Abstract
The invariant NKT (iNKT) cell lineage contains CD4+ and CD4- subsets. The mechanisms that control such subset differentiation and iNKT cell maturation in general have not been fully understood. RasGRP1, a guanine nucleotide exchange factor for T cell receptor-induced activation of the Ras-Erk1/2 pathway, is critical for conventional αβ T cell development but dispensable for generating regulatory T cells. Its role in iNKT cells has been unknown. Here we report severe decreases of iNKT cells in RasGRP1-/- mice through cell intrinsic mechanisms. In the remaining iNKT cells in RasGRP1-/- mice, there is a selective absence of the CD4+ subset. Furthermore, RasGRP1-/- iNKT cells are defective in T cell receptor induced proliferation in vitro. These observations establish that RasGRP1 is not only important for early iNKT cell development, but also for the generation/maintenance of the CD4+ iNKT cells. Our data provides genetic evidence that the CD4+ and CD4- iNKT cells are distinct sub-lineages with differential signaling requirements for their development.
INTRODUCTION
Natural killer T (NKT) cells are subsets of T cells co-expressing markers found on NK cells and T cells. While rare in number, NKT cells play important roles in immune responses and pathogenesis of disease (1-3). The invariant Vα14-Jα18 T cell receptor (iVα14TCR) expressing NKT (iNKT) cells represent the major subset within the NKT cell lineage and are the best characterized (4, 5). The iVα14TCR recognizes both endogenous and synthetic glycolipids such as iGB3 and α-galactosyl ceramide (α-GalCer), respectively, presented by CD1d (6, 7). Use of CD1d tetramers loaded with α-GalCer has provided a pivotal tool to define iNKT cells and has allowed for the delineation of iNKT cell development into multiple developmental stages. The earliest iNKT cells (stage 0) are defined as CD24+CD44-NK1.1-, and such cells are extremely rare in the thymus. As iNKT cells mature, they down-regulate CD24 expression and progress sequentially through stage 1 (CD24-CD44-NK1.1-), stage 2 (CD24-CD44+NK1.1-), and finally stage 3 (CD24-CD44+NK1.1+) (8, 9). Further from these stage definitions, iNKT cells can also be divided into CD4+ and CD4- subsets that may branch out at the stage 1 and represent two different sub-lineages of iNKT cells (10). However, genetic evidence supporting such sub-lineage definition remains quite rare.
The iVα14TCR is critical for iNKT cell development. Deficiency of the receptor or its ligand CD1d results in a failure to generate iNKT cells in mice (11-13). Upon TCR engagement, PLCγ1 plays a crucial role in TCR signaling by producing diacylglycerol (DAG) and inositol 1,4,5-tris-phosphate (IP3) second messengers (14). IP3 activates the Ca++-calcineurin-NFAT pathway, which has been recently demonstrated to be crucial for iNKT cell maturation via the transcription factor Egr2 (15). DAG activates the PKCθ-Carma1/Bcl10/Malt-IKK-NFκB pathway. The NFκB pathway is critical for iNKT cell ontogeny, as deficiencies of its different components have been shown to block iNKT cell development at various stages (16-21). DAG also associates with and activates RasGRP1, a guanine nucleotide releasing factor for Ras. RasGRP1 in turn activates the downstream Ras-Erk1/2-AP1 pathway. The RasGRP1-Ras-Erk1/2 pathway is important for positive selection of conventional αβ T (cαβT) cells (22, 23). While uncontrolled DAG-mediated signaling due to absence of diacylglycerol kinases α and ζ causes severe defect of iNKT cell development (24), the role of RasGRP1 in iNKT cell development remains unclear. In this report, we demonstrate severe decreases of iNKT cells and a selective absence of the CD4+ subset of iNKT cells in RasGRP1-/- mice. Our data not only reveals a critical role of RasGRP1 for early iNKT cell development, but also provide genetic evidence that the CD4+ and CD4- subsets of iNKT cells are indeed distinct sublineages since they have differential signaling requirements for their generation/maintenance.
MATERIALS AND METHODS
Mice
The C57BL6/J and TCRβ-/-δ-/- mice were all purchased from the Jackson Laboratory. The RasGRP1-/- mice were previously described (22) and were backcrossed onto B6 background for 9 generations. All mice were used according to a protocol approved by the Duke University Institute Animal Care and Use Committee. Thymocytes and splenocytes were prepared following standard procedures. Liver mononuclear cells were isolated according to a published protocol (18).
Antibodies and flow cytometry
Cells were stained with PE- or APC-CD1d-Tet (NIH tetramer core facility) and fluorescence-conjugated anti-mouse CD24, CD44, NK1.1, CD4, CD8, TCRβ, CD45.1, CD45.2, Thy1.1, Thy1.2, CD1d, CD150 (SLAM), and Ly108 (SLAMF6) antibodies (BioLegend) in PBS-2% FBS on ice for 30 minutes. Cell survival/death was determined by addition of the Live/Dead® Fixable Violet Dead Cell Stain (L/D, Invitrogen) during the staining according to the manufacturer’s protocol. Dead cells stain positive for L/D. For Ki67 expression, cells were permeabolized using the Foxp3 staining kit (eBioscience) after cell surface staining, followed by staining with unconjugated anti-Ki-67 (B56, BD Biosciences). An Alexa Fluor® 488-conjugated goat anti-mouse IgG (H+L) (Invitrogen) was used to detect anti-Ki67 antibody. Stained cells were collected on FACSCanto™ II (BD Biosciences) and analyzed using the Flowjo software.
