Abstract
In this brief report, we provide a portrait of the CEM-NET initiatives regarding molecular epidemiological studies and review how they have contributed to a better knowledge of the nature of drug-resistant Staphylococcus aureus and Streptococcus pneumoniae clones, as well as their routes and modes of geographic expansion.
Keywords: CEM-NET, Epidemiology, Staphylococcus aureus, Streptococcus pneumoniae
Introduction
The introduction of vast quantities of antibiotics into the environment – beginning around the middle of the last century – has triggered an “accelerated evolution” of drug-resistant bacterial lineages – propelled by the extensive use of antibacterial agents in clinical practice and prophylaxis both in the human and veterinarian environments and in agriculture as well. Characterization of these novel bacterial clones and tracking their geographic spread required the use of molecular biology techniques which underwent a parallel evolution in time – beginning with phage typing and serotyping through pulsed-field gel electrophoresis (PFGE) all the way to full genome sequencing (Oliveira et al., 2011).
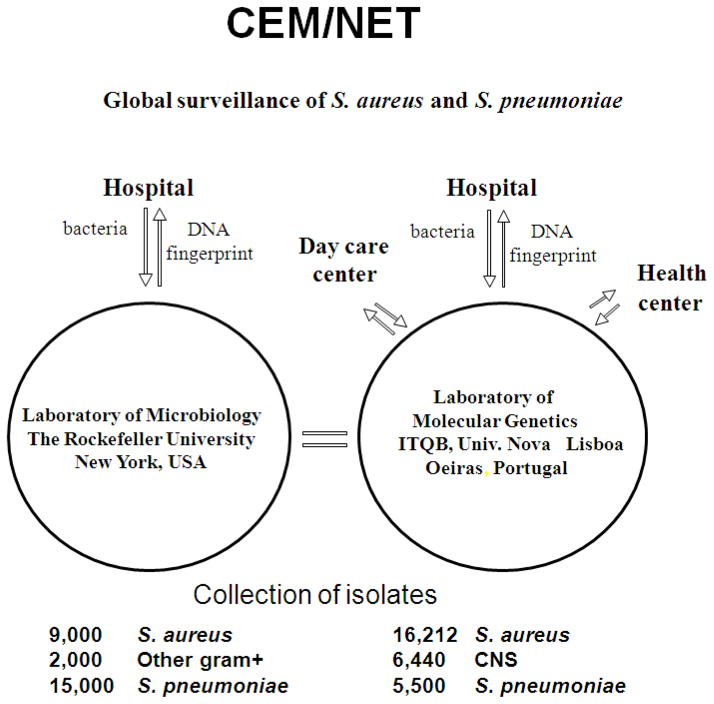
In this brief report, we shall provide a portrait of the CEM-NET initiative that probably represents the chronologically first organized international effort in molecular epidemiology of drug-resistant staphylococci and pneumococci. CEM-NET (for Centro de Epidemiologia Molecular) had its first international meeting in 1995. CEM-NET combined the interests and scientific skills of two university-based centers: the Laboratory of Microbiology and Infectious Disease at Rockefeller University in New York, USA (headed by Dr. Alexander Tomasz) and the Laboratory of Molecular Genetics at Instituto de Tecnologia Química e Biológica (ITQB), affiliated with the Universidade Nova de Lisboa in Portugal (headed by Dr. Herminia de Lencastre). The informal structure and mode of operation of CEM-NET is shown in Figure 1.
Fig. 1.
The structure and mode of operation of CEM-NET.
Studies on the molecular epidemiology of Streptococcus pneumoniae
An international meeting organized by CEM-NET in 1996 at the Laboratory of Molecular Genetics, ITQB was attended by 96 colleagues from 25 different countries and produced an update on the molecular biology of pneumococci and its diseases (Tomasz, 2000). This meeting also catalyzed the birth of ISPPD (International Symposium on Pneumococci and Pneumococcal Disease) (http://www2.kenes.com/isppd2012/Pages/home.aspx) – an international group of scientists and medical practitioners who have been gathering every 2nd year, with each time a different country serving as host. ISPPD has become an international forum for update and information exchange on pneumococcal diseases and their control – with specific emphasis on efforts to control these devastating diseases which account for over a million annual mortalities mostly in preschool age children. The last meeting of ISPPD in Tel Aviv (ISPPD 7) drew an attendance of over 2,000 participants (http://www2.kenes.com/isppd/Pages/Home.aspx).
