Abstract
Quinone reductases (QRDs) have two important functions in the basidiomycete Gloeophyllum trabeum, which causes brown rot of wood. First, a QRD is required to generate biodegradative hydroxyl radicals via redox cycling between two G. trabeum extracellular metabolites, 2,5-dimethoxyhydroquinone (2,5-DMHQ) and 2,5-dimethoxy-1,4-benzoquinone (2,5-DMBQ). Second, because 2,5-DMBQ is cytotoxic and 2,5-DMHQ is not, a QRD is needed to maintain the intracellular pool of these metabolites in the reduced form. Given their importance in G. trabeum metabolism, QRDs could prove useful targets for new wood preservatives. We have identified two G. trabeum genes, each existing in two closely related, perhaps allelic variants, that encode QRDs in the flavodoxin family. Past work with QRD1 and heterologous expression of QRD2 in this study confirmed that both genes encode NADH-dependent, flavin-containing QRDs. Real-time reverse transcription PCR analyses of liquid- and wood-grown cultures showed that qrd1 expression was maximal during secondary metabolism, coincided with the production of 2,5-DMBQ, and was moderately up-regulated by chemical stressors such as quinones. By contrast, qrd2 expression was maximal during fungal growth when 2,5-DMBQ levels were low, yet was markedly up-regulated by chemical stress or heat shock. The total QRD activity in lysates of G. trabeum mycelium was significantly enhanced by induction beforehand with a cytotoxic quinone. The promoter of qrd2 contains likely antioxidant, xenobiotic, and heat shock elements, absent in qrd1, that probably explain the greater response of qrd2 transcription to stress. We conclude from these results that QRD1 is the enzyme G. trabeum routinely uses to detoxify quinones during incipient wood decay and that it could also drive the biodegradative quinone redox cycle. However, QRD2 assumes a more important role when the mycelium is stressed.
Brown rot basidiomycetes cause a highly destructive type of wood decay and are important lignocellulose recyclers in forest ecosystems (7, 19, 32). During early brown rot, cellulose is rapidly oxidized and depolymerized, even though the porosity of sound wood is too low for enzymes such as cellulases to penetrate (6). This observation suggests that small oxidants rather than enzymes initiate brown rot. Numerous studies indicate that these oxidants include hydroxyl radicals produced via Fenton chemistry (H2O2 + Fe2+ + H+ → H2O + Fe3+ + •OH) (8, 9, 29). It follows that brown rot fungi require extracellular mechanisms to reduce Fe3+ and O2, the forms of iron and oxygen they generally encounter, if they are to degrade wood by this mechanism.
Recent work has shown that two brown rot fungi, Gloeophyllum trabeum and Postia placenta, generate a hydroquinone-driven Fenton system. They produce extracellular 2,5-dimethoxyhydroquinone (2,5-DMHQ), which rapidly reduces both Fe3+ and O2 to give 2,5-dimethoxy-1,4-benzoquinone (2,5-DMBQ) (5, 11, 14, 21). Experiments with G. trabeum have shown that it drives this chemistry by continuously reducing extracellular 2,5-DMBQ to extracellular 2,5-DMHQ, thus assuring a steady supply of electrons for the reduction of Fe3+ and O2 (11, 14). Therefore, the extracellular Fenton system of G. trabeum must be catalyzed by a quinone reductase (QRD).
There is an additional reason why QRDs are important enzymes in G. trabeum. 2,5-DMBQ and many other quinones are cytotoxic because they readily accept one electron from intracellular reductants to form semiquinones, which rapidly reduce O2 to produce superoxide (22). Moreover, the electron-deficient carbonyl groups of quinones make them good electrophiles that can alkylate proteins and DNA (2). Hydroquinones, by contrast, do not have these properties, and therefore organisms generally reduce quinones to detoxify them. G. trabeum is likely to require an efficient reductive system to detoxify the large amounts of 2,5-DMBQ that it produces during biodegradation.
In most organisms, flavin-containing QRDs have a major role in quinone detoxification. Animals produce DT diaphorase, an FAD-containing NAD(P)H:QRD that is part of an inducible set of enzymes, termed the phase 2 system, which protects the organism against chemical carcinogenesis (10, 26). In plants and fungi, quinone detoxification is evidently catalyzed by a different group of flavin mononucleotide-containing NAD(P)H:QRDs that are homologous to flavodoxin (1, 12, 16, 18). The genes that encode all of these QRDs are up-regulated by quinones and, in some cases, by other chemical stressors.
Since brown rot QRDs are required for both biodegradation and detoxification, it may eventually be possible to use QRD inhibitors to prevent the growth of these fungi on wood. There is a need for such relatively specific control agents because most of the wood preservatives in current use are highly toxic, general biocides. With this goal in mind, we recently characterized a flavodoxin NADH:QRD from G. trabeum that might detoxify 2,5-DMBQ, drive Fenton chemistry, or perhaps fill both roles (12). We have now identified an additional G. trabeum flavodoxin QRD. Both enzymes probably detoxify quinones, but they are expressed at different stages of growth and the genes that encode them are differentially regulated in response to chemical stress and heat shock.
MATERIALS AND METHODS
Chemicals and organism.
dl-4-Methylsulfinylbutyl isothiocyanate (DL-sulforaphane) was purchased from ICN Biomedicals (Aurora, Ohio). All other chemicals were obtained from Sigma/Aldrich (St. Louis, Mo.), including 2,6-dimethoxybenzoquinone (2,6-DMBQ), 2-methyl-1,4-naphthoquinone (menadione), butylated hydroxyanisole (BHA), and 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT).
Stock cultures of G. trabeum (ATCC 11539) were maintained on agar plates that contained (per liter) 10 g of glucose, 10 g of malt extract, 2 g of peptone, 2 g of yeast extract, 1 g of asparagine, 2 g of KH2PO4, 1 g of MgSO4 · 7H2O, and 0.001 g of thiamine.
G. trabeum in liquid medium.
Static liquid cultures of G. trabeum were grown at 31°C under air in a nitrogen-limited medium previously described (11). The medium was inoculated with arthrospores, which were collected from 14-day-old agar plates of the fungus by adding 10 ml of sterile, deionized, distilled water (ddH2O) to each plate and then filtering the suspension through sterile glass wool. For time-course experiments, 125-ml Erlenmeyer flasks, each containing 7 ml of medium, were inoculated with 1.0 × 106 spores. For experiments on the induction of QRD genes, 250-ml Erlenmeyer flasks, each containing 9 ml of medium, were inoculated with 1.3 × 106 spores.
The fungal biomass in time-course experiments was determined by drying harvested mycelial mats to constant weight at 55°C under vacuum. To determine the total 2,5-DMBQ plus 2,5-DMHQ concentration in these cultures, extracellular medium was harvested at 2-day intervals, oxidized with FeCl3, and analyzed for the quinone by high-performance liquid chromatography as described earlier (12).
