Abstract
DEPTOR is a recently identified inhibitor of the mTOR kinase that is highly regulated at the posttranslational level. In response to mitogens, we found that DEPTOR was rapidly phosphorylated on three serines in a conserved degron, facilitating binding and ubiquitylation by the F-box protein βTrCP, with consequent proteasomal degradation of DEPTOR. Phosphorylation of the βTrCP degron in DEPTOR is executed by CK1α, after a priming phosphorylation event mediated by either the mTORC1 or mTORC2 complexes. Blocking the βTrCP-dependent degradation of DEPTOR via βTrCP knockdown or expression of a stable DEPTOR mutant that is unable to bind βTrCP results in mTOR inhibition. Our findings reveal that mTOR cooperates with CK1α and βTrCP to generate an auto-amplification loop to promote its own full activation. Moreover, our results suggest that pharmacologic inhibition of CK1 may be a viable therapeutic option for the treatment of cancers characterized by activation of mTOR signaling pathways.
Introduction
The mammalian Target Of Rapamycin (mTOR) kinase controls many aspects of the response to growth factor and nutrient signaling (Laplante and Sabatini, 2009; Zoncu et al., 2011). mTOR Complex 1 (TORC1) and TORC2 share the mTOR and mLST8/GβL proteins, but the complexes also feature distinct subunits, with RAPTOR and PRAS40 in TORC1 and RICTOR, PROTOR, and mSin1 in TORC2 (Laplante and Sabatini, 2009). In general, TORC1 controls mRNA translation, ribosome biogenesis, cell growth, and autophagy through substrates such as S6K1 and 4E-BP1, while TORC2 controls cell proliferation, cell survival, and the cytoskeleton through substrates such as Akt, SGK1, and PGCα (Dancey, 2010; Guertin and Sabatini, 2007; Sengupta et al. 2010)
The pathways controlled by TORC1 and TORC2 are frequently activated in tumors by mutations in upstream signaling factors (e.g., growth factor receptors, PI3K regulators, or PTEN) and mTOR inhibitors have been used successfully in the treatment of several cancers (Dancey, 2010; Hay, 2005). However, direct activating mutations of mTOR have not been observed in cancer, and in some settings, mTOR has been shown to possess tumor suppressive properties, likely due to negative feedback loops that control the TORC1 or TORC2 pathways (Laplante and Sabatini, 2009).
Both mTOR complexes are directly inhibited by DEPTOR, which binds and inhibits mTOR through a PDZ domain (Peterson et al., 2009). DEPTOR is downregulated in many tumors, suggesting a tumor suppressor function, which is consistent with the activation of mTOR in many tumors. However, DEPTOR is overexpressed in multiple myeloma via transcription or copy number amplifications, and this overexpression is necessary for Akt activation and cell survival, which is likely mediated through the feedback inhibition of PI3K (Carrasco et al., 2006; Peterson et al., 2009). Notably, despite a general downregulation of DEPTOR across other tumor types, amplification of the genomic region containing the DEPTOR locus is an indicator of poor prognosis or tumor progression in tumor subsets from multiple cancers, including breast cancer, prostate cancer, lung cancer, and CML, and DEPTOR is overexpressed in many of these tumors (Chin et al., 2007; Joos et al., 2003; van Duin et al., 2005a; van Duin et al., 2005b).
The impact of DEPTOR in cancer makes it vital to understand the regulation of DEPTOR. DEPTOR activity appears to be regulated largely through the control of DEPTOR levels, which are tightly controlled both transcriptionally and posttranslationally in response to growth factor signaling (Peterson et al., 2009). While DEPTOR levels are high in the absence of serum, in response to serum, transcription of DEPTOR decreases and DEPTOR protein is rapidly phosphorylated on as many as 13 sites. Many of these phosphorylations are mTOR-dependent, and non-phosphorylated mutants of DEPTOR bind mTOR more efficiently than wild type DEPTOR, indicating that phosphorylation of DEPTOR inhibits binding to mTOR. mTOR activity also correlates with DEPTOR degradation, suggesting that these two processes are linked. However, the precise mechanisms for this regulation remain unclear.
