Abstract
Intact ribosomal RNAs (rRNAs) comprise the majority of somatic transcripts, yet appear conspicuously absent in spermatozoa, perhaps reflecting cytoplasmic expulsion during spermatogenesis. To discern their fate, total RNA retained in mature spermatozoa from three fertile donors was characterized by Next Generation Sequencing. In all samples, >75% of total sequence reads aligned to rRNAs. The distribution of reads along the length of these transcripts exhibited a high degree of non-uniformity that was reiterated between donors. The coverage of sequencing reads was inversely correlated with guanine-cytosine (GC)-richness such that sequences greater than ∼70% GC were virtually absent in all sperm RNA samples. To confirm the loss of sequence, the relative abundance of specific regions of the 28S transcripts in sperm was established by 7-Deaza-2′-deoxy-guanosine-5′-triphosphate RT–PCR. The inability to amplify specific regions of the 28S sequence from sperm despite the abundant representation of this transcript in the sequencing libraries demonstrates that approximately three-quarters of RNA retained in the mature male gamete are products of rRNA fragmentation. Hence, cleavage (not expulsion of the RNA component of the translational machinery) is responsible for preventing spurious translation following spermiogenesis. These results highlight the potential importance of those transcripts, including many mRNAs, which evade fragmentation and remain intact when sperm are delivered at fertilization.
Sequencing data are deposited in GEO as: GSE29160.
Keywords: next generation sequencing, rRNA, sperm, spermiogenesis
Introduction
Initial investigations of sperm-retained RNAs largely focused on mRNAs, partly due to several lines of evidence suggesting they were the dominant class of transcript in the cell (Kumar et al., 1993; Chiang et al., 1994; Wykes et al., 1997; Ostermeier et al., 2002, 2004). The ability of sperm to store and protect spermatogenic messenger RNAs (mRNA) provided the rationale for the presence of any RNA despite transcriptional and translational quiescence (Kierszenbaum and Tres, 1975; Grunewald et al., 2005; reviewed in Krawetz, 2005). Subsequent studies have since attempted to identify mRNAs common to sperm from fertile men, which may be of clinical importance (Wang et al., 2004; Platts et al., 2007; Lalancette et al., 2009; Linschooten et al., 2009). In contrast, little is known about the fate of ribosomal RNAs (rRNAs) during and after spermiogenesis. These transcripts are the most abundant in all cells yet until now, assumed to be absent from the mature male gamete (reviewed in Krawetz, 2005). Instead of exhibiting defined 18S and 28S rRNA peaks, electrophoretic analysis of sperm RNAs reveals an enrichment of short-length transcripts (Ostermeier et al., 2002; Gilbert et al., 2007; Das et al., 2010). Although intact mRNAs are detected in this pool of transcripts, full-length 28S and 18S rRNAs are not (Ostermeier et al., 2005). Failure to observe rRNAs in sperm has been presumed to be due to the large reduction in cytoplasmic volume accompanying morphogenesis. Indeed, expulsion of the translational machinery during sperm maturation is supported by detection of RNA and ribosomal proteins in the residual body/cytoplasmic droplet (Breucker et al., 1985; Cappallo-Obermann et al., 2011). Together, these and other historical observations led to the assumption that spermatozoa lack rRNAs (Miller et al., 2005). However, the fate of these transcripts has recently begun to be reevaluated, suggesting the presence of rRNA components (Cappallo-Obermann et al., 2011).
Interrogation of sperm RNAs by Next Generation Sequencing has permitted an in-depth analysis, until now, could not be attained. This technology has recently been used to show that the male gamete harbors many RNA species in addition to mRNAs (Johnson et al., 2011). Herein, we describe the composition of rRNAs in sperm. Sequencing of sperm total RNA suggests that though not intact, these RNAs may be present at levels approaching that observed in somatic cells. The depletion of specific subregions of the 28S transcript supports the conclusion that rRNA cleavage, not expulsion during cellular condensation, is the primary means of preventing spurious translation of mRNAs stored in mature spermatozoa and delivered at fertilization.