Enrichment of iNKT cells
Thymocytes were resuspended in 500 μl IMDM with 10% FBS (IMDM-10) and were sequentially added with 5μl Fc-blocker from the EasySep PE selection kit (Stem Cell Technologies) and 2.5 μl of PE-CD1d-Tetramer. After incubation at room temperature (RT) for 15 minutes, cells were washed once with IMDM-10. The cells were resuspended in 500 μl of IMDM-10 and mixed with 5 μl of EasySep PE selection cocktail. After incubation at RT for 15 minutes, 5 μl of EasySep nanoparticles were added and the mixture was incubated at RT for additional 15 minutes. After addition of IMDM-10 to a total volume of 2.5 ml, cells in FACS tubes were inserted into the EasySep magnet and let stand for 5 minute. The unbound cells were discarded and the bound cells were resuspended in 2.5 ml IMDM-10. After repeating magnetic enrichment for another time, the magnetic bounding fractions were collected for staining and FACS analysis.
iNKT cell proliferation
Thymocytes from WT and RasGRP1-/- mice were labeled with 10 μM CFSE at RT for 9 minutes as previously described (25). Cells were seed at 5 ×106 cells/ml in a 48-well plate plate and left unstimulated or stimulated with 125 ng/ml α-GalCer at 37°C for 72 hours. Cells were then stained for TCRβ and APC-conjugated CD1d-Tet before analyzed by flow cytometry.
Bone marrow reconstitution
Recipient TCRβ-/-δ-/- mice were sublethally irradiated (600 rad) one day before adoptive transfer. Ten million 1:1 mixed bone marrow (BM) cells from age- and sex-matched CD45.1+ B6 and CD45.2+ RasGRP1-/- mice were intravenously injected into the recipients. Alternatively, lethally irradiated (1100 rad) WT C57B6 mice were used as recipients and were reconstituted with Thy1.1+-C57B6 (WT) and Thy1.2+-RasGRP1-/- BM cells at 1:10 ratio. The resulting chimeric mice were analyzed 7 to 8 weeks later.
Real time PCR
Viable CD4+CD8+ double positive (DP) thymocytes and TCRβ+CD1dTet+ iNKT cells from age- and sex-matched control or RasGRP1-/- mice were sorted on MoFlo Cell Sorter (Beckman Coulter), with post-sort purity>98%, and lysed in Trizol (Invitrogen). Total RNAs were extracted, and cDNAs were obtained using the Superscript III First-Strand Synthesis System (Invitrogen). Realtime PCR was prepared using the RealMasterMix (Eppendorf) and performed on the Mastercycler® ep realplex2 system (Eppendorf). Primers used for different genes are listed as following. SAP: forward 5’-acgcctctgcagtatccagt-3’, reverse 5’-ttcttcatggtgcattcagg-3’; Fyn: forward 5’-caagccaagcagtgtttgaa-3’, reverse 5’-acattgcacacagcccatta-3’; RORγt: forward 5’-cgactggaggaccttctacg-3’, reverse 5’-ttggcaaactccaccacata-3’; RUNX1: forward 5’-gcaggacgaatcacactgaa-3’, reverse 5’-tggcatctctcatgaagcac-3’; cMyc: forward 5’-tgaaggctggatttcctttg-3’, reverse 5’-ttctcttcctcgtcgcagat-3’; HEB: forward 5’-aggtatggatgagcgtggag-3’, reverse 5’-agccttcgtgggttcctaat-3’; PLZF: forward 5’-tgcgcagctatatttgcagt-3’, reverse 5’-tgtggctcttgagtgtgctc-3’; RasGRP1: forward 5’-agcccaccttctgtgacaac -3’, revers3 5’-cttcttgcactcgaacacca-3’; RasGRP2: forward 5’-gggcttcgtacacaacttcc-3’, reverse 5’-gtggcagttcacaccacaag-3’.
Assessment of Vα-Jα recombination
Decreasing amounts of DNA template (100ng, 33ng, 11ng) from sorted viable RasGRP1+/- and RasGRP1-/- CD4+CD8+ thymocytes were used for semi-quantitative PCR. The forward primer for Vα14 segment was 5’-acactgccacctacatctgt-3’. The reverse primers for different Jα segments were: Jα2 5’-ggttgcaaatggtgccactt-3’; Jα18 5’-gtagaaagaaacctactcacca-3’; Jα56 5’-tgtcatcaaaacgtacctggt-3’. Primers for CD14 PCR (loading control) were: forward 5’-gctcaaactttcagaatctaccgac-3’, reverse agtcagttcgtggaggccggaaatc-3’.
Statistics
For statistic analysis, two-tail Student t-test was performed. *, p<0.05. **, p<0.01, ***, p<0.001.