Skp1/Cul1/F-box protein (SCF) ubiquitin ligase complexes control the degradation of many important regulatory proteins (Cardozo and Pagano, 2004). In mammals, there are 69 SCF ligases, each characterized by a different F-box substrate targeting subunit (Jin et al., 2004). In this study, we identify SCFβTrCP as the ubiquitin ligase for DEPTOR and demonstrate that SCFβTrCP mediates the mTOR- and CK1α-dependent degradation of DEPTOR.
Results
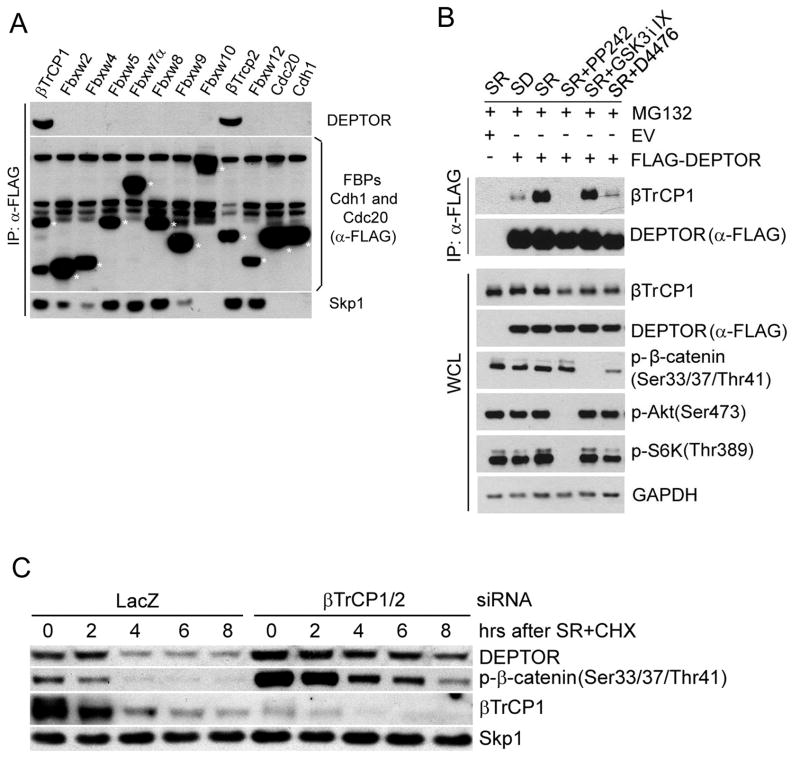
The expression of Cul1(1-252), a dominant negative Cul1 mutant that binds Skp1 and F-box proteins, but cannot recruit an E2 ubiquitin conjugating enzyme, results in the accumulation of SCF substrates (Piva et al., 2002; Yen and Elledge, 2008). To identify new SCF substrates, we transiently transfected Cul1(1-252) into HeLa cells and analyzed cell extracts for the levels of several regulators of cell proliferation by immunoblotting. The level of DEPTOR was increased compared to mock transfected controls (Fig. S1A), suggesting that DEPTOR is an SCF substrate. Therefore, we investigated which F-box protein targets DEPTOR to the SCF using a library of F-box protein cDNAs. Screening of the FBXW (F-box proteins with WD40 repeats) family proteins, as well as Cdc20 and Cdh1 (WD40 domain-containing subunits of an SCF-like ubiquitin ligase), revealed that endogenous DEPTOR specifically interacts with βTrCP1 and βTrCP2 (Fig. 1A), paralogous F-box proteins that share identical biochemical properties and substrates. (βTrCP will refer to both, unless specified.)
Figure 1. DEPTOR is a serum-dependent substrate of βTrCP.
A) DEPTOR binds βTrCP1 and βTrCP2. HEK293T cells were transfected with the indicated FLAG-tagged proteins. Forty-eight hours post-transfection, after a 16-hour serum starvation, cells were re-stimulated with media containing serum and MG132 for three hours, prior to harvesting for immunoprecipitations and immunoblotting as indicated. Asterisks indicate the position of exogenously expressed proteins.