Materials and Methods
RNA isolation and Next Generation Sequencing
Spermatozoal RNAs were isolated from ejaculates from three fertile donors, as described previously (Goodrich et al., 2007). Briefly, following the removal of contaminating somatic and residual cells by centrifugation through a 50% percoll gradient to obtain pure populations of spermatozoa, RNA was extracted from purified sperm using the RNEasy system (Qiagen, Valencia, CA, USA). Total testis RNAs were obtained from two commercial libraries (Applied Biosystems/Ambion, Austin, TX, USA, Lot# 054P010702031A; ClonTech, Mountain View, CA, USA, Lot# 3090051). RNA size and quality were assayed using the 2100 bioanalyzer with a RNA Pico chip (Agilent Technologies, Palo Alto, CA, USA). Separate RNA-seq libraries were constructed for each sample using the Illumina mRNA-Seq kit (Illumina, San Diego, CA, USA). Sperm libraries were prepared with and without poly[A+] selection. All libraries were subjected to paired-end sequencing using the Illumina Genome Analyzer GAIIx (Illumina, San Diego CA, USA) for 36 cycles. Image analysis and base calling were performed using the Firecrest and Bustard modules of the genome analyzer pipeline software (Illumina Pipeline software v. 1.3.0). Sequencing reads were aligned to 18S (NR_003286.2, 1869 nt length) and 28S (NR_003287.2, 5070 nt length) rRNA sequences. Alignments were performed with Novoalign (Novocraft Technologies SdnBhd, v. 2.05.43) using default parameters to align individual reads from a paired-end set separately. Alignment results were confirmed independently using the GERALD (Illumina). Only sequences uniquely aligning to the genome were considered since the addition of sequences that aligned to multiple regions had little effect. The number of sequence reads in each library was standardized as a function of the total number of reads obtained for that sample. To locally reduce non-uniformity, coverage at each position is shown as a sum of reads starting over the preceding 35 bases (Hansen et al., 2010).
RT–PCR analysis
Total testis and sperm RNAs (20 ng) were reverse-transcribed and amplified using the SuperScriptIII First-Strand Synthesis (Invitrogen, Carlsbad, CA, USA) and HotStarTaq (Qiagen) systems, essentially as described by the manufacturer. Following random hexamer priming, second-strand synthesis was performed at 55°C for 1 h. Each 60 µl PCR contained 6 µl of the reverse transcription reaction, 0.2 mM dNTPs, 0.6 units enzyme and 1 µM primer. Primer sequences are provided in Supplementary data, Table I. Cycling conditions were 94°C for 2 min, followed by 45 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 40 s. Use of dimethylsulfoxide (DMSO) and/or betaine in all reactions was not pursued as these additives failed to rescue amplification of guanine-cytosine (GC)-rich regions. However, these sequences could be amplified in the presence of 0.15 mM 7-Deaza-2′-deoxy-guanosine-5′-triphosphate (GTP) (Roche, Indianapolis, IN, USA) plus 0.05 mM dGTP in all reactions. Reaction aliquots were removed every five cycles and visualized with ethidium bromide. Quenching of intercalative dyes by the nucleotide analog necessitated gel densitometric analysis of product enrichment in place of quantitative real-time PCR (Pivonkova et al., 2010). Gel images were captured on the Typhoon 9210 (Molecular Dynamics) and were well within the optical linear range of response and thus analyzed with Quantity One version 4.5.2 (BioRad).