RESULTS
Critical role of RasGRP1 for iNKT cell development
The expression of RasGRP1 and the other RasGRP1 family members in iNKT cells has been unknown. We first examined their expression in iNKT cells. iNKT cells stained positive for both α-Galcer loaded CD1d tetramer (CD1d-Tet) and TCRβ in thymocytes from wild-type (WT) mice. These iNKT cells were sorted by FACS and mRNA levels of these genes were determined by real-time quantitative PCR. As shown in Fig 1A, both RasGRP1 and RasGRP2 can be detected in cαβT cells and iNKT cells. However, RasGRP3 and RasGRP4 were undetectable in iNKT cells (Data not shown).
Figure 1. Critical role of RasGRP1 for iNKT cell development.
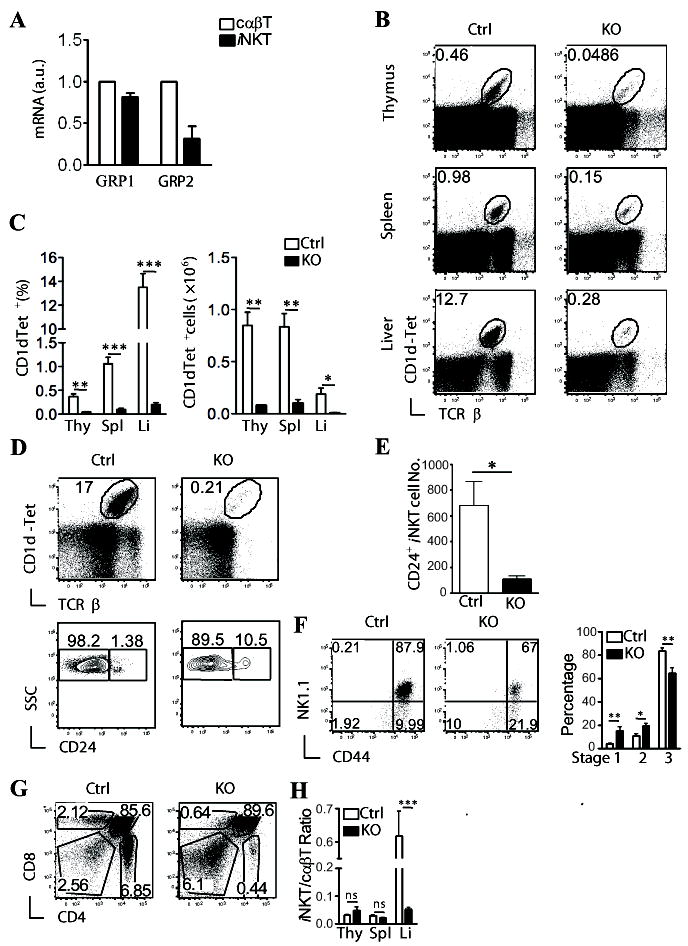
(A) Expression of RasGRP1 and RasGRP2 in WT iNKT cells. mRNA levels in FACS sorted iNKT cells and cαβT cells were determined by real-time PCR. (B-C) Thymocytes, splenocytes, and liver mononuclear cells from RasGRP1-/- (KO) and RasGRP1+/- (Ctrl) littermates were subjected to flow cytometry analysis. Data shown are representative of five mice per group. (B) Flow cytometry of cells stained with α-Galcer-loaded CD1d-tetramer (CD1d-Tet) and anti-TCRβ. (C) Percentage (left) and number (right) of live CD1d-Tet+TCRβ+ cells (mean ± SEM). (D) iNKT cells staining after enrichment for CD1d-Tet+ cells with magnetic beads. Thymocytes from WT and RasGRP1-/- mice were stained with PE-CD1d-Tet. iNKT cells were enriched with a PE-enrichment kit. Top panels, TCRβ and CD1d-Tet staining of enriched cells. Bottom panels, CD24 expression on gated TCRβ+CD1d-Tet+NK1.1- cells. (E). Total CD24+CD44-NK1.1- iNKT cell numbers in WT and RasGRP1-/- thymi (n=6). (F) Assessment of iNKT cell development within CD24- iNKT cells. Dot plots show expression of CD44 and NK1.1 on CD1d-Tet+CD24- gated live thymocytes. Bar figure shows percentages (mean ± SEM) of stages 1, 2, and 3 iNKT cells in RasGRP1-/- and control mice. (G) CD4 and CD8 staining of WT and RasGRP1-/- thymi. (H) Ratios of iNKT cells to cαβT cells in the thymus (Thy), spleen (Spl), and liver (Li) from WT and RasGRP1-/- mice. *p<0.05; **, p<0.01; ***, p<0.001 (Student t-test).