To monitor rates of quinone reduction by the intact fungus in time-course experiments, one G. trabeum mycelial mat (washed beforehand with 100 ml of ddH2O), 100 μM 2,6-DMBQ, and 40 mM sodium oxalate buffer (pH 4.5) were combined in a final volume of 20 ml and shaken at 120 rpm in a 50-ml beaker at 25°C for 25 min. Samples (0.5 ml) were withdrawn every 5 min, and the decrease in absorbance at 288 nm due to 2,6-DMBQ reduction was recorded (ɛ288 = 7.12 mM−1 cm−1).
G. trabeum on wood.
Solid-state cultures of G. trabeum were grown at 31°C under air on blocks of spruce wood (3 by 10 by 30 mm). The blocks were autoclaved twice (121°C at 15 lb/in2 for 1 h) and then dried under vacuum in a sterile desiccator jar. Each block was infused with 300 μl of autoclaved 20% (wt/vol) potato dextrose broth. For each culture, three blocks were then placed on a piece of sterile polypropylene mesh over 25 ml of potato dextrose agar in a 250-ml Erlenmeyer flask that had been inoculated 5 days beforehand with G. trabeum. An additional agar plug inoculum of the fungus was placed on top of each block. Under these conditions, the blocks became completely covered with mycelium within 6 days.
To determine the total 2,5-DMBQ plus 2,5-DMHQ concentration in solid-state cultures, triplicate colonized wood blocks were combined, pulverized in an electric coffee grinder, and stirred in ddH2O at 80°C for 30 min. The mixture was then filtered, oxidized, and assayed by high-performance liquid chromatography (12). Final quinone concentrations were calculated based on the dry weight of the wood samples.
Isolation of nucleic acids and cDNA preparation.
Genomic DNA (gDNA) was isolated from G. trabeum mycelium as described earlier (31). For total RNA purification, one colonized wood block or mycelia from three to seven liquid cultures were ground with a mortar and pestle under liquid nitrogen, and 80 mg of material was worked up with an RNeasy kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. The resulting RNA samples were then treated with RNase-free DNase (Promega, Madison, Wis.) as outlined by the manufacturer. DNA and RNA concentrations were determined spectrophotometrically with a GeneQuant RNA/DNA calculator (Amersham, Piscataway, N.J.).
First-strand cDNA was prepared from RNA by using avian myeloblastosis virus reverse transcriptase (Promega). The reaction mixture (20 μl) contained 1 μg of total RNA, 5 mM MgCl2, 10 mM Tris-HCl (pH 9.0), 50 mM KCl, a 1 mM concentration of each deoxynucleoside triphosphate, 1 U of recombinant RNase inhibitor/μl, 15 U of reverse transcriptase/μl, and 0.5 μg of RNA/μg of random primers. The temperature program for the reaction was as follows: 25°C for 10 min, 42°C for 15 min, 95°C for 5 min, and 5°C for 5 min. Finally, the reaction mixture was diluted 1:1 with RNase-free ddH2O and a 5-μl sample was used for real-time reverse transcription-PCR (RT-PCR) analyses as described below.
Gene identification and characterization.
Oligonucleotide primers were obtained from the University of Wisconsin Biotechnology Center. To characterize a portion of the glyceraldehyde-3-phosphate dehydrogenase gene (gpd) in G. trabeum, we designed degenerate primers that were based on cDNA sequence comparisons between the gpd genes of Phanerochaete chrysosporium (GenBank accession no. M81754), Schizophyllum commune (M81724), Lentinus edodes (AB013136), and Agaricus bisporus (M81728). A forward primer (5′-GGTCGYATYGGCCGYATYGT-3′) and reverse primer (5′-ATRACCTTKCCGACRGCCTT-3′) were used to amplify G. trabeum cDNA (629 bp) and gDNA (750 bp) products by PCR. The PCR mixtures (50 μl; Promega) contained 60 pmol of degenerate primers, and the temperature program was as follows: a denaturation step at 94°C for 3 min followed by 35 cycles that consisted of 94°C for 30 s, 54°C for 1 min, and 72°C for 2 min and finally an extension step at 68°C for 7 min. The amplified cDNA and gDNA fragments were cloned and sequenced.
To find qrd genes, we used conserved regions in G. trabeum QRD1 (GenBank accession no. AAL67860) and the previously reported P. chrysosporium QRD (AAD21025) to design a degenerate forward primer (5′-CARTGGAARGCNTTYTGGGAYDSNAC-3′) and a degenerate reverse primer (5′-CKNSWNCCRTCNSHNYYNGCRAANGT-3′) for PCR amplifications of G. trabeum gDNA. PCR conditions were as described above for gpd. We obtained a PCR product of 250 bp, which was cloned and sequenced to reveal the presence of both qrd1 and qrd2. Full-length qrd2 from the genomic sequence was obtained by using the Universal Genome Walker kit and Advantage GC Genomic Polymerase mix from Clontech (Palo Alto, Calif.) with the forward primer 5′-ATCGCCGCGATCGCGGTG-3′ and reverse primer 5′-CAACTTCCCCGCGCAGTGGAAGGTA-3′. A high-fidelity polymerase, Pfu (Stratagene, LaJolla, Calif.), was then used to PCR amplify the full-length gDNA clone by using the forward primer 5′-ACTGATGATTGCGCGATAAT-3′ and reverse primer 5′-CAGTCCTTCTATACATGCGA-3′. Total RNA from liquid- and wood-grown G. trabeum was used with Pfu to amplify cDNA by using the forward primer 5′-AAGGTCGCTATTGCTATCTAC-3′ and reverse primer 5′-GACCTTGGAGACGGTCTCGTA-3′. PCR conditions were as described above but with 10 pmol of each primer.
To clone the promoter region of each gene, we used the Universal Genome Walker kit with specific reverse primers for each gene as follows: qrd1A, 5′-CACTAGATTATGGACGCTCACCACA-3′; qrd1B, 5′-CTCTAGATTATGGACGCTTACCACC-3′; qrd2A, 5′-GAATACCATCACTCAAAGACCTCCCG-3′; and qrd2B, 5′-CATTGAGGTTGGTGAGCTGCG-3′.
Purification of DNA from agarose gels was done with a GENECLEAN Spin kit (Bio 101, Carlsbad, Calif.). Cloning was done with the pGEM-T Easy system (Promega), and either Escherichia coli DH5α or E. coli TOP10F' (Invitrogen, Carlsbad, Calif.) was used as the host for plasmids. Minipreps were done with a Wizard Plus SV Minipreps system (Promega). Sequencing was done on an ABI 3700 instrument at the University of Wisconsin Biotechnology Center, using BigDye dideoxy termination reactions. Sequence analysis was done with GCG Wisconsin Package software and with the ClustalW program (http://www.ebi.ac.uk/clustalw).