Results
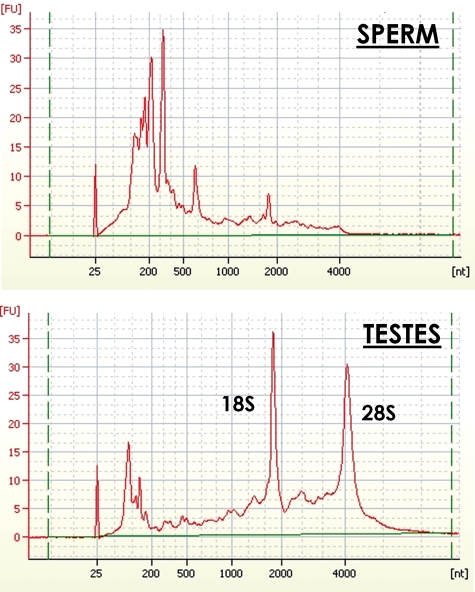
Despite robust transcription and translation during early spermiogenesis, it has been assumed that mature spermatozoa are depleted in rRNAs. This notion was supported by the transcriptionally silent state of the mature sperm, the expulsion of the majority of the cytoplasm as a droplet during maturation and the virtual complete absence of full-length 18S or 28S rRNAs following condensation (Fig. 1). As a hallmark of the mature male gamete, the lack of intact 18S and 28S rRNAs in sperm has been consistently observed in several species and is independent of extraction procedures (Ostermeier et al., 2002; Gilbert et al., 2007; Yang et al., 2009; Das et al., 2010). Whether depletion of these sequences is achieved by RNA cleavage and/or physical expulsion with bulk cytoplasm is not known. To discern the fate of these transcripts following spermiogenesis, total RNA retained in mature spermatozoa from three fertile donors was characterized by Next Generation Sequencing. The sperm RNA-seq libraries produced an average of 58.2 million sequence reads, of which ∼80% aligned to portions of the 18S and 28S rRNAs (Table I). This establishes the 18S and 28S rRNAs as the most abundant spermatozoal transcripts. However, as indicated earlier, the electrophoretic analysis of sperm total RNAs demonstrated that though highly abundant, these rRNAs are not intact (Fig. 1). The 5S and 5.8S RNAs corresponded to <0.1% of the total number of sequence reads and were not considered further. Similarly, the mitochondrial 16S and 12S rRNAs contributed to only 1.8 and 0.8% of total sequencing reads, respectively and were not considered further. This establishes the 18S and 28S rRNAs as representing the most abundant spermatozoal transcripts. As shown in Fig. 1, these abundant rRNAs do not electrophoretically resolve as discrete peaks. This is consonant with the recent proposal that high-quality sperm samples can be identified by a substantially lower ratio of 28S:18S rRNAs (0.11) than observed in somatic cells (Cappallo-Obermann et al., 2011). However, the total number of sequencing reads that mapped to either the 28S or 18S transcripts was proportional to their respective lengths and thus in the expected ratio of 2:1. As shown below, this apparent inconsistency reflects the fragmentation of the majority of the sperm RNAs. This highlights the significance of those mRNAs and small non-coding RNAs that are consistently found intact.
Figure 1.
Electrophoretic size distribution of RNAs in sperm and testis from human fertile donors. Total RNA from sperm and testes was heat-denatured in the presence of ETDA. RNAs were then resolved on an Agilent Bioanalyzer.
Table I.
Percentage of Next Generation Sequencing reads of sperm and testes RNAs mapping to 18S and 28S rRNA.
| Samples | Total readsa | Reads aligned to 18S rRNAa | % Aligned to 18S rRNA | Reads aligned to 28S rRNAa | % Aligned to 28S rRNA | % rRNA of total RNA |
|---|---|---|---|---|---|---|
| Donor 1 | 52.15 | 11.49 | 22.03 | 31.37 | 60.15 | 82.2 |
| Donor 2 | 64.45 | 14.85 | 23.1 | 41.66 | 64.75 | 87.8 |
| Donor 3 | 58.11 | 13.60 | 23.4 | 30.98 | 53.3 | 76.7 |
| Donor 1b | 22.99 | 0.24 | 1.03 | 0.86 | 3.74 | 4.8 |
| Testis CloneTechb | 20.96 | 0.21 | 0.99 | 0.67 | 3.2 | 4.17 |
| Testis Ambionb | 21.94 | 0.089 | 0.40 | 0.23 | 1.03 | 1.44 |
aValues denote the number of high-quality sequencing reads in millions.
bPoly[A+]-enriched samples (shaded).
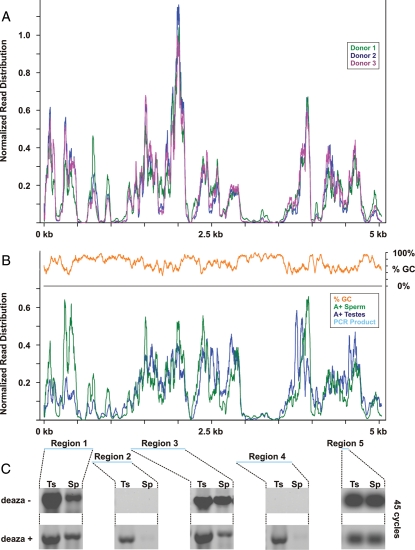
The distribution of sequencing reads across these transcripts revealed a consistent series of peaks and valleys reiterated between donors (Fig. 2A; Supplementary data, Fig. S1). Notably, several subregions of the 28S RNA were extremely read-poor suggesting their depletion in sperm. To determine whether the absence of select rRNA sequences was a previously undescribed feature of the mature male gamete, sperm and testes poly[A+]-enriched RNAs were compared by RNA-seq. As expected, poly[A+] selection diminished the representation of the 18S and 28S transcripts to <3% of that observed in the total RNAs. However, the distribution of reads across the transcripts was essentially identical to that observed in sperm prior to enrichment and strikingly concordant with that observed in the testes libraries after selection (Fig. 2B; Supplementary data, Figs S2–4, Table I).