To determine the role of RasGRP1for iNKT cell development, we analyzed mice deficient in RasGRP1. Thymocytes, splenocytes, and liver mononuclear cells were stained with CD1d-Tet, as well as other cell surface markers. As shown in Figure 1B-1C, CD1dTet+TCRβ+ cells were decreased about 6 – 11 fold in RasGRP1-/- mice as compared to RasGRP1+/- mice. To further determine whether RasGRP1 is required for generation of stage 0 CD24+ iNKT cells, we enriched iNKT cells from WT and RasGRP1-/- thymus using PE-CD1d-Tet and magnetic beads. As shown in Figure 1D-1E, the CD24+ iNKT cell number in RasGRP1-/- thymi was 6-fold lower than WT control, indicating that RasGRP1 is required for efficient generation of stage 0 iNKT cells. Further analysis of the CD1dTet+CD24- iNKT cells revealed relative enrichment of CD44-NK1.1- and CD44+NK1.1- populations, but a decrease of the CD44+NK1.1+ population in RasGRP1-/- mice (Fig. 1F). Together, these observations indicate that RasGRP1 is critical for early iNKT cell development and is also involved in promoting iNKT maturation at later stages. Similar to a previous report (22), there was a severe decrease of cαβT cells and about 50% decrease of total thymocyte number in RasGRP1-/- mice (Fig. 1G and data not shown). The ratios of iNKT cells to cαβT cells in RasGRP1-/- thymus and spleen were similar to those of WT controls (Fig. 1H). Thus, the defect of iNKT cell development caused by RasGRP1 deficiency was in parallel with that of cαβT cells. However, the iNKT cells to cαβT cell ratio was about seven-fold lower in RasGRP1-/- liver than in WT liver.
Cell intrinsic defect of developing RasGRP1-/- iNKT cells
Since RasGRP1 is expressed in multiple cell lineages and iNKT cells are positively selected by engagement of iVα14TCR with CD1d expressed on DP thymocytes, we further investigated whether the developmental defects of RasGRP1-/- iNKT cells are intrinsic. To this end, a 1:1 mixture of CD45.2+ RasGRP1-/- and CD45.1+CD45.2+ WT BM cells were adoptively transferred to reconstitute TCRβ-/-δ-/- hosts. The recipients were analyzed seven to eight weeks after reconstitution. Although a close to 1:1 ratio of WT and RasGRP1-/- BM cells were injected into the recipients (Fig. 2A), only 15% of thymocytes from the recipients and less than 40% of total splenocytes and liver mononuclear cells were derived from RasGRP1-/- origin (Fig. 2B-2C). There were progressive decreases of representation by RasGRP1-/- thymocytes as they mature from the CD4-CD8- double negative (DN), the CD4+CD8+ DP, to the CD4+CD8- or CD4-CD8+ single positive (SP) stage. The decrease was most severe within the RasGRP1-/- CD4 SP and CD8 SP populations. Further analysis of non-T cells (DN) from splenocytes in the recipients revealed a roughly equal contributions of WT and RasGRP1-/- origins, suggesting that RasGRP1 deficiency does not globally affect hematopoietic stem cell engraftment or early lymphoid progenitor cells and that RasGRP1 may promote cαβT cell development at multiple stages.
Figure 2. iNKT developmental defects in RasGRP1-/- mice are cell-intrinsic.
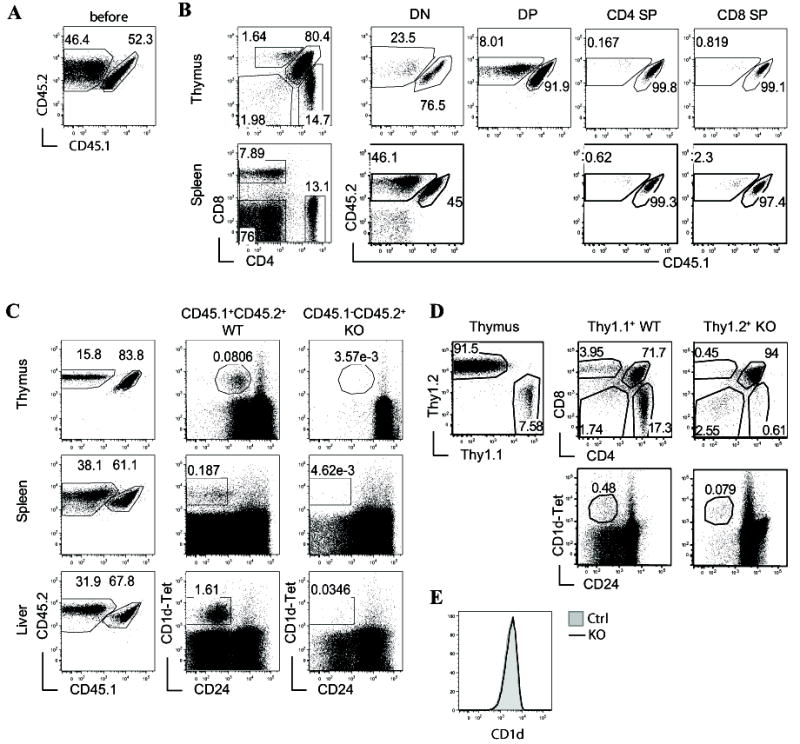
(A-C) Generation and analysis of sublethally irradiated TCRβ-/-δ-/- recipient mice reconstituted WT (CD45.1+CD45.2+) and RasGRP1-/- (CD45.1-CD45.2+) bone marrow (BM) at 1:1 ratio. Chimeric mice were analyzed 7-8 weeks after reconstitution. (A) Expression of CD45.1 and CD45.2 on mixed WT and RasGRP1-/- BM cells before adoptive transfer. (B) Analysis of cαβT cells and non-T cells in recipient mice. Left panels show CD4 and CD8 staining of thymocytes and splenocytes from recipient mice. Middle and right panels show CD45.1 and CD45.2 staining in the DN, DP, and SP populations based on CD4 and CD8 expression. (C) Analysis of iNKT cells in recipient mice. Left panels show CD45.1 and CD45.2 staining in the indicated organs from recipient mice. Middle and right panels show expression of CD1d-Tet and CD24 on gated CD45.1+CD45.2+ WT and CD45.1-CD45.2+ RasGRP1-/- cells. (D) Analysis of lethally irradiated WT C57B6 recipient mice reconstituted with WT (Thy1.1) and RasGRP1-/- (Thy1.2) BM cells at 1:10 ratio. Left panel, Thy1.1 and Thy1.2 staining of recipient thymocytes; Middle and right panels, CD24 and CD1d-Tet staining as well as CD4 and CD8 staining gated on Thy1.1+ and Thy1.2+ thymocytes. (E) CD1d expression on RasGRP1-/- and control CD4+CD8+ DP thymocytes. Data are representative of three (A-C) or two (D, E) experiments.