Northern hybridizations.
PCR products from cDNAs of qrd1, qrd2, and gpd were used to prepare 32P-labeled probes by using a Random Primed StripAble DNA Probe Synthesis kit (Ambion, Austin, Tex.) with [α-32P]dATP (3,000 Ci/mmol; Amersham). Total RNA (10 μg per lane) was resolved on agarose gels that contained 2% formaldehyde. The RNA was then transferred to a positively charged nylon membrane (Roche, Indianapolis, Ind.). Northern hybridization was done with a Northern Max kit (Ambion) and 106 cpm of [α-32P]dATP per ml of hybridization solution overnight at 42°C. The membranes were washed under low-stringency (25°C) and high-stringency (45°C) conditions according to the manufacturer's instructions and then exposed to XAR film (Kodak, Rochester, N.Y.) for 4.5 h at ambient temperature.
Real-time RT-PCR.
Relative quantitations of gene expression were done by using Taqman real-time RT-PCR on an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, Calif.). This relatively new technique is highly sensitive, and the simultaneous use of two specific primers and one specific probe for each gene confers high specificity when closely related genes are analyzed (3). The primers and probes (Table 1) were designed with PrimerExpress software from Applied Biosystems and were purchased from that company. The probes spanned the junction of two exons to ensure discrimination between cDNA and gDNA. Each probe was labeled at the 5′ end with the reporter dye FAM (6-carboxyfluorescein) and at the 3′ end with the quencher dye TAMRA (6-carboxytetramethylrhodamine).
TABLE 1.
Sequences of real-time RT-PCR primers and Taqman probes
| Gene | Primer or probe | Sequence (5′ → 3′) | Length (bp) |
|---|---|---|---|
| gpd | Forward | CGTCGTGGGTGTCAACTTAGAC | 22 |
| Reverse | GGCGCAAGGCAGTTGGT | 17 | |
| Probe | ACCCCAAGTACACTGTCGTCTCCAATGC | 28 | |
| qrd1 | Forward | CGAGGCCATCAAGAGTGGTATT | 22 |
| Reverse | GCCTTCACCAGGTTCAAAATCT | 22 | |
| Probe | CGCTTCCATCTTCCAGGTTGCAGAGACT | 28 | |
| qrd2 | Forward | TACTCGCTGTACGGCCACATC | 21 |
| Reverse | AGCGCTCTCGATACCGGAC | 19 | |
| Probe | CCAAGCTCGCCGAGGCTGTCA | 21 |
PCRs contained an 800 nM concentration of each primer, a 250 nM concentration of the Taqman probe, 5 μl of cDNA, and 12.5 μl of Taqman Universal PCR Mastermix (Applied Biosystems) in a final volume of 25 μl. The samples were placed in 96-well plates and amplified in the ABI Prism 7000 sequence detection system. Amplification conditions were as follows: 2 min at 50°C, 10 min at 95°C, and then 40 cycles that consisted of 15 s at 95°C and 1 min at 60°C.
We used the standard curve method according to the manufacturer's instructions (Applied Biosystems User Bulletin no. 2) to calculate relative expression levels for qrd1, qrd2, and gpd. We used gpd as the housekeeping gene because real-time RT-PCR analyses showed that its expression relative to total RNA did not change significantly under any of the conditions we employed. cDNA samples were prepared from 2-day-old liquid cultures, diluted serially 10-fold, and amplified by real time RT-PCR. The cycle threshold was plotted against the log10 of the cDNA dilution factor. The calculated slopes were within 2% of the theoretical slope, −log10 2, for all three genes.
These standard curves were used to determine the expression of qrd1, qrd2, and gpd in each sample, and the qrd values were normalized by dividing them by the gpd value. In the time-course experiment, the normalized values for qrd expression were then calibrated to the expression of qrd1 on day 2, which was set to 1. In induction experiments, the normalized values for qrd expression were expressed as n-fold differences relative to the expression values in the untreated controls, which were set to 1. The standard deviations for the final relative expression values were calculated as a composite of the error between triplicate cultures and the estimated deviation introduced by the calibration curve.
Induction of qrd1 and qrd2 transcripts in G. trabeum.
We tested the effects of potential inducers on qrd gene expression in 4- and 7-day G. trabeum liquid cultures. Each analysis was done with seven pooled mycelial mats. 2,6-DMBQ (50 or 500 μM) was added to cultures in 900 μl of ddH2O. BHA (500 μM) and DL-sulforaphane (500 μM) were added in 900 μl of ddH2O-dimethyl sulfoxide (9:1). Control cultures received ddH2O or ddH2O-dimethyl sulfoxide alone. The solutions were added to the surface of each culture, which was then swirled gently and incubated statically at 31°C for 1 h. Heat shock treatment of liquid- and wood-grown cultures was performed by incubating cultures statically at 45°C in a water bath for 1 h, and recovery from heat shock was accomplished by returning the cultures to 31°C in the growth incubator for 2 h. After induction or heat shock treatments, mycelial mats from liquid cultures were harvested, filtered by suction through a 0.45 μm-pore-size membrane, washed quickly with cold sterile ddH2O, and quick-frozen in liquid nitrogen. Colonized wood blocks were also quick-frozen in liquid nitrogen after heat shock treatment.
Induction of NADH:2,6-DMBQ reductase activity in G. trabeum by 2,6-DMBQ.
Four-day-old G. trabeum cultures were treated with 500 μM 2,6-DMBQ or with ddH2O for various times as described above. For each analysis, five mats were harvested, filtered by suction through a 0.45 μm-pore-size membrane to remove the growth medium, washed with 100 mM sodium citrate (pH 6.0) that contained 1 mM EDTA, and filtered again by suction to yield a moist pellet. The mycelia were then ground for 3 min with a chilled mortar and pestle in 2.0 ml of sand and 2.5 ml of extraction buffer that consisted of 100 mM sodium citrate (pH 6.0), 1 mM EDTA, and one tablet of Complete Protease Inhibitor (Roche) per 50 ml. An additional 0.5 ml of sand was then added, and the mycelia were ground for 3 min more. The mixture was centrifuged at 20,800 × g for 5 min at 4°C, after which the supernatant fraction was removed to a clean tube and centrifuged again as described above. The clarified supernatant fraction was assayed for NADH:2,6-DMBQ reductase activity by monitoring NADH oxidation spectrophotometrically as described earlier (12). One unit of activity was defined as the amount of QRD that oxidizes 1 μmol of NADH per min. Total protein in the extracts was assayed by using a Coomassie blue dye-binding assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin as the standard.
Heterologous expression of QRD2.
