Abstract
BACKGROUND
There has been substantial interest in assessing whether RNAs (mRNAs and sncRNAs, i.e. small non-coding) delivered from mammalian spermatozoa play a functional role in early embryo development. While the cadre of spermatozoal mRNAs has been characterized, comparatively little is known about the distribution or function of the estimated 24 000 sncRNAs within each normal human spermatozoon.
METHODS
RNAs of <200 bases in length were isolated from the ejaculates from three donors of proved fertility. RNAs of 18–30 nucleotides in length were then used to construct small RNA Digital Gene Expression libraries for Next Generation Sequencing. Known sncRNAs that uniquely mapped to a single location in the human genome were identified.
RESULTS
Bioinformatic analysis revealed the presence of multiple classes of small RNAs in human spermatozoa. The primary classes resolved included microRNA (miRNAs) (≈7%), Piwi-interacting piRNAs (≈17%), repeat-associated small RNAs (≈65%). A minor subset of short RNAs within the transcription start site/promoter fraction (≈11%) frames the histone promoter-associated regions enriched in genes of early embryonic development. These have been termed quiescent RNAs.
CONCLUSIONS
A complex population of male derived sncRNAs that are available for delivery upon fertilization was revealed. Sperm miRNA-targeted enrichment in the human oocyte is consistent with their role as modifiers of early post-fertilization. The relative abundance of piRNAs and repeat-associated RNAs suggests that they may assume a role in confrontation and consolidation. This may ensure the compatibility of the genomes at fertilization.
Keywords: microRNA, Piwi-interacting RNAs RNA, spermatozoa, small non-coding RNA
Introduction
Our understanding of the complexity of 18–30 nucleotides small non-coding RNAs (sncRNA) continues to be refined. Their constituency has expanded to range from the well-known microRNA (miRNA) to the processing products of tRNAs and small nucleolar RNAs (snoRNA) (Kawaji and Hayashizaki, 2008; Kawaji et al., 2008; Jung et al., 2010). snoRNAs are best known as guiding RNA methylation of pseudouridylation. Recently, tiRNAs (tiny) that arise from sites of transcription initiation have been described (Taft et al., 2009, 2010), which may epigenetically mark CTCF (an evolutionarily conserved zinc finger protein) association (Taft et al., 2011). These CTCF sites are particularly interesting in light given their correlation with the human sperm histone regions (Arpanahi et al., 2009). It is clear that this collection of sncRNAs may function in varied processes that include gene expression, chromatin remodeling and the protection of the genome against transposition. The miRNA family is the best characterized of the sncRNAs. They were first identified in human (Ostermeier et al., 2005) then confirmed in mouse (Amanai et al., 2006; Yan et al., 2008) and porcine spermatozoa (Curry et al., 2009). The effect of miRNAs is generally post-transcriptional mediated degradation, through their interaction with the 3′ untranslated region (UTR).
There has been great interest in assessing whether both mRNAs and sncRNAs delivered from mammalian spermatozoa play a functional role in early embryo development [reviewed in (Krawetz, 2005; Lalancette et al., 2008)]. Proposed functions include developmental modifiers, as in the case of miRNAs or piwi-interacting RNA (piRNA) protectors of the genome, by masking repetitive and transposable elements or as part of confrontation and consolidation when the genomes first meet. The role of miRNAs as epigenetic modifiers is becoming appreciated (Kim et al., 2008; Valeri et al., 2009; Khraiwesh et al., 2010) in which disequilibrium is implicated in a diverse range of physiological responses ranging from non-obstructive azoospermia (Lian et al., 2009) to paramutation (Rassoulzadegan et al., 2006). In addition, the germ cell-specific 24–30 nucleotides piRNAs protect the germline from invasive elements as part of the repetitive element silencing pathway. For example, Miwi2-deficient mice arrest spermatogenesis at the leptotene stage accompanied by expression of the usually silent LINE and IAP transposable elements (Carmell et al., 2007). To date, the primary strategy that has been employed to assess function is the use of co-injection. Like spermatozoal mRNAs, the role of sncRNAs in early embryo development remains controversial (Amanai et al., 2006; Rassoulzadegan et al., 2006; Grandjean et al., 2009) and this is partly because of their unknown composition. To begin to resolve this issue, we have characterized the distribution and relative abundance of the major classes of sncRNAs consistently found within normal human spermatozoa. Their unique composition reveals their likely influence in a variety of processes, which could include a role in confrontation and consolidation when the genomes first meet to their subsequent epigenetic modification.
Materials and Methods
Sperm preparation
Sperm samples were obtained from three fertile donors who provided informed written consent. The study was coordinated under the auspices of a protocol approved by the Wayne State University Human Investigation Committee. The samples were labeled anonymously as AS062, AS064 and AS066 processed for freezing as described previously (Goodrich et al., 2007). A total of three ejaculates were obtained from each donor. Each sample was rapidly thawed and washed twice in supplemented hepes buffered saline (HBS) (0.145 M NaCl, 5 mM KCl, 1 mM MgSO4, 10 mM HEPES pH7.4, 10 mM glucose) by centrifugation at 200g. Spermatozoa were subsequently purified by centrifugation at 200g for 15 min through a 50% isotonic Percoll cushion supplemented with HBS (World Health Organization, 1999). The pelleted spermatozoa were washed in HBS and concentration was determined before processing for RNA extraction.
