Scholar]
- Jewett AI, Shea JE (2010) Reconciling theories of chaperonin accelerated folding with experimental evidence. Cell Mol Life Sci 67: 255–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220 [DOI] [PubMed] [Google Scholar]
- Koike-Takeshita A, Shimamura T, Yokoyama K, Yoshida M, Taguchi H (2006) Leu309 plays a critical role in the encapsulation of substrate protein into the internal cavity of GroEL. J Biol Chem 281: 962–967 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Masters M, Blakely G, Coulson A, McLennan N, Yerko V, Acord J (2009) Protein folding in Escherichia coli: the chaperonin GroE and its substrates. Res Microbiol 160: 267–277 [DOI] [PubMed] [Google Scholar]
- McLennan N, Masters M (1998) GroE is vital for cell-wall synthesis. Nature 392: 139. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540 [DOI] [PubMed] [Google Scholar]
- Niwa T, Ying BW, Saito K, Jin W, Takada S, Ueda T, Taguchi H (2009) Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc Natl Acad Sci USA 106: 4201–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noivirt-Brik O, Unger R, Horovitz A (2007) Low folding propensity and high translation efficiency distinguish in vivo substrates of GroEL from other Escherichia coli proteins. Bioinformatics 23: 3276–3279 [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21: 3435–3438 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- Sakikawa C, Taguchi H, Makino Y, Yoshida M (1999) On the maximum size of proteins to stay and fold in the cavity of GroEL underneath GroES. J Biol Chem 274: 21251–21256 [DOI] [PubMed] [Google Scholar]
- Shimamura T, Koike-Takeshita A, Yokoyama K, Masui R, Murai N, Yoshida M, Taguchi H, Iwata S (2004) Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure 12: 1471–1480 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T (2001) Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751–755 [DOI] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M (2008) Essential role of the chaperonin folding compartment in vivo. EMBO J 27: 1458–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125: 903–914 [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS (2009) Do viral proteins possess unique biophysical features? Trends Biochem Sci 34: 53–59 [DOI] [PubMed] [Google Scholar]
- Van Dyk TK, Gatenby AA, LaRossa RA (1989) Demonstration by genetic suppression of interaction of GroE products with many proteins. Nature 342: 451–453 [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Gatenby AA, Lorimer GH (1992) Purified chaperonin 60 (groEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci 1: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388: 741–750 [DOI] [PubMed] [Google Scholar]
- Ying BW, Taguchi H, Kondo M, Ueda T (2005) Co-translational involvement of the chaperonin GroEL in the folding of newly translated polypeptides. J Biol Chem 280: 12035–12040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.