Recombinant QRD2 was expressed in Pichia pastoris by using Invitrogen's Easy Select Pichia Expression kit. P. pastoris strain X-33 and the media used to grow it are described in the Invitrogen manual. QRD2 was PCR amplified with Pfu from a plasmid that carried the cDNA by using the forward primer 5′-GGAATTCGCGAATGTGCTTTCCATCGAGACGCC-3′ and the reverse primer 5′-GTCCTTTTGCGGCCGCGAACTTGACCTTGGAGACGG-3′. The amplified qrd2 coding region was then digested with EcoRI and NotI, gel purified, and cloned into plasmid pPICZ B (Invitrogen) as a translational fusion to a myc epitope and His6 tag. Positive clones were verified by PCR and DNA sequence analysis. Transformations into P. pastoris were carried out by electroporation according to the manufacturer's instructions. Positive integrates were able to use methanol as the sole carbon source and were confirmed by PCR.
The transformants were grown on minimal glycerol medium plus histidine and then scaled up on minimal methanol medium plus histidine as described in the Invitrogen manual. Cells were grown for 54 h, frozen, suspended in breaking buffer (50 mM sodium phosphate [pH 7.4], 1 mM EDTA, 5% glycerol, 1 mM phenymethylsulfonyl fluoride), placed in an ice-jacketed glass bead homogenizer (Bead-Beater; Biospec, Bartlesville, Okla.), and disrupted according to the manufacturer's instructions. The suspension was centrifuged at 12,000 × g for 10 min at 4°C, and the supernatant fraction was stored at 4°C until use.
Nickel-affinity chromatography of recombinant QRD2 was done at 4°C under nondenaturing conditions on a Ni-CAM HC Resin column (Sigma) according to the manufacturer's instructions. QRD2 was eluted with a buffer that consisted of 250 mM imidazole and 0.3 M NaCl in 50 mM NaHPO4 at pH 8.0. Fractions with QRD activity were then dialyzed against 20 mM NaHPO4 (pH 7.0) that contained 20% (vol/vol) glycerol.
Electrophoresis of recombinant QRD2.
Recombinant QRD2 was subjected to polyacrylamide gel electrophoresis (PAGE) on 10% nondenaturing gels (Ready Gel Precast Gels; Bio-Rad) and stained for total protein with Coomassie blue R-250.
To detect the enzyme by immunoblotting, the proteins on the gel were transferred electrophoretically to polyvinylidene difluoride membranes (Bio-Rad) at 100 V for 1 h with 25 mM Tris buffer (pH 8.3) that contained 192 mM glycine. The membranes were soaked in 10 ml of BLOTTO (13) for 1 h with shaking at 25°C and then washed and incubated with anti-myc antibody conjugated to alkaline phosphatase (Invitrogen) according to the manufacturer's instructions. The QRD2 band was detected by immersing the membrane in 1-Step Nitro Blue Tetrazolium-bromochloroindolyl phosphate mixture (Pierce, Rockford, Ill.) until a blue-black color precipitated in 5 to 15 min, after which the membrane was rinsed with ddH2O.
The NADH:QRD reductase activity of recombinant QRD2 was detected in the gels by observing the NADH- and menadione-dependent reduction of MTT. After electrophoresis, the gels were immediately soaked in 50 mM Tris, pH 7.5, that contained 0.3 mg of MTT/ml and 1 mM NADH, with or without 30 μM menadione (30). The gels were shaken gently at 25°C in the dark until a blue band at the location of QRD2 developed in 30 to 60 min. The reaction was stopped with 5% acetic acid.
Nucleotide sequence accession numbers.
The G. trabeum gDNA (including promoter region) and cDNA sequences have been deposited in the GenBank database under the following accession numbers: qrd1A promoter, AY286072; qrd1B gDNA, AY286073; qrd2A gDNA, AY286074; qrd2B gDNA, AY286075; qrd2A cDNA, AY286076; qrd2B cDNA, AY286077; gpd gDNA, AY286078; gpd cDNA, AY286079.
RESULTS
Identification of qrd genes in G. trabeum.
We designed degenerate PCR primers that were based on two conserved regions in the previously reported QRD genes of G. trabeum (12) and P. chrysosporium (1), and we used these to amplify G. trabeum gDNA. When these sequences were extended, we obtained full-length genomic clones of two distinct G. trabeum genes, one of which matched the sequence we originally reported (12). Amplifications with gene-specific primers and a high-fidelity polymerase then showed that each gene exists in two closely related forms that may be allelic (see below). We named these qrd1A (the originally reported sequence), qrd1B, qrd2A, and qrd2B.
The sequences for qrd1A and qrd1B differ by seven nucleotides and are 99.3% identical. Two of the changes occur in introns, of which qrd1 has four. One of the five changes in exons occurs in a putative leader sequence, whereas the other four result in amino acid changes in the predicted mature QRD1 proteins. The sequences for qrd2A and qrd2B differ by 14 nucleotides and are 98.6% identical. Three of the changes are in introns, of which qrd2 has three. One of the 11 changes in exons results in an amino acid change in a putative leader sequence. The remaining 10 nucleotide changes are located in the region that encodes the mature QRD2 protein, but none of them results in an amino acid change. Therefore, there are potentially two QRD1 proteins, but only one QRD2 protein.
We used qrd1- and qrd2-specific primers to PCR amplify cDNA from G. trabeum cultures that had been grown in liquid medium (2, 6, and 14 days) and on wood (8 days). Forty clones of each sample were sequenced, and the sequences were aligned with those of the qrd genes. The results showed that qrd1A mRNA was present in all samples, whereas qrd1B mRNA was not detected in any sample. By contrast, qrd2A and qrd2B clones were obtained with equal frequency from all samples. Therefore, qrd2A and qrd2B are very likely alleles. The relationship between qrd1A and qrd1B is more ambiguous, but the high similarity of their sequences suggests that they also may be alleles. So far, we have failed to induce G. trabeum to fruit and thus produce diagnostic qrd segregants.
Analysis of the qrd promoters.
We sequenced the qrd promoters by gene walking with reverse primers that were specific for each of the four qrd genes. Amplifications with gene-specific primers and a high-fidelity polymerase yielded sequences that extended about 800 bp upstream from the ATG codons. Both qrd1 genes contain a TATA box, whereas neither qrd2 gene does (Table 2). All of the genes contain CAAT boxes, and in the two qrd2 genes this element is very close to the ATG codon, as is generally the case for genes without TATA boxes (17). The core promoters (−40 bp to + 40 bp) for qrd1A and qrd1B differ by eight nucleotides, whereas those of qrd2A and qrd2B are identical. In addition, qrd1B lacks a palindrome that is present upstream of qrd1A, qrd2A, and qrd2B (Table 2).
TABLE 2.