B) HEK293T cells were transfected with an empty vector (EV) or FLAG-tagged DEPTOR. Forty-eight hours post-transfection, after a 24-hour serum starvation, cells were pretreated with the indicated drugs for two hours and then stimulated with serum-containing media (SR) for three hours, prior to harvesting for immunoprecipitations and immunoblotting as indicated. (WCL, whole cell lysate).
C) During a 72-hour serum starvation, T98G cells were transfected with siRNAs targeting either LacZ or βTrCP1 and βTrCP2. Cells were subsequently re-stimulated with media containing serum and cycloheximide (SR+CHX), and samples were harvested at the indicated time points for immunoblotting.
Significantly, the binding of DEPTOR to transiently-expressed βTrCP was dependent on the substrate binding domain, as demonstrated by the inability of a previously-established substrate binding point mutant, βTrCP2(R434A), or a WD40 repeat deletion mutant, βTrCP2(ΔWD40) (Suzuki et al., 2000; Wu et al., 2003), to bind endogenous DEPTOR (Fig. S1B). Serum starvation of HeLa cells induces accumulation of DEPTOR, whereas serum stimulation results in DEPTOR degradation (Peterson et al., 2009). After confirming these results in T98G cells (Fig. S1C), we found that serum stimulation induced a significant increase in the binding of DEPTOR to endogenous βTrCP1 (Fig. 1B).
To investigate the hypothesis that βTrCP controls the degradation of DEPTOR in serum-stimulated cells, we reduced the expression of both βTrCP1 and βTrCP2 in T98G and HeLa cells using a validated siRNA (Dehan et al., 2009; Dorrello et al., 2006; Fong and Sun, 2002; Guardavaccaro et al., 2008; Peschiaroli et al., 2006). Fig. 1C and Fig. S1D show that silencing βTrCP increased the DEPTOR half-life upon stimulation with serum, demonstrating that βTrCP controls DEPTOR stability.
Next, we mapped the βTrCP binding motif in human DEPTOR. Using deletion mutants, the binding motif was mapped to a region of DEPTOR between amino acids 241 and 340 (Fig. S2A). βTrCP binds substrates via phosphorylated residues in conserved degradation motifs (degrons), typically including the consensus sequence DpSGXXpS or similar variants, such as pS/TpSGXXpS (Fig. S2B). The βTrCP binding region of DEPTOR contains a conserved 286SSGYFS291 motif, matching other βTrCP substrate degrons. To investigate whether DEPTOR binds βTrCP via this motif, we generated serine to alanine mutants and tested their binding to endogenous βTrCP1. Single mutations of Ser286, Ser287, and Ser291 to Ala or a triple mutation of Ser286/287/291 to Ala inhibited the interaction between DEPTOR and βTrCP1, although the mutations did not affect DEPTOR binding to endogenous mTOR (Fig. 2A).
Figure 2. The DEPTOR degron is controlled by phosphorylation.
A) Ser286, Ser287, and Ser291 are required for the interaction of DEPTOR with βTrCP. HEK293T cells were transfected with an empty vector (EV) or the indicated HA-DEPTOR constructs. Forty-eight hours post-transfection, after a 24-hour serum starvation, cells were re-stimulated with media containing serum and MG132 for three hours, prior to harvesting for HA immunoprecipitation and western blotting as indicated. (WCL, whole cell lysate).
B) The DEPTOR degron requires phosphorylation to bind βTrCP1. HEK293T lysates were used in binding reactions with beads coupled to a peptide containing the sequence SSGYFS (lane 2) or the phospho-motif SpSpSGYFpS (lane 3). Beads were washed with lysis buffer, and bound proteins were eluted and subjected to SDS-PAGE and immunoblotting.
C) In vivo phosphorylation of DEPTOR on Ser286/287/291/299 is induced by mitogens. HEK293T cells were transfected with the indicated HA-tagged DEPTOR constructs. Following serum deprivation (SD) for 24 hours, cells were stimulated with serum (SR) for three hours, in the presence or absence of PP242 or D4476, as indicated. Whole cell lysates (WCL) were immunoprecipitated and immunoblotted as indicated.