Lisbon Day Care Center Initiative
The main – if not the only – ecological reservoir of S. pneumoniae on this planet is the human nasopharynx, primarily in preschool age children, a very large proportion of whom are recruited into Day Care Centers in the developed world. A systematic – annual – screening of S. pneumoniae carried by children attending Day Care Centers in Lisbon, Portugal began in 1996 by the European arm of CEM-NET (laboratory of Molecular Genetics, Lisbon). This ongoing surveillance – supported by the European community and Fundaçãopara a Ciência e a Tecnologia (FCT) (de Lencastre et al., 1999) has produced numerous important insights into evolutionary changes in the natural flora of pneumococci in response to such global interventions as use of antimicrobial agents (Sa-Leao et al., 2009) or the introduction of conjugate anti-pneumococcal vaccine (Frazao et al., 2005). A website containing an enormous body of invaluable information on the epidemiology and molecular types of these pneumococcal isolates has been produced (Silva et al., 2003).
The CEM-NET fellowship program
The CEM-NET fellowship program funded by the Gulbenkian Foundation and FCT allowed working visits to the Laboratory of Molecular Genetics, Lisbon, by interested colleagues from different European and South American countries and Turkey and North Africa. The purpose of these visits was to provide a hands-on laboratory experience in a tutorial type of environment to learn molecular typing techniques and characterize staphylococcal isolates brought along by the visitors from their countries. The visiting colleagues worked in close collaboration with members of the Laboratory of Molecular Genetics and most of these visits resulted in collaborative publications.
Collaborative studies with the Pan American Health Organization (PAHO)
Colleagues from Brazil, Argentina, Mexico, Colombia, Uruguay, brought along a large collection of penicillin-resistant S. pneumoniae isolates to a workshop of CEM-NET organized at the Rockefeller University. The isolates were recovered from pediatric disease through the South American network of Regional Vaccines Systems (SIREVA). The isolates were characterized by molecular typing techniques. Part of the purpose of this collaborative workshop was to teach molecular typing techniques to our South American colleagues. A major conclusion of this study was recognition of the dominance of a few pandemic penicillin-resistant S. pneumoniae clones which were responsible for the overwhelming majority of antibiotic-resistant pediatric disease in these countries (Tomasz et al., 1998).
The aim of a second collaborative study with PAHO – also organized at the USA branch of CEM-NET, was to teach multilocus sequence typing (MLST) and also examine the nature of penicillin-susceptible S. pneumoniae isolates causing pediatric disease in South America – in the same areas where the former study with resistant isolates was performed (Zemlickova et al., 2005). In contrast to the penicillin-resistant isolates, the susceptible strains showed enormous genetic variation in sequence type (Figs. 2, 3).
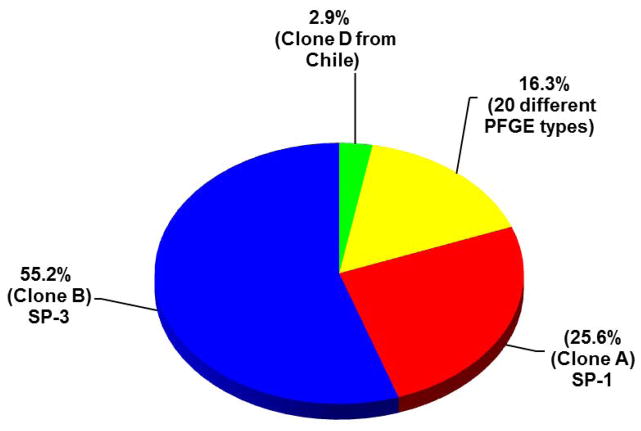
Fig. 2.
Molecular typing of penicillin-resistant S. pneumoniae isolates from South America. Of the 172 penicillin resistant isolates collected in 5 South American countries in the SIREVA initiative and typed by molecular typing techniques 137 (80%) of the isolates belonged to two dominant sequence types represented by the epidemic clones SP-1 and SP-3.