Putative transcription elements in the qrd1A and qrd2AB promotersa
| Element | Sequence | Positionb in:
|
|
|---|---|---|---|
| qrd1 | qrd2 | ||
| TATA box | TATAA | 58 | —d |
| CAAT box | CCAAT | 43,c 273 | 17c |
| Palindrome | ACTAGT | 698 | 796 |
| HSE | CGAACGTTCTCGAAA | — | 300 |
| XRE | GCGTG | — | 116,c 336,c 465c |
| ARE | GCGACCGTGC | — | 155 |
| STRE | CCCCT | 634, 643 | 307, 443 |
| MRE | TGCACGC | 286 | 338, 341,c 693c |
| TRE | TCACTCAA | — | 754 |
The elements were identified by using the FindPatterns program of GCG Wisconsin Package software.
Position upstream from the start codon (ATG).
Reverse orientation of the element.
—, not found.
The promoters of the three transcribed genes (qrd1A, qrd2A, and qrd2B) all contain putative stress response elements (STRE) and metal response elements (MRE). No other response elements are evident in the qrd1A promoter, but both of the qrd2 promoters contain consensus sequences for the xenobiotic element (XRE) and heat shock element (HSE), as well as sequences that differ by one nucleotide from the consensuses for the antioxidant response element (ARE) and AP1-binding element (TRE). The qrd2A and qrd2B promoters differ by seven nucleotides, but none of these changes occurs in a response element.
Properties of QRD2.
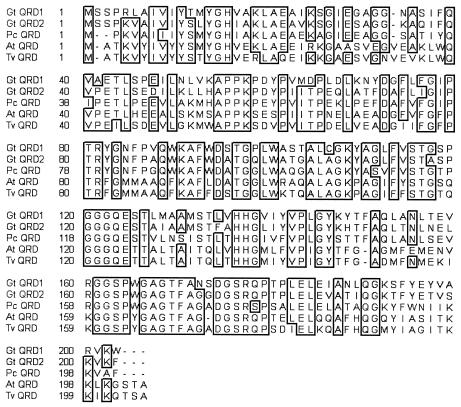
The deduced polypeptide sequence of G. trabeum QRD2 contains 243 amino acids, including a putative leader sequence of 40 amino acids similar to one found in G. trabeum QRD1, P. chrysosporium QRD, and some related sequences (1, 12). The mature QRD2 protein (without the leader) is 69% identical to the previously described P. chrysosporium QRD (AAD21025) (1) and 67% identical to G. trabeum QRD1 (AAL67860) (12). It is also similar to plant quinone reductases in this flavodoxin family, e.g., 56% identical to an Arabidopsis thaliana QRD (BAA97523) (16) and 54% identical to a Triphysaria versicolor QRD (AAG53945) (18) (Fig. 1).
FIG. 1.
Amino acid sequence comparison between G. trabeum QRD1 (Gt QRD1, AAL67860), G. trabeum QRD2 (Gt QRD2), P. chrysosporium QRD (Pc QRD, AAD21025), A. thaliana QRD (At QRD, BAA97523), and T. versicolor QRD (Tv QRD, AAG53945). Boxed areas indicate conserved sequences.
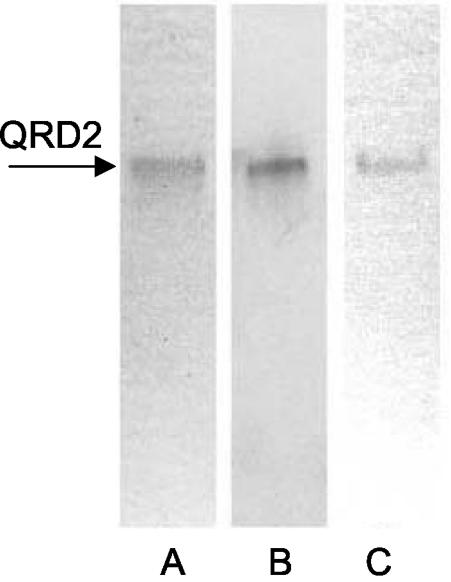
We expressed QRD2 with a C-terminal myc epitope and His6 tag in P. pastoris and purified it by nickel-affinity chromatography. Most of the QRD activity was lost on the column, but sufficient enzyme was recovered to assess its basic properties. Sodium dodecyl sulfate-PAGE showed that the enzyme was apparently pure and that it had a subunit molecular mass of 25 kDa, in agreement with the predicted value of 24.9 kDa (data not shown). Nondenaturing PAGE revealed a single band by Coomassie blue staining or immunoblotting against an anti-myc antibody. Activity staining of the native gel with NADH, menadione, and the tetrazolium dye MTT established that QRD2 is an NADH:quinone reductase (Fig. 2). Additional experiments showed that QRD2, like QRD1, shows low specificity for quinones but exhibits much higher activity with NADH than with NADPH (12). The visible absorption spectrum of recombinant QRD2 was that of a typical flavoprotein, with maxima at 450 and 380 nm (data not shown).
FIG. 2.
Native PAGE of recombinant QRD2. Lanes: A, 0.5 μg of enzyme stained with Coomassie blue; B, 0.5 μg of enzyme detected by immunoblotting against an anti-myc antibody; C, 2.5 μg of enzyme stained for QRD activity with NADH, menadione, and MTT. No staining occurred when menadione was omitted.
Expression of qrd1 and qrd2 by G. trabeum.
We used real-time RT-PCR to compare qrd transcript levels in G. trabeum cultures (Fig. 3B). This method is quantitative when transcripts that arise from a single qrd gene are compared. However, it is only semiquantitative for comparisons between qrd1 and qrd2 transcripts. The problem is that two criteria must be met for a quantitative comparison: the RT efficiencies for the qrd1 and qrd2 mRNAs must be the same, and the PCR efficiencies for the resulting qrd1 and qrd2 cDNAs must be the same. It is evident that the PCR efficiencies were near 100% for both cDNAs because plots of cycle threshold versus log dilution of cDNA gave slopes of −log10 2 (Applied Biosystems User Bulletin no. 2). However, it is possible that the RT efficiencies were not the same.
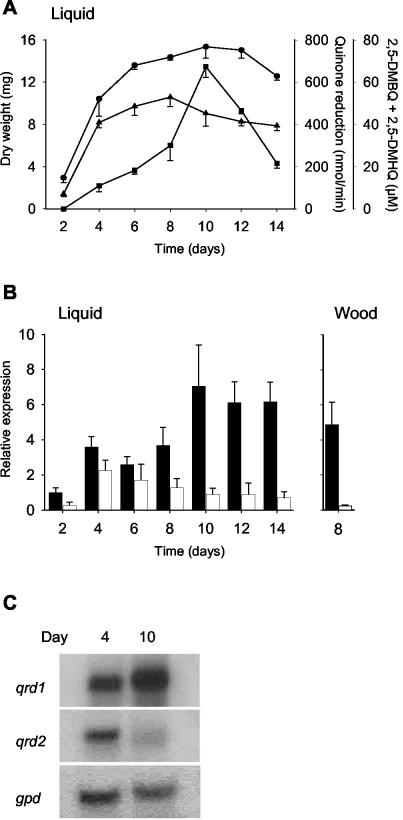
FIG. 3.