In the thymi of recipient mice, CD45.1+CD45.2+ WT CD1dTet+ iNKT cells could be easily detected. However, the CD45.2+ RasGRP1-/- CD1dTet+ iNKT cells were virtually undetectable in the recipients (Fig. 2C). Similarly, few RasGRP1-/- iNKT cells were observed in the spleen and liver as well. Due to under representation of RasGRP1-/- thymocytes in chimeric mice reconstituted with WT and RasGRP1-/- BM cells at 1:1 ratio, we further generated and analyzed chimeric mice reconstituted with BM cells from WT (Thy1.1) and RasGRP1-/- (Thy1.2) mice at 1:10 ratio. As shown in figure 2D, Thy1.1 and Thy1.2 staining of thymocytes from recipients displayed a close to expected ratio of cells originated from WT to RasGRP1-/- BM cells. Severe decreases of Thy1.2+ RasGRP1-/- iNKT cells as well as CD4 and CD8 SP cαβT cells were observed as compared with their Thy1.1+ WT counterparts in the recipients. Together, these observations indicate that the developmental defects of RasGRP1-/- iNKT cells and cαβT cells are cell-intrinsic and cannot be rescued by WT thymocytes. CD1d expression on cortical thymocytes are critical for iNKT cell development (26, 27). No obvious difference was observed between RasGRP1-/- and control thymocytes (Fig. 2E). Together, these data indicate that RasGRP1 deficiency does not affect CD1d expression, and rule out defective presentation by cortical thymocytes as a cause of defective iNKT cell development in RasGRP1-/- mice.
Increased death of iNKT cells in the absence of RasGRP1
Insufficient Vα14-Jα18 recombination has been shown to cause a severe developmental block early in iNKT development in some mouse models (28, 29). We detected similar levels of Vα14 to Jα18, Jα2, or Jα56 recombination in RasGRP1+/- and RasGRP1-/- CD4+CD8+ DP thymocytes (Fig. 3A), ruling out the possibility that the deficiency of RasGRP1 somehow inhibited Vα14-Jα18 recombination. Beside iVα14TCR, homotypic interactions of cell surface receptors Slamsf1 and Slamsf6 on thymocytes also play an essential role in iNKT development (30). No differences in the surface expression of these receptors were detected between RasGRP1+/- and RasGRP1-/- thymocytes (Fig. 3B). However, a significantly higher rate of cell death was observed in the RasGRP1-/- CD1d-Tet+TCRβ+ iNKT cells as well as CD1d-Tet-TCRβ+ cαβT cells than in the RasGRP1+/- controls, suggesting that RasGRP1 is important for iNKT and cαβT cell survival, and increased death of these cells may contribute to the developmental defects in RasGRP1-/- mice (Fig. 3C). Ki67 expression is usually correlated with cell division. Freshly isolated RasGRP1-/- iNKT cells displayed a higher level of Ki67 staining compared with control iNKT cells (Fig. 3D-3E), suggesting increased homeostatic proliferation of RasGRP1-/- iNKT cells in vivo, likely due to the T cell lymphopenic environment in these mice. We further used CFSE-dilution assay to examine whether RasGRP1 regulates TCR induced iNKT cell activation. As shown in Figure 3F, WT but not RasGRP1-/- iNKT cells proliferated following α-GalCer stimulation for 72 hours in vitro (Fig. 3F). Together, these data suggest that RasGRP1 is important for iVα14TCR-induced iNKT cell activation.
Figure 3. Increased death of iNKT cells in the absence of RasGRP1.