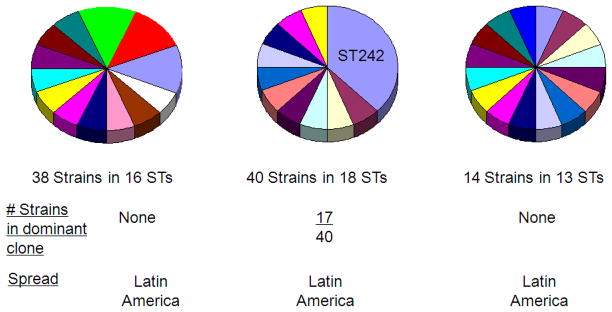
Fig. 3.
Molecular typing of penicillin-susceptible S. pneumoniae isolates from South America. 185 penicillin-susceptible S. pneumoniae isolates recovered from pediatric disease in 5 South American countries were typed for serotype and for sequence type. In contrast to the penicillin-resistant isolates (see Fig. 2), the susceptible strains showed a high degree of genetic diversity.
The CEM-NET laboratories in Lisbon and New York provided some of the S. pneumoniae isolates for a recent collaborative study with colleagues at the Sanger Institute, the aim of which was to reconstruct stages in the pathway of evolution of one of the multidrug-resistant clones of S. pneumoniae through the use of full genome sequencing (Croucher et al., 2011). The disease-causing potential of the PMEN-1 clone was compared to the disease potential by penicillin-susceptible pneumococcal strains of the same serotype – 23F – in a small case control study conducted in several New York hospitals (Roberts et al., 2001).
Studies on methicillin-resistant Staphylococcus aureus (MRSA)
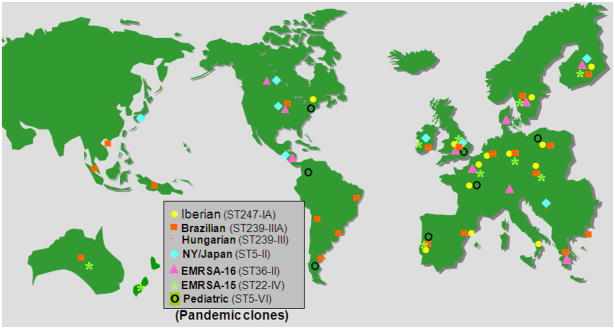
“RESIST” – an acronym for the first international surveillance of MRSA that used molecular typing techniques – was conducted by the CEM-NET laboratories. Over 4,500 MRSA isolates recovered from 84 hospitals in 20 countries were typed by molecular techniques (Santos Sanches et al., 2000) (Table 1). A major finding of this study was the recognition that as few as 5–6 globally spread MRSA clones were responsible for over 80% of MRSA disease in hospitals in each of the participating countries (Oliveira et al., 2002, Aires de Sousa and de Lencastre, 2004) (Fig. 4).
Table 1.
International surveillance of MRSA
| Region | Country | Isolation dates | No. of cities | No. of hospitals | No. of isolates studied |
|---|---|---|---|---|---|
| Europe | Belgium | 1992, 1995 | 2 | 2 | 4 |
| Czech Republic | 1996–2001 | 8 | 22 | 159 | |
| Denmark | 1964–1970, 1986–1992 | 17 | 21 | 59 | |
| Germany | 1992–1993 | 2 | 2 | 2 | |
| Greece | 1992–2000 | 1 | 1 | 129 | |
| Hungary | 1993–1998 | 10 | 24 | 285 | |
| Italy | 1993–1995 | 1 | 12 | 53 | |
| Poland | 1990–1998 | 8 | 18 | 270 | |
| Portugal | 1990–2003 | 5 | 9 | 815 | |
| Spain | 1989–1993 | 1 | 1 | 189 | |
| Turkey | 1993–1997 | 1 | 1 | 99 | |
| UK | 1990–1993 | 1 | 10 | 13 | |
| North America | USA | 1994–1998 | 5 states | 44 | 1033 |
| Latin America | Argentina | 1994–1998 | 3 | 13 | 237 |
| Brazil | 1992–1998 | 3 | 11 | 314 | |
| Chile | 1997–1998 | 2 | 7 | 118 | |
| Colombia | 1996–1998 | 2 | 5 | 76 | |
| Mexico | 1997–2003 | 2 | 2 | 317 | |
| Uruguay | 1997–1998 | 2 | 5 | 102 | |
| Asia | China | 1998–1999 | 1 | 1 | 14 |
| Japan | 1997–1998 | 1 | 1 | 143 | |
| Taiwan | 1998–1999 | 1 | 1 | 118 |
A total of 4549 MRSA isolates were collected in an international collaborative study from over 80 hospitals located in 20 countries and the isolates were classified according to molecular type using pulse-field gel electrophoresis, multilocus sequence typing and SCCmec typing.