Expression of G. trabeum qrd1 and qrd2 as a function of growth conditions and time in culture. (A) Rate of 2,6-DMBQ reduction by one intact mycelial mat from a liquid culture (triangles), concentration of 2,5-DMBQ plus 2,5-DMHQ in one liquid culture (squares), and dry weight of one mycelial mat from a liquid culture (circles). Error bars show the standard deviations for triplicate samples. (B) Relative expression of qrd1 (black bars) and qrd2 (white bars) in liquid cultures and in spruce wood blocks, as determined by Taqman real-time RT-PCR. Gene expression was normalized to gpd expression and calibrated to the value for qrd1 on day 2, which was assigned a value of 1. Error bars show the standard deviations for triplicate samples. (C) Relative expression of qrd1, qrd2, and gpd in 4- and 10-day-old liquid cultures, as determined by Northern hybridization.
Nevertheless, it is evident that any error thus introduced is small, because comparisons between qrd1 and qrd2 transcript levels by Northern blotting (Fig. 3C), which does not involve an RT step, gave results similar to those obtained by real-time RT-PCR (Fig. 3B). Since the real-time RT-PCR method was evidently no less quantitative than Northern blotting for our purposes and since it is by far the more sensitive technique (3), we used it to compare qrd1 and qrd2 transcript levels.
Analyses of G. trabeum grown on nitrogen-limited liquid medium showed that the peak level of qrd2 transcripts (i.e., mRNAs made from either qrd2A or qrd2B) was relatively low and occurred during the growth phase (Fig. 3A and B). By contrast, qrd1 transcripts (i.e., mRNAs made from qrd1A) were more abundant and peaked after mycelial growth ceased, i.e., during secondary metabolism. The rate at which the mycelium reduced quinones and the extracellular concentration of 2,5-DMHQ plus 2,5-DMBQ also peaked during secondary metabolism, in agreement with previous work (28). That is, the properties that are associated with extracellular Fenton chemistry by G. trabeum (11, 14) coincided with a high level of qrd1 transcripts and a low level of qrd2 transcripts.
When G. trabeum was grown on spruce wood under conditions that lead to significant decay (9), qrd1 transcript levels after 8 days were much higher than qrd2 transcript levels, which they exceeded by a factor of about 20 (Fig. 3B). 2,5-DMBQ was present in the colonized wood at a concentration of 0.9 μmol/g (dry weight). These results provide the first evidence that G. trabeum expresses its hydroquinone-driven Fenton system not only on glucose (11, 14) and cellulose (5) but also on wood.
Induction of qrd1 and qrd2 in G. trabeum.
Since G. trabeum's principal natural quinone, 2,5-DMBQ, is too insoluble to prepare concentrated stock solutions, we used a structurally related quinone, 2,6-DMBQ, to test whether the qrd genes were induced by quinones. Previous work has shown that G. trabeum reduces 2,5- and 2,6-DMBQ at the same rate and that the two resulting hydroquinones drive Fenton chemistry equally well (11, 14).
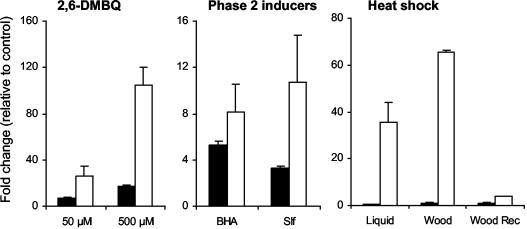
Both qrd genes were induced when 2,6-DMBQ was added to 4-day-old nitrogen-limited liquid cultures of G. trabeum, with greater induction occurring at a higher quinone concentration (Fig. 4). Transcript levels were already elevated 15 min after administration of the quinone, peaked at 1 h, and declined to near baseline levels by 5 h (data not shown). The response of qrd2 to the quinone was much stronger than that of qrd1, with transcript levels transiently exceeding 100 times those found in uninduced mycelia. Similar results were obtained when the experiment was repeated with 7-day-old cultures (data not shown).
FIG. 4.
Induction of qrd1 (black bars) and qrd2 (white bars) in G. trabeum, as determined by Taqman real-time RT-PCR. Gene expression in treated samples is given as the fold change relative to expression in untreated controls. 2,6-DMBQ, 4-day-old liquid cultures were treated with 50 or 500 μM 2,6-DMBQ for 1 h; phase 2 inducers, 4-day-old liquid cultures were treated with 500 μM dl-sulforaphane (Slf) or 500 μM BHA for 1 h; heat shock, 4-day-old liquid cultures or 8-day-old wood block cultures were treated at 45°C for 1 h. The bars labeled Wood Rec (recovery) show relative gene expression after heat-shocked wood block cultures had been returned to the normal incubation temperature of 31°C for 2 h. Error bars show the standard deviations for triplicate samples.
BHA and sulforaphane, two known inducers of mammalian phase 2 genes (10, 23, 26), gave moderate elevations of G. trabeum qrd transcript levels when they were added to 4-day-old cultures (Fig. 4) or 7-day-old cultures (data not shown) on liquid medium. As with 2,6-DMBQ, these inducers had a greater effect on qrd2 transcripts than they did on qrd1 transcripts. A variety of other phase 2 inducers, including H2O2, paraquat, 1-nitrocyclohexene, and β-naphthoflavone, failed to induce either of the G. trabeum genes.
Heat shock (45°C for 1 h) induced qrd2 markedly but had no effect on qrd1 transcription in 4-day-old cultures (Fig. 4) or 7-day-old cultures (data not shown) on liquid medium. The effect on qrd2 was so large that we repeated the experiment with wood-grown cultures, in which qrd1 transcripts were normally dominant (Fig. 3B), to see whether qrd2 transcripts would become quantitatively important. On wood, qrd2 transcripts were increased nearly 80-fold by heat shock, making them significantly more abundant than qrd1 transcripts under this type of stress. When the colonized wood was returned to 31°C, transcripts reverted to near their preinduced levels after 2 h (Fig. 4).
Induction of QRD activity in G. trabeum.
To determine whether greater transcription of the qrd genes would have an observable effect on translation, we assayed NADH:2,6-DMBQ oxidoreductase activity in lysates of 4-day-old mycelia from liquid cultures that had been induced with 2,6-DMBQ or treated with an equivalent volume of water. Since the activities of the two enzymes, QRD1 and QRD2, are indistinguishable, this approach was only able to yield a composite value of QRD activities from all sources in the mycelium. An immunochemical approach based on non-cross-reacting monoclonal antibodies to QRD1 and QRD2 would have been better, but so far we have been unable to produce enough recombinant QRD2 to obtain antibodies.