D) Ser293 and Ser299 are required for the interaction of DEPTOR with βTrCP. HEK293T cells were transfected with FLAG-βTrCP1 and either an empty vector (EV) or the indicated HA-DEPTOR constructs. Forty-eight hours post-transfection, after a 24-hour serum starvation, cells were re-stimulated with media containing serum and MG132 for three hours, prior to harvesting for immunoprecipitations and immunoblotting as indicated. (WCL, whole cell lysate).
To confirm the role of phosphorylation in the interaction of DEPTOR with βTrCP, we used immobilized, synthetic peptides containing the candidate degron sequence to test binding to βTrCP1. While a peptide containing phosphorylated Ser286, Ser287, and Ser291 efficiently bound βTrCP1 (but not Fbxw5 or Fbxw9), a corresponding, non-phosphorylated peptide was unable to bind βTrCP1 (Fig. 2B). Accordingly, λ-phosphatase treatment of βTrCP1 immunoprecipitates abolished the interaction with DEPTOR (Fig. S2C). These results, together with the analysis of point mutants (Fig. 2A), the crystal structure of the βTrCP1-β-catenin complex (Wu et al., 2003), and the modeling of the βTrCP1-BimEL degron interaction (the BimEL and DEPTOR degrons are identical, as shown in Figure S2B) (Dehan et al., 2009) indicate that phosphorylation of all three serine residues in the DEPTOR degron (Ser286, Ser287, and Ser291) is necessary for - and directly mediates - the interaction with βTrCP.
To further investigate DEPTOR phosphorylation, we used a phospho-specific antibody against the pSpSGYFpS degron motif. This antibody recognized wild type DEPTOR, but not a DEPTOR(S286/287/291A) mutant (Fig. 2A). Additionally, DEPTOR point mutants displayed decreasing levels of detection, suggesting that all three serines are phosphorylated and contribute to recognition by this antibody. Significantly, we found that DEPTOR was phosphorylated on its degron in HEK293T cells in response to stimulation with serum, but it was poorly phosphorylated in serum-starved HEK293T cells (Fig. 2C).
Several βTrCP substrates, such as β-catenin, Cdc25A, Emi1, Snail, Wee1, and YAP, are phosphorylated on their degrons only after an initial phosphorylation event that either allows binding to or exposure of a previously masked site for a second kinase (Frescas and Pagano, 2008; Hunter, 2007). To investigate whether a similar mechanism controls phosphorylation of the DEPTOR degron, we mutated a number of residues flanking the degron. Mutation of Ser279, Ser280, Ser292, Thr295, Ser297, and Ser298 to Ala, singly or in combination, did not inhibit DEPTOR binding to βTrCP1 (Fig. 2A and S2D). In contrast, mutation of Ser293, Ser299, or both strongly reduced the interaction between βTrCP and DEPTOR, even in serum-stimulated cells (Fig. 2D and Fig. S2D). Additionally, a DEPTOR phospho-mimic mutant, in which Ser286, Ser287, and Ser291 in the degron are mutated to Asp [DEPTOR(S286/287/291D)], retains the ability to bind βTrCP1 even when Ser293 and Ser299 are mutated to Ala (Fig. 2D). The ability of the phospho-mimic degron mutant of DEPTOR to bind βTrCP, together with the phospho-peptide experiment in Fig. 2B, demonstrates that Ser293 and Ser299 are dispensable for binding a pre-phosphorylated degron and suggest that phosphorylation of Ser293 and Ser299 may function to prime the phosphorylation of Ser286, Ser287, and Ser291.
We also used a phospho-specific antibody generated against a DEPTOR peptide C-terminal to the degron, with Ser299 phosphorylated. This antibody recognized wild type DEPTOR, but not DEPTOR(S299A) or DEPTOR(S293/299A) (Fig. 2C and data not shown). We found that DEPTOR was phosphorylated on Ser299 in HEK293T cells stimulated with serum, but this site was poorly phosphorylated in serum-starved cells (Fig. 2C). Interestingly, of the five serines in DEPTOR that are important for binding to βTrCP, four (Ser286, Ser287, Ser293, and Ser 299) have been previously identified as phosphorylation sites (Peterson et al., 2009; Villen et al., 2007). Additionally, a different study also identified these four serines as sites of phosphorylation that are enriched after proteasome inhibition (Gao et al. 2011).