Fig. 4.
International spread of a few pandemic clones of MRSA.
A sentinel system of hospitals in Portugal
A sentinel system of hospitals in Portugal was organized by the European branch of CEM-NET. Periodic surveillance of MRSA clones over a several years period has allowed documentation of the “ebb and flow” of major epidemic clones over time (Aires-de-Sousa et al., 2008).
Tracking MRSA clones in 12 major hospitals in New York City and in hospitals in the Tri-State Area of New Jersey, Pennsylvania and Connecticut
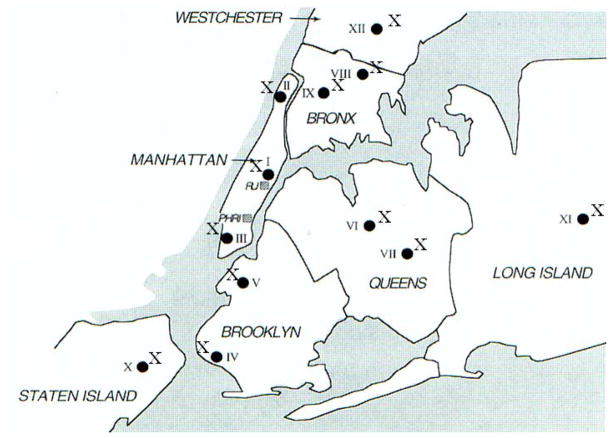
This molecular epidemiological study documented the wide-spread dominance of the New York/Japan MRSA clone (sequence type 5) as a major cause of hospital-borne MRSA disease (Roberts et al., 1998, 2000) (Fig. 5).
Fig. 5.
Spread of the New York/Japan MRSA clone (ST-5) in New York City Hospitals. Location of participating hospitals in the various boroughs of New York City and the presence of the ST-5 MRSA clone (X) is indicated. The locations of Rockefeller University (RU) and the Public Health Research Institute (PHRI, participating in the study) are also indicated (grey squares).
Carriage of MRSA among healthy individuals in the community
A study of major epidemiological importance was carried out by the European branch of CEM-NET in 1998. The study addressed the important question of exactly where the ecological reservoirs of MRSA are in a country, Portugal, where the frequency of hospital-associated MRSA disease was as high as 50%. A carefully selected cohort of young healthy individuals (air force recruits, high school students and university students) was selected for the study. Participants had no physical contact with hospitals and never used antibiotics. Screening of colonization sites (nostrils and armpits) of over 1,009 individuals identified – as expected – about 30% as carriers of S. aureus but the carriage rate of MRSA was as low as 5/5010 isolates (Sa-Leao et al., 2001). Low carriage rate of MRSA among healthy individuals was subsequently confirmed by a study in 2010 (Tavares et al., 2010). A study by the CDC in 2003/2004 found carriage rate of 1.5% in a general population sample from the USA (Gorwitz et al., 2008). These observations really indicate that before the appearance of community-acquired MRSA in the early 2000s, MRSA clones were restricted to the hospital environment.
Molecular epidemiology of S. aureus disease in Africa
Two studies conducted by the European branch of CEM-NET examined the nature of S. aureus carriage in a former Portuguese colony in Africa: Cape Verde islands. Most interestingly, the frequency of S. aureus was extremely high in the two hospitals screened, but no MRSA could be identified (Aires De Sousa et al., 2000) probably reflecting the lack of availability of antimicrobial agents. On the other hand, the carriage of the PVL toxin was unusually high among these MSSA isolates (Aires-de-Sousa et al., 2006).