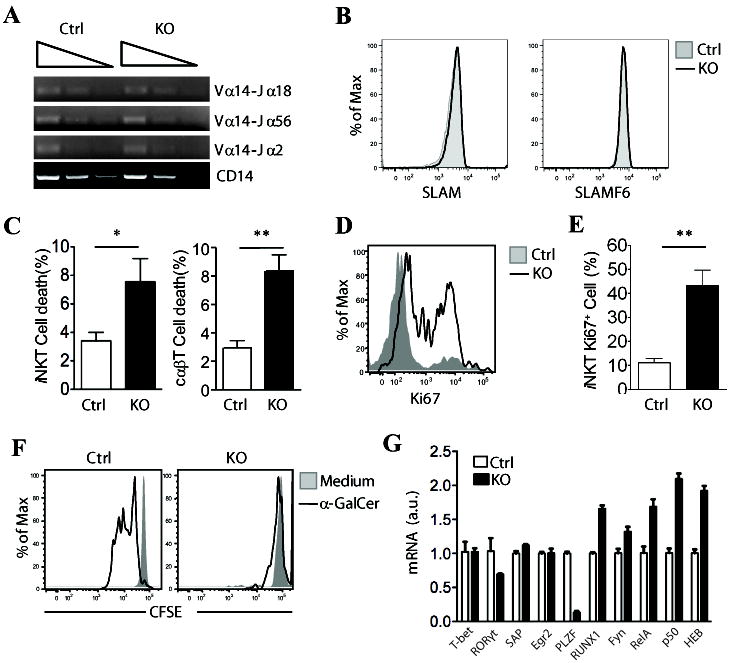
(A) Semi-quantitative PCR analysis of sorted CD4+CD8+ thymocytes from RasGRP1+/- and RasGRP1-/- mice with primers for Vα14-Jα2, Vα14-Jα18, Vα14-Jα56, and CD14 (loading control). (B) Expression of CD1d, SLAM (CD150), and SLAMF6 (Ly108) on CD4+CD8+ thymocytes from RasGRP1+/- and RasGRP1-/- mice. Data shown are representative of three mice per group. (C) Percentages of cell death of CD1d-Tet+TCRβ+ iNKT cells and CD1d-Tet-TCRβ+ cαβT cells from thymus (mean ± SEM, n=4). (D, E) Increased Ki67 expression in RasGRP1-/- iNKT cells. Ki67 expression in iNKT cells gated from WT and RasGRP1-/- thymocytes were determined by intracellular staining. (D) Overlay of histogram for Ki67 expression of gated iNKT cells; (E) Mean ± SEM of Ki67+ iNKT cells from WT and RasGRP1-/- thymus (n=5). (F) Impaired proliferation of RasGRP1-/- iNKT cells in response to α-Galcer stimulation in vitro. CFSE-labeled WT and RasGRP1-/- thymocytes were left unstimulated or stimulated with 125 ng/ml α-Galcer at 37°C for 72 hours. Cells were then stained for APC-CD1d-Tet and TCRβ. Overlaid histograms show CFSE levels in gated WT and RasGRP1-/- iNKT cells. (G) Real-time PCR analysis of mRNA expression of various proteins in sorted CD4+CD8+ thymocytes from RasGRP1+/- and RasGRP1-/- mice. *p<0.05; **, p<0.01 (Student t-test).
Signaling proteins SAP (31, 32) and Fyn (33, 34), as well as several transcription factors, such as RORγt (29, 35), Runx1 (35), cMyc (36), and HEB (28), are all critical for early iNKT cell development (37). No obvious differences in mRNA expression of these molecules were detected between RasGRP1+/- and RasGRP1-/- DP thymocytes (Fig. 3G), ruling out that RasGRP1 deficiency may affect iNKT cell development through modulating mRNA expression of these molecules. However, expression of PLZF, a transcription factor critical for the development of CD44+ iNKT cells (38, 39), was much lower in RasGRP1-/- iNKT cells than in RasGRP1+/- control, which might contribute to the relative enrichment of CD24-CD44- iNKT cells in RasGRP1-/- mice.
Selective absence of CD4+ iNKT cells in RasGRP1-/- mice
It has been demonstrated that, while stage 0 iNKT cells all express CD4 (8), the presence of CD4- iNKT cells can be observed in thymus at later stages or in the periphery. While accumulating evidence has revealed that the CD4+ and CD4- iNKT cells are functionally distinct (10, 40, 41), the developmental relationship between these two subsets is not well understood. Recently published data show that the CD4-NK1.1- cells appeared to be precursors of the CD4-NK1.1+ iNKT cells in the thymus. A revised model of thymic iNKT development was proposed in which the CD4- and CD4+ subsets represent two distinct sub-lineages of iNKT cells, whose divergence appears to occur at stage 1 when the CD4- iNKT cells are first observed (10). However, genetic evidence supporting such lineage definition is rare and mechanisms directing such lineage differentiation are not well defined. Strikingly, a dramatic decrease in the percentage of CD4+ subset was observed in the RasGRP1-/- iNKT cells in thymus, spleen, and liver (Fig. 4A-4B). When assessing the CD4 expression pattern at each iNKT developmental stage in RasGRP1+/- thymus, there is a progressive increase of the CD4- subset as the iNKT cells mature. About 10%, 25% and 40% of stage 1, stage 2 and stage 3 iNKT cells are CD4-. However, in RasGRP1-/- iNKT cells, about 90% of stage 2 and stage 3 iNKT cells were CD4-, yet no difference in CD4+/CD4- ratio was observed at stage 1 as compared to RasGRP1+/- (Fig. 4C-4D). Thus, besides promoting early iNKT cell development, RasGRP1 is selectively required for the maturation and/or maintenance of the CD4+CD44+ iNKT cells.
Figure 4. Selective absence of CD4+ iNKT cells in RasGRP1-/- mice.