To identify the kinase(s) involved in the phosphorylation and degradation of DEPTOR, we performed a candidate search using pharmacologic inhibition. We found that D4476 (a CK1 inhibitor) and PP242 (an mTOR inhibitor) counteracted the destabilizing effect of serum on DEPTOR, whereas GSK3i IX (a GSK3 inhibitor), U0126 (a MEK inhibitor), and API-2 (an Akt inhibitor) had no effect (Fig. S3A-B). Importantly, D4476 and PP242, but not U0126 and GSK3i IX, inhibited the interaction between DEPTOR and βTrCP and the phosphorylation of the DEPTOR degron (Fig. 1B, Fig. 2C, and data not shown). In agreement with the involvement of mTOR in DEPTOR degradation, we observed that low doses of rapamycin (an mTORC1 inhibitor) and high doses of wortmannin (a PI3K inhibitor that, at high concentrations, inhibits mTOR) induced DEPTOR stabilization (Fig. S3B and S3C). We also found that knockdown of mTOR or CK1α (but not CK1δ or CK1ε) resulted in accumulation of DEPTOR (Fig. 3A-B). Furthermore, silencing of either RAPTOR or RICTOR inhibited the serum-dependent destabilization of DEPTOR, although to a lesser extent than mTOR depletion (Fig. 3A), indicating that both mTORC1 and mTORC2 control DEPTOR turnover.
Figure 3. mTOR and CK1α are required for DEPTOR degradation.
A) Inhibition of TORC1 and TORC2 blocks DEPTOR degradation. During a 72-hour serum starvation, T98G cells were transfected with siRNAs targeting LacZ, RAPTOR, RICTOR, or mTOR. Cells were subsequently re-stimulated with media containing serum (SR), and samples were harvested at the indicated time points for immunoblotting.
B) Silencing of CK1α, but not CK1δ or CK1ε, induces DEPTOR accumulation. HeLa cells were infected with an empty lentivirus (EV) or lentiviruses containing shRNA targeting either CK1α, CK1δ, or CK1ε. Seven days post-infection, samples were harvested at the indicated time points for immunoblotting.
C) Differential sensitivity of wild type DEPTOR and DEPTOR mutants to inhibition of CK1 and mTOR. HEK293T cells were transfected with the indicated HA-DEPTOR constructs. Forty-eight hours post-transfection, after a 24-hour serum starvation, cells were re-stimulated with media containing serum and MG132 for three hours, in the presence or absence of PP242 or D4476, as indicated. Cell lysates were immunoprecipitated and immunoblotted as indicated. (WCL, whole cell lysate).
D) In vitro ubiquitylation assays of recombinant DEPTOR [WT or DEPTOR(S287/287/291A),] were conducted in the presence of the indicated proteins. Samples were incubated at 30°C for 90 minutes. The bracket on the left side of the top panel marks a ladder of bands corresponding to polyubiquitylated DEPTOR.
E) In vitro phosphorylation of DEPTOR by mTOR promotes DEPTOR-CK1α interaction. Recombinant, purified GST-DEPTOR, GST-DEPTOR(S293/299A), or GST alone were incubated with ATP in the presence or absence of purified, recombinant mTOR. Reaction products were then diluted, incubated with FLAG-tagged CK1α, purified with GST sepharose 4B beads, and immunobloted as indicated.
We then used phospho-mimic mutants of DEPTOR to study the hierarchy of mTOR- and CK1α-mediated phosphorylation of DEPTOR. The binding of wild type DEPTOR to endogenous βTrCP is inhibited by either PP242 or D4476 (Fig. 1B and Fig. 3C), but DEPTOR phosho-mimic mutants are differentially responsive to these inhibitors. The binding of DEPTOR (S286/287/291D) to βTrCP is not inhibited by either D4476 or PP242 (Fig. 3C). In contrast, the binding of DEPTOR (S293/299D) to βTrCP is not inhibited by PP242, but is still inhibited by D4476 (Fig. 3C). These findings suggest that mTOR phosphorylates Ser293 and Ser299 to promote degron phosphorylation by CK1α. Accordingly, while PP242 inhibited the phosphorylation of DEPTOR on both Ser299 and the degron, D4476 was able to inhibit the phosphorylation of degron, but not of Ser299 (Fig. 2C). Finally, CK1α-mediated stimulation of the DEPTOR-βTrCP interaction was inhibited by PP242 (Fig. S3D).