Update of MRSA clones in Europe for Year 2006
The CEM-NET laboratory participated in a major molecular epidemiological study in which 25 European countries and over 500 hospitals provided MRSA isolates for a typing effort that used spa typing to identify the distribution and dominance of MRSA clones in Europe (Grundmann et al., 2010).
“Cosmopolitan” MRSA clones
“Cosmopolitan” MRSA clones that can exist both in hospitals and in the community and the likely route of transmission between these two environments have recently been identified by sampling handrails in public bus lines through which visitors travel to and from hospitals in Oporto (Simoes et al., 2011).
Evolutionary history of the “Brazilian” clone of MRSA
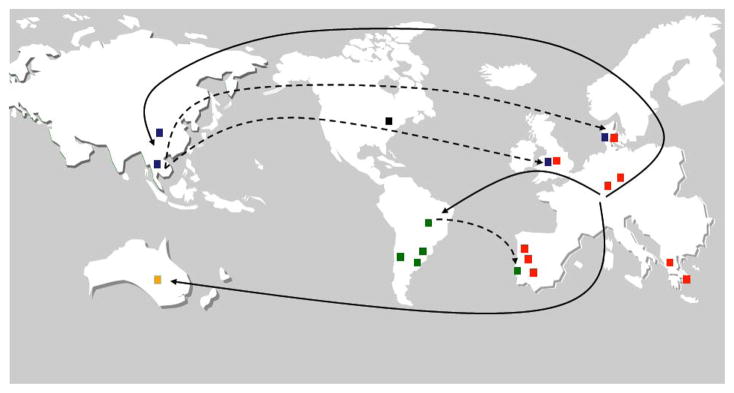
Evolutionary history of the “Brazilian” clone of MRSA was deciphered in a collaborative study between the Laboratory of Molecular Genetics, a hospital in Thailand and the Sanger Institute. Complete genome sequencing was performed on chronologically and geographically characterized isolates leading to the identification of the most likely “birthplace” as well as routes of travel of this important MRSA clone (Fig. 6) (Harris et al., 2010).
Fig. 6.
“Birthplace” and migrations of the Brazilian MRSA clone ST239. Figure drawn by S. Gardete and H. de Lencastre from data in described by Harris et al., 2010. Red, Europe; Green, South America; Black, North America; Blue, Asia; Yellow, Australia.
Micro evolution of an MRSA lineage in a patient undergoing antibiotic therapy
Consecutive isolates of an MRSA – belonging to New York/Japan clone – were recovered from the blood stream of a patient during various times after the onset of extensive chemotherapy with the antibiotic vancomycin. The isolates included the fully susceptible first isolate recovered from the blood stream at the time of hospital admission and the vancomycin-resistant isolate recovered from the patient at the end of chemotherapy. All isolates were shown to be isogenic which prompted us to determine the full genome sequence of the initial and final isolates. The point mutations in 33 chromosomal genes identified in the ultimate vancomycin-resistant isolate were shown to appear in a sequential manner in isolates with intermediate resistance level during earlier times of chemotherapy. Using PCR sequencing it was possible to construct a genetic pathway through which this particular MRSA clone had evolved (Mwangi et al., 2007). While the precise contributions of the 33 mutations to stages of antibiotic resistance remains to be determined, this study appears to have identified steps in the evolution of a single drug-resistant MRSA lineage – in vivo – in the realistic setting of a patient undergoing chemotherapy.
Conclusion and outlook
Molecular epidemiology studies have already produced a vast amount of important information about the nature of drug-resistant clones, their routes and modes of geographic expansion, clearly documenting the importance of close collaboration between epidemiologists, who can provide bacterial isolates with well-documented information concerning dates and place of isolation, and molecular biologists, who can characterize these isolates by full-genome sequencing to reconstruct with even more precision the pathways through which a particular pathogenic clone has evolved.