Figure 2.
The abundance of specifically cleaved rRNA in sperm compared with those in testes. The normalized distribution of sequencing reads across the 28S rRNA are shown for (A) sperm total RNAs from three fertile donors and (B) sperm and testis poly[A+]-enriched RNA-seq libraries. For all libraries, sequencing coverage was negatively correlated with GC content (B, orange). Regions subsequently interrogated by RT–PCR for total testes (Ts) and sperm (Sp) RNAs are shown in blue and their corresponding reaction products after 45 cycles presented below (C). Note that amplification of Regions 1, 3 and 5 was not affected by replacement of dGTP with nucleotide analog 7-deaza-GTP though signal from these products was uniformly reduced, compare deaza- and deaza+. Note that Regions 2 and 4 could only be amplified when the deaza analog was present.
Paradoxically, despite the presence of intact rRNAs in testis and their absence from sperm (Fig. 1), specific regions of the 18S and 28S transcripts were underrepresented. Approximately half of 28S rRNA is ≥70% GC (36 nt window and 1 nt step). Sequences exceeding this threshold were markedly reduced or absent from both the testis and sperm libraries (Fig. 2B, orange line). This trend was also observed along the 18S transcript (∼10% ≥ 70% GC; Supplementary data, Fig. S4). The ∼340 nt 28S RNA region bounded by nucleotide coordinates 3000–3500 underscores the relationship between GC richness and suppressed representation in the sequencing libraries. Thus, the simple comparison of 18S and 28S sequencing coverage in sperm and testis RNA-seq libraries was not sufficient to reveal potential sites of cleavage. It has been suggested that the underrepresentation of extremely GC-rich genomic sequences may reflect PCR amplification bias. In these cases, loss of coverage was circumvented by either altering cycling conditions or forgoing enrichment (Kozarewa et al., 2009; Aird et al., 2011; Kozarewa and Turner, 2011). However, adapting this approach to the rRNAs did not normalize coverage across the 18S and 28S rRNAs (Mamanova et al., 2010). The persistent absence of GC-rich subregions within these transcripts suggests that lack of sequencing coverage may reflect the tendency of these highly structured RNAs to intramolecular base-pair (Wellauer and Dawid, 1973; Wakeman and Maden, 1989). Stably folded RNAs can occlude primers and reverse transcriptase, leading to their exclusion from further analysis, thus marginalizing their representation during sequencing (Zhang et al., 2001; Mortazavi et al., 2008). Although this concern is generally mitigated by fragmentation preceding cDNA synthesis, it is evident that some transcripts cannot be resolved using current sequencing protocols (Mortazavi et al., 2008).