To test whether CK1 and mTOR can phosphorylate DEPTOR on its degron, we performed in vitro kinase assays using recombinant, bacterially-expressed, purified DEPTOR and kinases. CK1 phosphorylated the degron of DEPTOR, as shown by western blotting with the phospho-specific antibody (Fig. S3E-F). In contrast, mTOR alone was unable to induce phosphorylation of DEPTOR on Ser286, Ser287, and Ser291. Importantly, incubation with mTOR enhanced the CK1-dependent phosphorylation of DEPTOR, likely due to mTOR-dependent phosphorylation of Ser293 and Ser299, since no mTOR-dependent enhancement of phosphorylation was observed with DEPTOR(S293/299A). Finally, mTOR, but not CK1, was able to phosphorylate DEPTOR on Ser299 in vitro (Fig. 3D-E). Accordingly, in vivo phosphorylation of Ser293 and Ser299 is inhibited by torin (a highly specific mTOR inhibitor; Peterson et al., 2009), mTOR phosphorylates Ser293 and Ser299 in vitro, and pre-phosphorylation of DEPTOR by mTOR enhances its CK1-dependent in vitro phosphorylation (Gao et al., 2011).
Finally, we reconstituted the ubiquitylation of DEPTOR in vitro. Wild type DEPTOR, but not DEPTOR(S286/287/291A) or DEPTOR(S293/299A), was ubiquitylated only when both βTrCP1 and CK1 were present in the reaction (Fig. 3D and Fig. S3G). Moreover, in agreement with the phosphorylation data, mTOR enhanced the βTrCP1- and CK1-dependent ubiquitylation of DEPTOR.
The above results indicate that mTOR promotes phosphorylation of the DEPTOR degron by CK1α, so we further investigated potential molecular mechanisms. Fig. S3D shows that CK1α and DEPTOR bind and that treatment of HEK293T cells with PP242 inhibits this binding. Additionally, purified, recombinant mTOR strongly stimulates the in vitro binding of CK1α to wild type DEPTOR, but not to DEPTOR(S293/299A) (Fig. 3E), suggesting that phoshorylation of Ser293 and Ser299 in DEPTOR by mTOR generates a binding site for CK1α, promoting DEPTOR phosphorylation by CK1α.
Altogether, these data strongly support a model in which, in response to mitogenic stimulation, mTOR phosphorylates DEPTOR on Ser293 and Ser299, promoting the CK1α-mediated phosphorylation of DEPTOR on Ser286, Ser287, and Ser291, facilitating βTrCP binding, SCFβTrCP-mediated ubiquitylation, and consequent degradation. Therefore, inhibition of βTrCP-mediated degradation of DEPTOR should lead to increased DEPTOR levels and decreased mTOR activity. This hypothesis was tested in three ways. First, T98G cells were transfected with siRNAs against LacZ or βTrCP and were synchronized in G0/G1 by serum starvation (Fig. 4A). Following re-stimulation with serum, DEPTOR levels rapidly decreased in the siLacZ transfected cells, but they decreased much less in the siβTrCP cells. In accordance with the increased DEPTOR levels in the βTrCP knockdown samples, the induction of phosphorylated S6K1 (Thr389) was severely blunted, demonstrating a decrease in mTOR activation. Second, to confirm that the observed effect of βTrCP knockdown on mTOR activity was mediated through increased DEPTOR levels, we transiently transfected either wild type DEPTOR or DEPTOR(S286/287/291A) into HeLa cells, which were subsequently serum starved for 24 hours, before re-stimulation with serum. As predicted, in contrast to wild type DEPTOR, DEPTOR(S286/287/291A) was not degraded when cells were exposed to serum, and the mTOR-mediated phosphorylation of S6K1 in response to serum was strongly inhibited (Fig. S4A). Virtually identical results were obtained using retroviruses that expresses DEPTOR (S286/287/291A) at near physiological levels in T98G cells (Fig. 4B). Significantly, after serum stimulation, cells expressing DEPTOR(S286/287/291A) displayed reduced cell size relative to cells expressing wild-type DEPTOR or containing an empty virus (Fig. 4C and Fig. S4B).