We found that the QRD specific activity in lysates of G. trabeum cultures was elevated more than threefold by pretreatment with 500 μM 2,6-DMBQ (Table 3). The QRD activity per volume of extract was also elevated by the same factor, thus showing that none of the specific activity increase was attributable merely to lower total protein levels in induced samples. Induction was first observable at 2 h of induction, after transcript levels had peaked. It is not clear why QRD induction at the enzyme level was considerably less than qrd induction at the RNA level in these experiments, but similar discrepancies have been reported in other systems and are thought to reflect posttranscriptional regulation (27).
TABLE 3.
NADH:QRD activity in G. trabeum mycelial lysates after induction of cultures with 500 μM 2,6-DMBQ
| Length of induction (h) | Treatment | Activity/vol of extract
|
Sp act
|
||
|---|---|---|---|---|---|
| U/ml ± SDa | Fold change | U/mg of protein ± SDa | Fold change | ||
| 1b | H2O | 4.91 ± 0.68 | 1.0 | 7.56 ± 0.99 | 1.0 |
| 2,6-DMBQ | 4.19 ± 0.91 | 0.9 | 7.34 ± 0.74 | 1.0 | |
| 2b | H2O | 4.56 ± 0.87 | 1.0 | 7.04 ± 0.99 | 1.0 |
| 2,6-DMBQ | 7.93 ± 0.39 | 1.7 | 10.86 ± 0.27 | 1.5 | |
| 2.5c | H2O | 3.42 ± 1.21 | 1.0 | 4.80 ± 0.46 | 1.0 |
| 2,6-DMBQ | 10.06 ± 1.33 | 2.9 | 13.18 ± 0.69 | 2.7 | |
| 3.5c | H2O | 3.52 ± 0.23 | 1.0 | 5.44 ± 0.37 | 1.0 |
| 2,6-DMBQ | 11.08 ± 0.18 | 3.1 | 17.44 ± 1.34 | 3.2 | |
Standard deviation for triplicate samples.
Experiment 1.
Experiment 2.
DISCUSSION
QRD1.
Our results provide three indications that QRD1 rather than QRD2 is the enzyme that catalyzes routine intracellular quinone reductions by G. trabeum: (i) qrd1 transcripts predominated in G. trabeum cultures that were grown in a liquid medium that elicits high levels of quinone biosynthesis and biodegradative Fenton chemistry; (ii) levels of qrd1 transcripts rather than qrd2 transcripts paralleled levels of 2,5-DMBQ plus 2,5-DMHQ in these liquid-grown cultures; and (iii) qrd1 transcripts predominated during early wood decay by G. trabeum, when holocellulose cleavage is most rapid (15) and the need for 2,5-DMBQ redox cycling therefore greatest.
These expression patterns may indicate that QRD1 catalyzes the extracellular 2,5-DMHQ-driven Fenton system of G. trabeum, while simultaneously ensuring that no toxic 2,5-DMBQ accumulates intracellularly. Alternatively, the cycle may be driven by an uncharacterized reductase located on the mycelial surface, and the sole function of QRD1 may be in surveillance against any quinones that happen to penetrate the mycelium while the cycle is operating. On the face of it, a quinone redox cycle that avoids any intracellular steps seems a safer and more efficient way to drive extracellular Fenton chemistry. However, so far we have not found significant membrane-bound quinone reductase activity in G. trabeum (12), and QRD1, despite its intracellular location, remains a reasonable candidate for the enzyme that catalyzes the cycle.
qrd1A transcription was transiently enhanced by 2,6-DMBQ, an analogue of 2,5-DMBQ, the principal quinone that G. trabeum produces. This result suggested that QRD1 might be regulated similarly to mammalian phase 2 detoxification enzymes, which are also up-regulated by quinones. There are many phase 2 inducers, all of which share the common property of being good electrophiles that react readily with sulfhydryl groups. This feature may provide a mechanism for interaction between these inducers and regulatory proteins (10, 23, 24, 26). We found that some phase 2 inducers enhanced qrd1A transcription, but most did not. The qrd1A promoter's lack of ARE or XRE, which are required for induction of the mammalian phase 2 genes (10, 24), likely contributed to this result. The basis for qrd1A induction by 2,6-DMBQ remains unclear, but the putative STRE that we found may play a role.
It is not yet clear why qrd1B was not transcribed in liquid medium or on wood, but it may be significant that its core promoter, a region required for RNA polymerase to bind (4), differs considerably from that of qrd1A. Another possible contributor is the palindromic sequence absent in the qrd1B promoter region. This palindrome was earlier shown to be required for transcription of a phase 2 metalloproteinase in mouse fibroblasts (25). It is present not only upstream of G. trabeum qrd1A, qrd2A, and qrd2B but also upstream of a related A. thaliana quinone reductase gene that is induced by auxin (16). It remains to be determined whether qrd1B is a functional gene that is regulated differently from qrd1A or whether it is simply a qrd1 allele that has been rendered nonfunctional by deleterious mutations.
QRD2.
G. trabeum apparently does not express QRD2 routinely at high levels during lignocellulose degradation but can deploy it to detoxify quinones when the mycelium is under stress. The marked induction of qrd2 by heat shock supports a role for QRD2 in the general stress response of G. trabeum (20). The very strong induction of qrd2 by 2,6-DMBQ, a compound of known cytotoxicity (22), also supports this conclusion. The transient nature of qrd induction by 2,6-DMBQ is probably attributable to the fact that this quinone does not persist in cultures, because G. trabeum rapidly reduces it to nontoxic 2,6-dimethoxyhydroquinone. Similarly, G. trabeum maintains its natural quinone, 2,5-DMBQ, predominantly in its nontoxic reduced form (11).
The qrd2 promoter contains an HSE sequence that probably accounts for the induction of qrd2 by heat shock. The auxin-responsive A. thaliana quinone reductase gene (16) also contains this HSE sequence upstream of its coding region, but has apparently not been tested for its response to heat shock. In addition, the G. trabeum qrd2 promoter contains several likely XRE sequences and a possible ARE sequence that probably explain why qrd2 responded much more strongly to 2,6-DMBQ than did qrd1. Mammalian phase 2 gene promoters also contain some of these response elements (10, 24), but since qrd2, like qrd1, failed to respond to most phase 2 inducers, it appears that the regulatory mechanisms differ somewhat in fungal and animal systems. Further progress in our understanding of the G. trabeum qrd promoters will require the development of a transformation system with which one can manipulate their response elements.
Acknowledgments
We are indebted to Carl Houtman for calculating the real-time RT-PCR results and for explaining the mathematics of this technique to us. Patricia Ortiz-Bermúdez kindly assisted with the QRD assays on mycelial homogenates. Thanks are also due to Dan Cullen and Luis Larrondo for valuable advice and a critical reading of the manuscript.