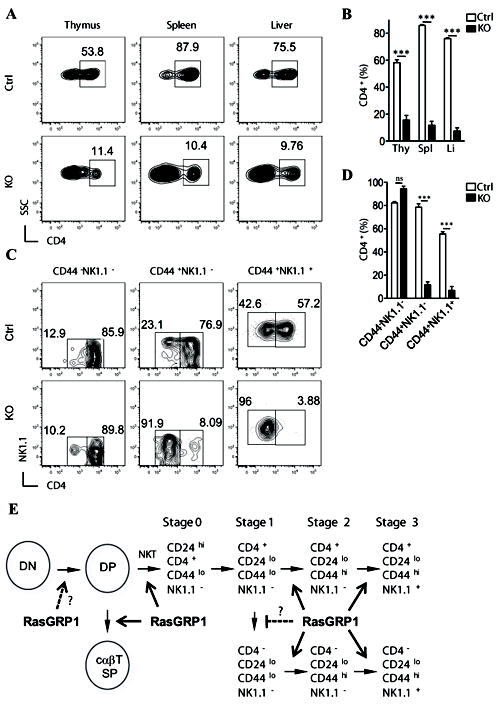
(A) Expression of CD4 on CD1d-Tet+CD24- gated cells from thymus, spleen, and liver. (B) Percentage of CD4+ in the CD1d-Tet+CD24- iNKT cells the indicated organs (n=4). (C) Expression of CD4 and NK1.1 on CD1d-Tet+CD24- gated thymocytes with various cell surface phenotypes. (D) Percentage of CD4+ iNKT cells in different stages (n=4). (E) Schematic illustration of RasGRP1 for cαβT and iNKT cell development. RasGRP1 plays a critical role for positive selection of both cαβT and iNKT cells. RasGRP1 is crucial for the generation/maintenance of CD4+ iNKT cells. In addition, RasGRP1 may promote late stage iNKT cell maturation. ***, p<0.001 (Student t-test).
Discussion
RasGRP1 promotes positive selection of cαβT cells, particularly those expressing TCR with low affinity to self-peptide-MHC complex (22). Positive selection of thymocytes with relative high affinity to self-peptide-MHC complex, including regulatory T cells and some innate CD8 T cells, is less dependent on RasGRP1 (42, 43). We have demonstrated here that RasGRP1 plays crucial roles in iNKT cell development and is important for the generation and/or maintenance of CD4+ iNKT cells (Fig. 4E). At present, it is still unclear how RasGRP1 promotes αβT and iNKT cell maturation. The increased death of RasGRP1 deficient cαβT cells and iNKT cells suggests that RasGRP1 may promote normal development of iNKT and cαβT cells by enhancing their survival. Of note, in addition to activating the Ras-Erk1/2 pathway in thymocytes following TCR engagement (22), we have recently found that RasGRP1 is also critical for TCR-induced activation of PI3K/Akt and the mammalian target of rapamycin (mTOR) (44). Both PI3K/Akt and mTOR are important regulators for cell survival, growth, and metabolism (45-47). It is likely that RasGRP1 may promote iNKT cells and cαβT cell maturation through multiple mechanisms.
The CD4+CD44+ iNKT cells are selectively or more severely affected than the CD4-CD44+ iNKT cells by RasGRP1 deficiency, suggesting that these two subsets of cells may signal differently. In RasGRP1 deficient thymocytes, TCR-induced activation of Ras/Erk1/2, PI3K/Akt, and mTOR is greatly decreased but not completely abolished (44). The exact differences of these signaling events between the CD4+iNKT T cells and CD4-iNKT T cells, as well as the effect of RasGRP1 deficiency on the activation of these signaling pathways in iNKT cells, are hard to assess since these cells are rare. At present, it is unclear whether the CD4-CD44+ iNKT cells are independent or less dependent of one or multiple signaling pathways downstream of RasGRP1 or they utilize other guanine nucleotide exchange factors such as Sos to activate these downstream signaling molecules. However, RasGRP1 promotes Sos to induce Ras activation (48). TCR induced Ras/Erk1/2 activation in RasGRP1-/- iNKT cells is likely decreased and the CD4+CD44+ and the CD4+CD44- iNKT cells probably have differential requirement for the Ras/Erk1/2 pathway. In addition to RasGRP1, deficiency of the transcription factor GATA-3 also cause a severe decrease of CD4+ iNKT cells in mice (49). Together, these observations provide genetic evidence that the CD4+ and CD4- iNKT cells are distinct sublineages with differential signaling/transcription factor requirements for their development. Further studies are required to determine whether RasGRP1 and GATA3 may regulate each other to promote CD4+ iNKT cell development.
It is important to note that our data appear to contradict a previous report that the Ras-Mek1/2-Erk1/2 pathway is dispensable for NKT cell development (50). In that study, dominant negative Ras and Mek1, specifically expressed in thymocytes, cause severe decreases of CD4+CD8- and CD4-CD8+ single positive thymocytes. However, NK1.1+TCRβ+ T cells were reported to be normal. Since CD1d-Galcer tetramer was not available at that time, the effects of dnRas/dnMek1 on iNKT cell development remain unclear. However, we did observe sharp decrease of NK1.1+TCRβ+ cells in RasGRP1-/- mice as well (data not shown). The discrepancy between these two studies could result from less complete abolishment of the Ras-Erk1/2 signaling in thymocytes of the dnRas/dnMek transgenic mice than in the RasGRP1-/- mice, some unknown effects of dnRas/dnMek1 transgenes on the cells, or variegate expression pattern due to the integration site effects on the transgenes. Additionally, RasGRP1 deficiency and dnRas or dnMek1 may differentially affect signaling pathways such as the PI3K/Akt, mTOR, and other yet to be identified signaling pathways that may play different roles for iNKT cell development.