Figure 4. Failure to degrade DEPTOR results in mTOR activation defects.
A) During a 72-hour serum starvation, T98G cells were transfected with siRNAs targeting either LacZ or βTrCP1 and βTrCP2. Cells were subsequently re-stimulated with media containing serum (SR), and samples were harvested at the indicated time points for immunoblotting as indicated.
B) T98G cells were infected with either an empty virus (EV), a retrovirus expressing wild type DEPTOR, or DEPTOR(S286/287/291A). After 72 hours of serum deprivation, cells were re-stimulated with serum (SR) for the indicated times, harvested, and analyzed by immunoblotting.
C) The experiment was performed as in (B), and cell size was determined by FACS (forward scatter) in cells deprived of serum (SD) and 24 hours after serum addition (SR).
Discussion
Proper regulation of mTOR activity is essential to blocking tumorigenesis, and deregulation of the mTOR pathway at the level of DEPTOR appears common (Peterson et al., 2009). Our study demonstrates that DEPTOR is regulated at the posttranslational level by the SCFβTrCP ubiquitin ligase, in an mTOR- and CK1α-dependent manner, generating a positive feedback loop that facilitates full activation of mTOR. The mTOR-dependence of this auto-amplification loop is reminiscent of the SCFSkp2-mediated degradation of the CDK1/2 inhibitor p27 following phosphorylation of p27 by CDK1 or CDK2 (Frescas and Pagano, 2008), suggesting a common mechanism for the regulation of kinase inhibitors. Conversely, the observed effects of CK1α on DEPTOR demonstrate a non-canonical mechanism for CK1, which typically requires priming phosphorylation at the -3 position. The negative charge of phosphorylated Ser293 and Ser299 may function as acidic-like C-terminal residues to prime the phosphorylation of Ser286/287/91, similar to other CK1 substrates (Marin et al., 2003).
Finally, our results show that pharmacologic inhibition of CK1 increases DEPTOR levels and inhibits mTOR signaling, suggesting that CK1 inhibition may be a viable therapeutic option for the treatment of cancers characterized by low DEPTOR levels and activation of mTOR. Paradoxically, although mTOR activity is required for DEPTOR degradation, multiple myelomas appear to retain both high mTOR activity and high DEPTOR levels. It remains to be determined whether the elevated levels of DEPTOR in multiple myelomas result solely from transcriptional regulation or whether the βTrCP-mediated degradation of DEPTOR is also perturbed.
Experimental Procedures
Extract generation and western blotting
Extract preparation, immunoprecipitation, and western blotting were performed as previously described (Dehan et al., 2009; Dorrello et al., 2006). The rabbit polyclonal phospho-specific antibody was generated using a peptide containing the phospho-degron sequence SpSpSGYFpS. Fbxw5, Fbxw9, and cyclin A rabbit polyclonal antibodies were generated/characterized by the Pagano laboratory. Commerical mouse antibodies used: S6K(Thr389) (Cell Signaling), CK1ε (BD), β-Catenin (BD), Actin (Sigma), FLAG (Sigma), HA (Covance), GST (Invitrogen), and Claspin (Peschiaroli et al., 2006). Commercial rabbit antibodies used: βTrCP1 (Cell Signaling), Akt (Ser473) (Cell Signaling), Akt (Cell Signaling), S6K (Cell Signaling), GAPDH (Cell Signaling), mTOR (Cell Signaling), DEPTOR (Millipore), DEPTOR(Ser299) (Cell Signaling), CK1α (Cell Signaling), RICTOR (Bethyl), RAPTOR (Millipore), β-Catenin (Ser33/37/Thr41) (Cell Signlaing), and Skp1 (Invitrogen).