Juxtaposing the absence of intact 18S and 28S rRNA in the mature male gamete with their abundance in the sperm sequencing data suggested that those transcripts are fragmented during spermiogenesis. To localize potential sites of cleavage, primers were designed to five specific subregions along the 28S rRNA transcript (Fig. 2B, blue horizontal bars; Supplementary data, Table SI) and relative levels of each specific segment was compared by RT–PCR. The intact testis template served as a baseline to evaluate the relative depletion of these regions in sperm. Several products were readily detected at varying levels in both testis and sperm (Region 1, 3–733 nt; Region 3, 1301–2108 nt and Region 5, 4441–4548 nt; Fig. 2C, deaza−). As expected, sequences with a GC content exceeding 70% were not amplified in either testis or sperm (Region 2, 716–1319 nt; Region 4, 2871–3699 nt). These directly overlap the highly folded regions of the 28S rRNA and represent the largest gaps in sequencing coverage along this transcript (Fig. 2B; Wellauer and Dawid, 1973; Wakeman and Maden, 1989). Initial efforts to minimize RNA secondary structure, including increasing reaction temperatures, the addition of DMSO and/or betaine, did not rescue amplification of these regions (data not shown). However, partial substitution of 7-deaza-GTP for dGTP in all reactions resulted in robust amplification of all sequences from testis RNAs (Fig. 2C, deaza+). Regions 1 and 3 were more abundant in testis than in sperm, while Region 5 exhibited comparable levels in both cell-types. Even after 45 cycles of amplification, Regions 2 and 4 could not be detected in sperm (Fig. 2C, deaza-). In contrast, amplification of these products from the testis template proceeded to saturation and was easily detected after cycle 30. These results establish the absence of specific regions of the 28S rRNA in sperm in accordance with the view that they harbor preferential sites of cleavage. Further, the requirement of 7-deaza-GTP to resolve these sequences by RT–PCR shows that highly base-paired GC-rich regions are underrepresented and thus excluded from cDNA synthesis. This is consistent with the use of denaturants like CH3HgOH to ensure the complete reverse transcription and synthesis of cDNAs from GC-rich RNAs. (Krawetz et al., 1986).
Discussion
Complimenting Next Generation Sequencing analysis of sperm RNAs with RT–PCR interrogation of select regions of the 28S rRNA demonstrated that these transcripts are highly abundant though fragmented in the mature male gamete. Percoll selection prior to RNA isolation ensured that sequencing reads were representative of RNAs from within purified sperm and not somatic contaminants nor transcripts within the residual body. The presence of intact full-length RNAs in sperm and the absence of specific subregions of the 28S transcript support the view that failure to resolve full-length rRNAs from the male gamete is not due to misdirected nuclease activity. Rather, selective cleavage of the rRNAs likely ensures translational cessation, which clearly cannot be achieved by cytoplasmic expulsion alone.
The regions of the 28S rRNA that are preferentially cleaved in sperm have been implicated in rRNA fragmentation in other systems. Region 4 highlighted in Fig. 2 is a likely target for initial nuclease attack, since it is positioned on the surface of intact ribosomes (Han et al., 1994; Holmberg et al., 1994) and is expected to contain an exposed loop that can be preferentially cleaved prior to association with the ribosomal proteins (Leffers and Andersen, 1993; Houge et al., 1995). In comparison, Region 2, the other sequence absent from sperm, appears to be positioned internally within the ribosome (Han et al., 1994; Holmberg et al., 1994). In this case, the initial cleavage of external sequences may induce a conformational change, thereby exposing internal regions to nuclease digestion or rendering it amenable to autocatalysis (Houge et al., 1995). Regardless, the quantity of cleavage products remaining in sperm indicates that fragmentation of these transcripts does precede their complete degradation. Further, evidence of post-transcriptional polyadenylation that is proposed to target rRNA endonucleolytic cleavage products for degradation in human somatic cells was not apparent in the sequenced sperm rRNAs (Slomovic et al., 2006). Complete abolition of these transcripts should not be necessary in the male gamete as long as translational cessation is assured.
The abundance of rRNA remnants in mature sperm indicates that expulsion through the cytoplasmic droplet is not complete. Accordingly, this mechanism is not sufficient to prevent spurious protein synthesis that is only assured by fragmentation. This is of particular significance, considering that several spermatozoal mRNAs and small non-coding RNAs evade cleavage and are delivered by sperm to the oocyte. On the one hand, some, like the protamines, typify those necessary for repackaging the sperm nucleus and must remain silent to avoid deleterious changes to the chromatin structure (Avendano et al., 2009; Johnson et al., 2011) of the zygote. This would be assured by the destruction of spermatozoal ribosomes enabling the clearance of detrimental transcripts following fertilization. On the other hand, paternal RNAs required for early development may be protected from such pathways prior to engaging the maternal translational machinery. Perhaps, this provides a means to ensure compatibility of the gametes, as part of the consolidation confrontation pathway (Bourc'his and Voinnet, 2010).
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
G.D.J. performed the experiments and C.L. created the sperm RNA sequencing library. E.S., G.D.J. and S.A.K. designed the experiments, analyzed the data and wrote the paper. M.D. and R.H. provided the samples and contributed to the discussion of the data and editing of the manuscript.