Acknowledgments
We thank Nancy Martin in Duke Cancer Center Flow Cytometry Core Facility for providing sorting services, Li Xu for technical support, the NIH Tetramer Core Facility for providing the CD1d tetramer, and Tommy O’Brien and Dr. Kim Nichols for critical reviewing the manuscript.
Abbreviations
- α-Galcer
α-galactosyl ceramide
- DAG
Diacylglycerol
- iNKT
invariant natural killer T cell
- iVα14TCR
invariant Vα14-Jα18 T cell receptor
- cαβT
conventional αβ T cells
- DP
double positive
- SP
single positive
- Li
Liver
- Spl
spleen
- Thy
thymus
Footnotes
This study is supported by funding from the National Institute of Health (R01AI076357, R01AI079088, and R21AI079873), the American Cancer Society, and the American Heart Association to X-P.Z.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerundolo V, Kronenberg M. The role of invariant NKT cells at the interface of innate and adaptive immunity. Semin Immunol. 2010;22:59–60. doi: 10.1016/j.smim.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 5.Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–364. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 7.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 8.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci U S A. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 12.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 13.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 14.Zhong XP, Guo R, Zhou H, Liu C, Wan CK. Diacylglycerol kinases in immune cell function and self-tolerance. Immunol Rev. 2008;224:249–264. doi: 10.1111/j.1600-065X.2008.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, Glimcher LH. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, Joyce S. NF-κB controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004;172:2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- 17.Stanic AK, Bezbradica JS, Park JJ, Van Kaer L, Boothby MR, Joyce S. Cutting edge: the ontogeny and function of Vα14Jα18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. J Immunol. 2004;172:4667–4671. doi: 10.4049/jimmunol.172.8.4667. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activation. Proc Natl Acad Sci U S A. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elewaut D, Shaikh RB, Hammond KJ, De Winter H, Leishman AJ, Sidobre S, Turovskaya O, Prigozy TI, Ma L, Banks TA, Lo D, Ware CF, Cheroutre H, Kronenberg M. NIK-dependent RelB activation defines a unique signaling pathway for the development of Vα14 iNKT cells. J Exp Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallabhapurapu S, Powolny-Budnicka I, Riemann M, Schmid RM, Paxian S, Pfeffer K, Korner H, Weih F. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur J Immunol. 2008;38:3508–3519. doi: 10.1002/eji.200737830. [DOI] [PubMed] [Google Scholar]
- 21.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 23.Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity. 2002;17:617–627. doi: 10.1016/s1074-7613(02)00451-x. [DOI] [PubMed] [Google Scholar]
- 24.Shen S, Wu J, Srivatsan S, Gorentla BK, Shin J, Xu L, Zhong XP. Tight Regulation of Diacylglycerol-Mediated Signaling Is Critical for Proper Invariant NKT Cell Development. J Immunol. 2011;187:2122–2129. doi: 10.4049/jimmunol.1100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong XP, Hainey EA, Olenchock BA, Jordan MS, Maltzman JS, Nichols KE, Shen H, Koretzky GA. Enhanced T cell responses due to diacylglycerol kinase zeta deficiency. Nat Immunol. 2003;4:882–890. doi: 10.1038/ni958. [DOI] [PubMed] [Google Scholar]
- 26.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A. 2005;102:5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 34.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Vα14 iNKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Mycko MP, Ferrero I, Wilson A, Jiang W, Bianchi T, Trumpp A, MacDonald HR. Selective requirement for c-Myc at an early stage of Vα14 iNKT cell development. J Immunol. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 37.Das R, Sant’Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human Vα24 natural killer T cells. J Exp Med. 2002;195:637–641. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priatel JJ, Chen X, Huang YH, Chow MT, Zenewicz LA, Coughlin JJ, Shen H, Stone JC, Tan R, Teh HS. RasGRP1 regulates antigen-induced developmental programming by naive CD8 T cells. J Immunol. 2010;184:666–676. doi: 10.4049/jimmunol.0803521. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Priatel JJ, Chow MT, Teh HS. Preferential development of CD4 and CD8 T regulatory cells in RasGRP1-deficient mice. J Immunol. 2008;180:5973–5982. doi: 10.4049/jimmunol.180.9.5973. [DOI] [PubMed] [Google Scholar]
- 44.Gorentla BK, Wan CK, Zhong XP. Negative regulation of mTOR activation by diacylglycerol kinases. Blood. 2011;117:4022–4031. doi: 10.1182/blood-2010-08-300731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong XP, Shin J, Gorentla BK, O’Brien T, Srivatsan S, Xu L, Chen Y, Xie D, Pan H. Receptor signaling in immune cell development and function. Immunol Res. 2011;49:109–123. doi: 10.1007/s12026-010-8175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol Cell Biol. 2007;27:2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 50.Alberola-Ila J, Hogquist KA, Swan KA, Bevan MJ, Perlmutter RM. Positive and negative selection invoke distinct signaling pathways. J Exp Med. 1996;184:9–18. doi: 10.1084/jem.184.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]