Plasmids, siRNAs, and shRNAs
DEPTOR mutants were generated using QuikChange Site-Directed Mutagenesis Kits (Stratagene). All cDNAs were completely sequenced. Transient transfections of HEK293T cells were performed using PEI. Additional cell lines were transfected using Lipofectamine 2000 (Invitrogen). siRNAs were transfected using Metafectene Pro (Biontex). The LacZ and βTrCP siRNAs were previously validated and described (Dehan et al., 2009; Dorrello et al., 2006; Fong and Sun, 2002). RAPTOR, RICTOR, mTOR siRNAs, and CK1α were from Sigma (SASI_Hs02_00366683, SASI_Hs01_00048380, SASI_Hs01_00203144, and TRCN0000006042, respectively). The Precision-LentiORF shRNA vector targeting CK1δ and CK1ε contained the sequence GGGCTTCTCCTATGACTAC. Retrovirus-mediated gene transfer was previously described (Guardavaccaro et al., 2008; Peschiaroli et al., 2006).
Cell lines, serum starvation, and drug treatments
Human Embryonic Kidney 293T (HEK293T), HeLa, and T98G cells (ATCC) were used as indicated. All cell lines were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% Fetal Bovine Serum (Hyclone) and antibiotics. All cells were starved for the indicated time periods in 0.1% serum. The following drugs were used: MG132 (Peptides International; 10μM), cycloheximide (Sigma; 100μg/mL), PP242 (Sigma; 2.5 μM), D4476 (Calbiochem; 50 μM), GSK3i (Calbiochem; 5 μM), Rapamycin (LC labs, 200nM), Akt inhibitor API-2 (Tocris Bioscience, 1μM), and U0126 (Calbiochem; 10 μM).
Supplementary Material
Highlights.
In response to mitogens, DEPTOR is degraded via SCFβTrCP.
Binding of DEPTOR to βTrCP requires phosphorylation of the DEPTOR degron.
mTOR and CK1α are required for the phosphorylation of the DEPTOR degron.
Failure to degrade DEPTOR results in mTOR activation defects.
Acknowledgments
The authors thank D. Foster, D. Sabatini, and W. Wei for reagents; W. Harper and W. Wei for sharing unpublished results. MP is grateful to T.M. Thor for continuous support. JRS is supported by the Mr. and Mrs. William G. Campbell Postdoctoral Fellowship in Memory of Carolyn Cabott from the American Cancer Society. This work was funded by grants from the National Institutes of Health (R01-GM057587, R37-CA076584, and R21-AG032560) and the Multiple Myeloma Research Foundation to MP. MP is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, Naderi A, Roberts I, Barbosa-Morais NL, Garcia MJ, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- Dancey J. mTOR signaling and drug development in cancer. Nat Rev Clin Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Fong A, Sun SC. Genetic evidence for the essential role of beta-transducin repeat-containing protein in the inducible processing of NF-kappa B2/p100. J Biol Chem. 2002;277:22111–22114. doi: 10.1074/jbc.C200151200. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tan M, Fukushima H, Locasale WJ, Liu P, Zhai B, Wan L, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR drives its own activation via SCFb-TRCP-dependent degradation of the mTOR inhibitor DEPTOR. Co-submitted to Mol Cell. 2011 doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, Pagano M. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos S, Granzow M, Holtgreve-Grez H, Siebert R, Harder L, Martin-Subero JI, Wolf J, Adamowicz M, Barth TF, Lichter P, Jauch A. Hodgkin’s lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer. 2003;103:489–495. doi: 10.1002/ijc.10845. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc Natl Acad Sci U S A. 2003;100:10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R, Liu J, Chiarle R, Podda A, Pagano M, Inghirami G. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol Cell Biol. 2002;22:8375–8387. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K. Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J Biol Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- van Duin M, van Marion R, Vissers K, Watson JE, van Weerden WM, Schroder FH, Hop WC, van der Kwast TH, Collins C, van Dekken H. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005a;44:438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
- van Duin M, van Marion R, Watson JE, Paris PL, Lapuk A, Brown N, Oseroff VV, Albertson DG, Pinkel D, de Jong P, et al. Construction and application of a full-coverage, high-resolution, human chromosome 8q genomic microarray for comparative genomic hybridization. Cytometry A. 2005b;63:10–19. doi: 10.1002/cyto.a.20102. [DOI] [PubMed] [Google Scholar]
- Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.