Funding
This work was supported, in part, by a pilot grant to SAK and RH from Harvard School of Public Health; National Institutes of Health (grant number ES017285); National Institute of Environmental Health Sciences (grant number ES009718); the Presidential Research Enhancement Program in Computational Biology and the Charlotte B. Failing Professorship (S.A.K.).
Supplementary Material
References
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. doi:10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendano C, Franchi A, Jones E, Oehninger S. Pregnancy-specific {beta}-1-glycoprotein 1 and human leukocyte antigen-E mRNA in human sperm: differential expression in fertile and infertile men and evidence of a possible functional role during early development. Hum Reprod. 2009;24:270–277. doi: 10.1093/humrep/den381. doi:10.1093/humrep/den381. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. doi:10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- Breucker H, Schafer E, Holstein AF. Morphogenesis and fate of the residual body in human spermiogenesis. Cell Tissue Res. 1985;240:303–309. doi: 10.1007/BF00222339. [DOI] [PubMed] [Google Scholar]
- Cappallo-Obermann H, Schulze W, Jastrow H, Baukloh V, Spiess A-N. Highly purified spermatozoal RNA obtained by a novel method indicates an unusual 28S/18S rRNA ratio and suggests impaired ribosome assembly. Mol Hum Reprod. 2011 doi: 10.1093/molehr/gar037. (2011) first published online May 18 doi:10.1093/molehr/gar037. [DOI] [PubMed] [Google Scholar]
- Chiang MH, Steuerwald N, Lambert H, Main EK, Steinleitner A. Detection of human leukocyte antigen class I messenger ribonucleic acid transcripts in human spermatozoa via reverse transcription-polymerase chain reaction. Fertil Steril. 1994;61:276–280. [PubMed] [Google Scholar]
- Das PJ, Paria N, Gustafson-Seabury A, Vishnoi M, Chaki SP, Love CC, Varner DD, Chowdhary BP, Raudsepp T. Total RNA isolation from stallion sperm and testis biopsies. Theriogenology. 2010;74:1099–1106. doi: 10.1016/j.theriogenology.2010.04.023. 106e1–2 doi:10.1016/j.theriogenology.2010.04.023. [DOI] [PubMed] [Google Scholar]
- Gilbert I, Bissonnette N, Boissonneault G, Vallee M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133:1073–1086. doi: 10.1530/REP-06-0292. doi:10.1530/REP-06-0292. [DOI] [PubMed] [Google Scholar]
- Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53:161–167. doi: 10.1080/01485010701216526. doi:10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Glander HJ, Anderegg U. Mature human spermatozoa do not transcribe novel RNA. Andrologia. 2005;37:69–71. doi: 10.1111/j.1439-0272.2005.00656.x. doi:10.1111/j.1439-0272.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- Han H, Schepartz A, Pellegrini M, Dervan PB. Mapping RNA regions in eukaryotic ribosomes that are accessible to methidiumpropyl-EDTA.Fe(II) and EDTA.Fe(II) Biochemistry. 1994;33:9831–9844. doi: 10.1021/bi00199a004. doi:10.1021/bi00199a004. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. doi:10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg L, Melander Y, Nygard O. Probing the structure of mouse Ehrlich ascites cell 5.8S, 18S and 28S ribosomal RNA in situ. Nucleic Acids Res. 1994;22:1374–1382. doi: 10.1093/nar/22.8.1374. doi:10.1093/nar/22.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houge G, Robaye B, Eikhom TS, Golstein J, Mellgren G, Gjertsen BT, Lanotte M, Doskeland SO. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol. 1995;15:2051–2062. doi: 10.1128/mcb.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011;141:21–36. doi: 10.1530/REP-10-0322. doi:10.1530/REP-10-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. Structural and transcriptional features of the mouse spermatid genome. J Cell Biol. 1975;65:258–270. doi: 10.1083/jcb.65.2.258. doi:10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I, Turner DJ. Amplification-free library preparation for paired-end Illumina Sequencing. Methods Mol Biol. 2011;733:257–266. doi: 10.1007/978-1-61779-089-8_18. doi:10.1007/978-1-61779-089-8_18. [DOI] [PubMed] [Google Scholar]
- Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free illumina sequencing-library preparation facilitates improved mapping and assembly of (G + C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. doi:10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6:633–642. doi: 10.1038/nrg1654. doi:10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- Krawetz SA, Connor W, Cannon PD, Dixon GH. A vector-primer-cloner-sequencer plasmid for the construction of cDNA libraries: evidence for a rat glyceraldehyde-3-phosphate dehydrogenase-like mRNA and a ferritin mRNA within testis. DNA. 1986;5:427–435. doi: 10.1089/dna.1986.5.427. doi:10.1089/dna.1986.5.427. [DOI] [PubMed] [Google Scholar]
- Kumar G, Patel D, Naz RK. c-MYC mRNA is present in human sperm cells. Cell Mol Biol Res. 1993;39:111–117. [PubMed] [Google Scholar]
- Lalancette C, Platts AE, Johnson GD, Emery BR, Carrell DT, Krawetz SA. Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med. 2009;87:735–748. doi: 10.1007/s00109-009-0485-9. doi:10.1007/s00109-009-0485-9. [DOI] [PubMed] [Google Scholar]
- Leffers H, Andersen AH. The sequence of 28S ribosomal RNA varies within and between human cell lines. Nucleic Acids Res. 1993;21:1449–1455. doi: 10.1093/nar/21.6.1449. doi:10.1093/nar/21.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linschooten JO, Van Schooten FJ, Baumgartner A, Cemeli E, Van Delft J, Anderson D, Godschalk RW. Use of spermatozoal mRNA profiles to study gene-environment interactions in human germ cells. Mutat Res. 2009;667:70–76. doi: 10.1016/j.mrfmmm.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Mamanova L, Andrews RM, James KD, Sheridan EM, Ellis PD, Langford CF, Ost TW, Collins JE, Turner DJ. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. doi:10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Ostermeier GC, Krawetz SA. The controversy, potential and roles of spermatozoal RNA. Trends Mol Med. 2005;11:156–163. doi: 10.1016/j.molmed.2005.02.006. doi:10.1016/j.molmed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. doi:10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–777. doi: 10.1016/S0140-6736(02)09899-9. doi:10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. doi:10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Diamond MP, Dix DJ, Krawetz SA. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil Steril. 2005;83:1687–1694. doi: 10.1016/j.fertnstert.2004.12.046. doi:10.1016/j.fertnstert.2004.12.046. [DOI] [PubMed] [Google Scholar]
- Pivonkova H, Horakova P, Fojtova M, Fojta M. Direct voltammetric analysis of DNA modified with enzymatically incorporated 7-deazapurines. Anal Chem. 2010;82:6807–6813. doi: 10.1021/ac100757v. doi:10.1021/ac100757v. [DOI] [PubMed] [Google Scholar]
- Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–773. doi: 10.1093/hmg/ddm012. doi:10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. doi:10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman JA, Maden BE. 28 S ribosomal RNA in vertebrates. Locations of large-scale features revealed by electron microscopy in relation to other features of the sequences. Biochem J. 1989;258:49–56. doi: 10.1042/bj2580049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhou Z, Xu M, Li J, Xiao J, Xu ZY, Sha J. A spermatogenesis-related gene expression profile in human spermatozoa and its potential clinical applications. J Mol Med. 2004;82:317–324. doi: 10.1007/s00109-004-0526-3. doi:10.1007/s00109-004-0526-3. [DOI] [PubMed] [Google Scholar]
- Wellauer PK, Dawid IB. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Nat Acad Sci USA. 1973;70:2827–2831. doi: 10.1073/pnas.70.10.2827. doi:10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes SM, Visscher DW, Krawetz SA. Haploid transcripts persist in mature human spermatozoa. Mol Hum Reprod. 1997;3:15–19. doi: 10.1093/molehr/3.1.15. doi:10.1093/molehr/3.1.15. [DOI] [PubMed] [Google Scholar]
- Yang CC, Lin YS, Hsu CC, Wu SC, Lin EC, Cheng WTK. Identification and sequencing of remnant messenger RNAs found in domestic swine (Sus scrofa) fresh ejaculated spermatozoa. Anim Reprod Sci. 2009;113:143–155. doi: 10.1016/j.anireprosci.2008.08.012. doi:10.1016/j.anireprosci.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Pan HY, Gao SJ. Reverse transcription slippage over the mRNA secondary structure of the LIP1 gene. Biotechniques. 2001;31:1286. doi: 10.2144/01316st02. , 8, 90, passim. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.