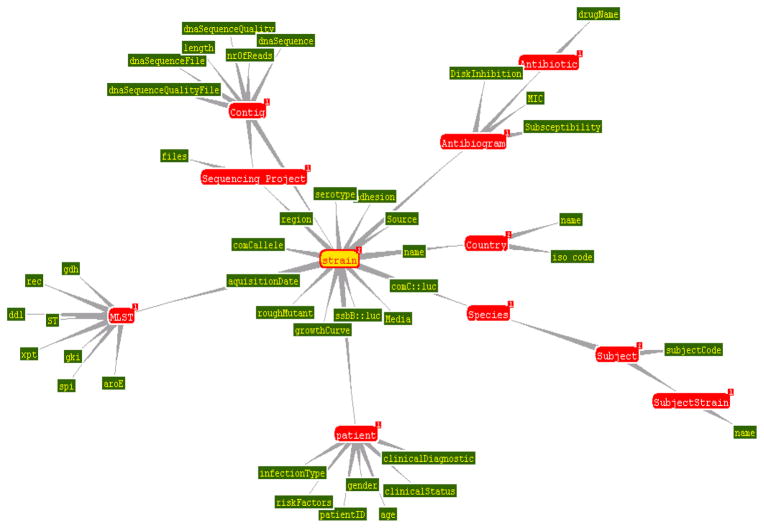
A special database containing all molecular, microbiological and demographic information about the S. aureus isolates in the CEM-NET collection is currently under development (Fig. 7) (Deus et al., 2008).
Fig. 7.
The structure of a Semantic Web management model (S3DB) for integrative biomedical informatics. Figure was drawn by Jonas Almeidato to illustrate the structure of the model.
Acknowledgments
This work was supported by grant from the US Public Health Service 2 RO1 AI457838-11 awarded to Alexander Tomasz and a grant awarded to Herminia de Lencastre from the Fundação Calouste Gulbenkian (Ref. P-99911) entitled “Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) in Portugal: a pilot study focusing on an emerging public health concern”. All figures originating from published papers were reproduced with the permission of the publisher.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aires-de-Sousa M, Conceicao T, de Lencastre H. Unusually high prevalence of nosocomial Panton-Valentine leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J Clin Microbiol. 2006;44:3790–3793. doi: 10.1128/JCM.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires-de-Sousa M, Correia B, de Lencastre H. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J Clin Microbiol. 2008;46:2912–2917. doi: 10.1128/JCM.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires de Sousa M, de Lencastre H. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol. 2004;40:101–111. doi: 10.1016/S0928-8244(03)00370-5. [DOI] [PubMed] [Google Scholar]
- Aires De Sousa M, Santos Sanches I, Ferro ML, De Lencastre H. Epidemiological study of staphylococcal colonization and cross-infection in two West African hospitals. Microb Drug Resist. 2000;6:133–141. doi: 10.1089/107662900419447. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H, Santos Sanches I, Brito-Avo A, Sa-Leao R, Saldanha J, Kristinsson KG, Tomasz A. Carriage and antibiotic resistance of respiratory pathogens and molecular epidemiology of antibiotic-resistant Streptococcus pneumoniae colonizing children in day-care centers in Lisbon: the Portuguese day-care center initiative. Clin Microbiol Infect. 1999;5(Suppl 4):S55–S63. doi: 10.1111/j.1469-0691.1999.tb00858.x. [DOI] [PubMed] [Google Scholar]
- Deus HF, Stanislaus R, Veiga DF, Behrens C, Wistuba II, Minna JD, Garner HR, Swisher SG, Roth JA, Correa AM, Broom B, Coombes K, Chang A, Vogel LH, Almeida JS. A Semantic Web management model for integrative biomedical informatics. PLoS One. 2008;3:e2946. doi: 10.1371/journal.pone.0002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazao N, Brito-Avo A, Simas C, Saldanha J, Mato R, Nunes S, Sousa NG, Carrico JA, Almeida JS, Santos-Sanches I, de Lencastre H. Effect of the seven-valent conjugate pneumococcal vaccine on carriage and drug resistance of Streptococcus pneumoniae in healthy children attending day-care centers in Lisbon. Pediatr Infect Dis J. 2005;24:243–252. doi: 10.1097/01.inf.0000154326.77617.3e. [DOI] [PubMed] [Google Scholar]
- Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, Tomasz A. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci USA. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H, Tomasz A. Evolution of molecular techniques for the characterization of MRSA clones. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Discovery and Development. Springer; New York: 2011. [Google Scholar]