This work was supported by grant DE-FG02-94ER20140 to K.E.H. from the Department of Energy.
REFERENCES
- 1.Akileswaran, L., B. J. Brock, J. L. Cereghino, and M. H. Gold. 1999. 1, 4-Benzoquinone reductase from Phanerochaete chrysosporium: cDNA cloning and regulation of expression. Appl. Environ. Microbiol. 65:415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolton, J. L., M. A. Trush, T. M. Penning, G. Dryhurst, and T. J. Monks. 2000. Role of quinones in toxicology. Chem. Res. Toxicol. 13:135-159. [DOI] [PubMed] [Google Scholar]
- 3.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 4.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, R., K. A. Jensen, Jr., C. J. Houtman, and K. E. Hammel. 2002. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett. 531:483-488. [DOI] [PubMed] [Google Scholar]
- 6.Flournoy, D. S., T. K. Kirk, and T. L. Highley. 1991. Wood decay by brown-rot fungi: changes in pore structure and cell wall volume. Holzforschung 45:383-388. [Google Scholar]
- 7.Gilbertson, R. L., and L. Ryvarden. 1986. North American polypores. Fungiflora, Oslo, Norway.
- 8.Goodell, B., J. Jellison, J. Liu, G. Daniel, A. Paszczynski, F. Fekete, S. Krishnamurthy, L. Jun, and G. Xu. 1997. Low molecular weight chelators and phenolic compounds isolated from wood decay fungi and their role in the fungal biodegradation of wood. J. Biotechnol. 53:133-162. [Google Scholar]
- 9.Hammel, K. E., A. N. Kapich, K. A. Jensen, Jr., and Z. C. Ryan. 2002. Reactive oxygen species as agents of wood decay by fungi. Enzyme Microb. Technol. 30:446-453. [Google Scholar]
- 10.Jaiswal, A. K. 2000. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 29:254-262. [DOI] [PubMed] [Google Scholar]
- 11.Jensen, K. A., Jr., C. J. Houtman, Z. C. Ryan, and K. E. Hammel. 2001. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 67:2705-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, K. A., Jr., Z. C. Ryan, A. Vanden Wymelenberg, D. Cullen, and K. E. Hammel. 2002. An NADH:quinone oxidoreductase active during biodegradation by the brown-rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 68:2699-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, D. A., J. W. Gautsch, J. R. Sportsman, and J. H. Elder. 1984. Improved techniques utilizing nonfat dry milk for analysis of proteins and nucleic acids transferred to nitrocellulose. Gene Anal. Tech. 1:3. [Google Scholar]
- 14.Kerem, Z., K. A. Jensen, and K. E. Hammel. 1999. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven fenton reaction. FEBS Lett. 446:49-54. [DOI] [PubMed] [Google Scholar]
- 15.Kleman-Leyer, K., E. Agosin, A. H. Connor, and T. K. Kirk. 1992. Changes in molecular size distribution of cellulose during attack by white rot and brown rot fungi. Appl. Environ. Microbiol. 58:1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski, M. J., K. A. Dreher, M. A. Gehring, S. Abel, A. L. Gensler, and I. M. Sussex. 2002. FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol. 128:578-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matvienko, M., A. Wojtowicz, R. Wrobel, D. Jamison, Y. Goldwasser, and J. I. Yoder. 2001. Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 25:375-387. [DOI] [PubMed] [Google Scholar]
- 19.McFee, W. W., and E. L. Stone. 1966. The persistence of decaying wood in the humus layers of northern forests. Soil Sci. Soc. Am. Proc. 30:513-516. [Google Scholar]
- 20.Nover, L. 1991. Inducers of HSP synthesis: heat shock and chemical stressors, p. 5-40. In L. Nover (ed.), Heat shock response. CRC Press, Boca Raton, Fla.
- 21.Paszczynski, A., R. Crawford, D. Funk, and B. Goodell. 1999. De novo synthesis of 4, 5-dimethoxycatechol and 2, 5-dimethoxyhydroquinone by the brown rot fungus Gloeophyllum trabeum. Appl. Environ. Microbiol. 65:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pethig, R., P. R. C. Gascoyne, J. A. McLaughlin, and A. Szent-Gyorgyi. 1983. Ascorbate-quinone interactions: electrochemical, free radical, and cytotoxic properties. Proc. Natl. Acad. Sci. USA 80:129-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prestera, T., W. D. Holtzclaw, Y. Zhang, and P. Talalay. 1993. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc. Natl. Acad. Sci. USA 90:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prestera, T., and P. Talalay. 1995. Electrophile and antioxidant regulation of enzymes that detoxify carcinogens. Proc. Natl. Acad. Sci. USA 92:8965-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz, L., E. Berra, M. M. Municio, I. Dominguez, J. Lozano, T. Johansen, J. Moscat, and M. T. Diazmeco. 1994. Zeta-PKC plays a critical role during stromelysin promoter activation by platelet-derived growth factor through a novel palindromic element. J. Biol. Chem. 269:10044-10049. [PubMed] [Google Scholar]
- 26.Talalay, P., J. W. Fahey, W. D. Holtzclaw, T. Prestera, and Y. Zhang. 1995. Chemoprotection against cancer by Phase 2 enzyme induction. Toxicol. Lett. 82/83:173-179. [DOI] [PubMed] [Google Scholar]
- 27.van den Brink, J. M., P. J. Punt, R. F. M. van Gorcom, and C. A. M. J. J. van den Hondel. 2000. Regulation of expression of the Aspergillus niger benzoate para-hydroxylase cytochrome P450 system. Mol. Gen. Genet. 263:601-609. [DOI] [PubMed] [Google Scholar]
- 28.Varela, E., T. Mester, and M. Tien. 2003.. Culture conditions affecting biodegradation components of the brown-rot fungus Gloeophyllum trabeum. Arch. Microbiol. 180:251-256. [DOI] [PubMed] [Google Scholar]
- 29.Wood, P. M. 1994. Pathways of production of Fenton reagent by wood-rotting fungi. FEMS Microbiol. Rev. 13:313-320. [Google Scholar]
- 30.Wrobel, R. L., M. Matvienko, and J. I. Yoder. 2002. Heterologous expression and biochemical characterization of an NAD(P)H:quinone oxidoreductase from the hemiparasitic plant Triphysaria versicolor. Plant Physiol. Biochem. 40:265-272. [Google Scholar]
- 31.Yatzkan, E., and O. Yarden. 1995. Inactivation of a single type-2A phosphoprotein phosphatase is lethal in Neurospora crassa. Curr. Genet. 28:458-466. [DOI] [PubMed] [Google Scholar]
- 32.Zabel, R. A., and J. J. Morell. 1992. Wood microbiology: decay and its prevention. Academic Press, San Diego, Calif.