- Oliveira DC, Tomasz A, de Lencastre H. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002;2:180–189. doi: 10.1016/s1473-3099(02)00227-x. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Chung M, de Lencastre H, Hargrave J, Tomasz A, Nicolau DP, John JF, Jr, Korzeniowski O. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb Drug Resist. 2000;6:245–251. doi: 10.1089/mdr.2000.6.245. [DOI] [PubMed] [Google Scholar]
- Roberts RB, de Lencastre A, Eisner W, Severina EP, Shopsin B, Kreiswirth BN, Tomasz A. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. MRSA Collaborative Study Group. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Tomasz A, Corso A, Hargrave J, Severina E. Penicillin-resistant Streptococcus pneumoniae in metropolitan New York hospitals: case control study and molecular typing of resistant isolates. Microb Drug Resist. 2001;7:137–152. doi: 10.1089/10766290152045011. [DOI] [PubMed] [Google Scholar]
- Sa-Leao R, Nunes S, Brito-Avo A, Frazao N, Simoes AS, Crisostomo MI, Paulo AC, Saldanha J, Santos-Sanches I, de Lencastre H. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin Microbiol Infect. 2009;15:1002–1007. doi: 10.1111/j.1469-0691.2009.02775.x. [DOI] [PubMed] [Google Scholar]
- Sa-Leao R, Sanches IS, Couto I, Alves CR, de Lencastre H. Low prevalence of methicillin-resistant strains among Staphylococcus aureus colonizing young and healthy members of the community in Portugal. Microb Drug Resist. 2001;7:237–245. doi: 10.1089/10766290152652783. [DOI] [PubMed] [Google Scholar]
- Santos Sanches I, Mato R, de Lencastre H, Tomasz A. Patterns of multidrug resistance among methicillin-resistant hospital isolates of coagulase-positive and coagulase-negative staphylococci collected in the international multicenter study RESIST in 1997 and 1998. Microb Drug Resist. 2000;6:199–211. doi: 10.1089/mdr.2000.6.199. [DOI] [PubMed] [Google Scholar]
- Silva S, Gouveia-Oliveira R, Maretzek A, Carrico J, Gudnason T, Kristinsson KG, Ekdahl K, Brito-Avo A, Tomasz A, Sanches IS, de Lencastre H, Almeida JJ. EURISWEB – Web-based epidemiological surveillance of antibiotic-resistant pneumococci in day care centers. BMC Med Inform Decis Mak. 2003;3:9. doi: 10.1186/1472-6947-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes RR, Aires-de-Sousa M, Conceicao T, Antunes F, da Costa PM, de Lencastre H. High prevalence of EMRSA-15 in Portuguese public buses: a worrisome finding. PLoS One. 2011;6:e17630. doi: 10.1371/journal.pone.0017630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares DA, Sa-Leao R, Miragaia M, de Lencastre H. Large screening of CA-MRSA among Staphylococcus aureus colonizing healthy young children living in two areas (urban and rural) of Portugal. BMC Infect Dis. 2010;10:110. doi: 10.1186/1471-2334-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. Streptococcus pneumoniae – Molecular Biology and Mechanisms of Disease – Update for the 1990s. Mary Ann Liebert, Inc; New York: 2000. Streptococcus pneumoniae: Functional anatomy; pp. 9–23. [Google Scholar]
- Tomasz A, Corso A, Severina EP, Echaniz-Aviles G, Brandileone MC, Camou T, Castaneda E, Figueroa O, Rossi A, Di Fabio JL. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American countries: an overview. PAHO/Rockefeller University Workshop Pan American Health Organization Microb. Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]
- Zemlickova H, Crisostomo MI, Brandileone MC, Camou T, Castaneda E, Corso A, Echaniz-Aviles G, Pasztor M, Tomasz A. Serotypes and clonal types of penicillin-susceptible Streptococcus pneumoniae causing invasive disease in children in five Latin American countries. Microb Drug Resist. 2005;11:195–204. doi: 10.1089/mdr.2005.11.195. [DOI] [PubMed] [Google Scholar]