RNA extraction
The previously published protocol using the RNeasy mini kit was used to initially isolate RNAs >200 bases (Goodrich et al., 2007). Spermatozoal RNAs <200 bases were subsequently recovered using the RNeasy Plus Mini kit and RNeasy MinElute Cleanup Kit, Protocol 2. Briefly, sperm cells were homogenized in 600 μl of RLT supplemented with beta-mercaptoethanol buffer. Ethanol (430 μl, 100%) was added and the mixture was centrifuged through an RNeasy column. Following centrifugation, each flow through containing RNAs of <200 bases was combined, to which 0.65 volume of 100% ethanol was added. Small RNAs were then purified using MinElute RNA and eluted using 2 × 14 μl nuclease-free water. The large RNAs retained on the RNAeasy column were then processed according to the RNeasy protocol and eluted using 2 × 50 μl nuclease-free water as described previously. Each fraction was treated with RNAse-Free DNase I and assessed for absence of genomic DNA contamination by real-time PCR before preparation of the sequencing libraries.
Preparation of small RNA libraries and sequencing
Fractions of small RNAs from each donor were pooled to yield a total of 40 ng small RNA (<200 bases) and used to prepare the sequencing libraries using Illumina's Small RNA DGE v1.0 kit (Morin et al., 2006). The resulting size distributions of the libraries were verified on Agilent's 2100 Bioanalyzer. Ten picomoles were sequenced on the Illumina's GAII sequencer. One lane was used for each library. All sequencing data has been deposited into GEO [GSE21191; GSM530234; GSM530235; GSM530236].
Data analysis
Adaptor trimming and alignment to the human genome (Hg19) was performed with Novoalign version 2.05.33MT (Novocraft technologies, Malaysia) with the following options: -a -l 15 -h 90 -t 30 -r N. Each sequence read was classified into one of four categories by Novoalign: failed quality control, mapped to more than one location on the genome, did not map to the genome or mapped uniquely to the genome. Reads not mapping to the genome but containing 10 nt of the primer sequence TACAGTCCGA represented 51, 36 and 29% of the total number of reads in AS062, AS064 and AS066, respectively.
Sequence assembly
Sequence reads considered as uniquely mapping to the genome were annotated by Novoalign with base quality calls, chromosome, strand and offset (start position). The end position is determined using the length of the read. Because of the different qualities of the sequencing reads, some identical sequences were mapped to different locations on the genome. To eliminate this discrepancy and simplify the analysis, all sets of identical sequencing reads for which there was at least one conflicting mapping location were removed. In addition, all individual reads that required more than two mismatches for alignment were rejected. This filtering was carried out on a per set basis, and resulted in the removal of 15, 5 and 6% of the original uniquely aligned sequence reads for AS062, AS064 and AS066, respectively.
The remaining sequence reads were organized into contigs using the information on chromosome, strand and starting and ending positions, also done on a per set basis. Contigs were assembled in three different ways to test for discreetness. In the first approach, sequencing reads could join a contig only if there was positional overlap. The other two approaches not only extended the first but also allowed reads to join a contig if the nearest position to the contig was ≤4 or ≤20 nucleotides away. The use of different rules for forming contigs had little impact on the number of contigs formed, implying that all sequences in a given contig had overlap in the majority of cases. The contigs formed allowing the joining of sequences four bases away, in addition to overlap, were used for further analyses. Contigs with only one sequence read were denoted as singletons. The ratios between the total number of contigs and total number of sequencing reads were: 0.54 for AS062, 0.27 for AS064 and 0.28 for AS066. The three sets of contigs were then used to count the number of reads from each set that are also represented in the others. For a given set, each individual read was compared with the assembly of contigs of the other two sets. Strict overlap was required. This resulted in the partitioning of the reads according to presence in one or both of the other sets. The contigs were also compared with count the number of unique loci also represented in the others.
Sequence annotation
For each sample, sequences corresponding to different known RNA classes were identified by comparison of genomic locations obtained from Ensembl BioMart (Hubbard et al., 2009) and piRNA bank (Sai Lakshmi and Agrawal, 2008). Sequences were also compared with UCSC CpG island and UCSC Repeat Masker tracks (Fujita et al., 2011), as well as histone regions (Hammoud et al., 2009). Updated coordinates for piRNA bank and histone regions were calculated using the UCSC liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver) to convert from Hg18 to Hg19. Reads were associated with these genomic elements if there was at least 25% read overlap. Those miRNAs identified by BioMART (Hubbard et al., 2009; Flicek et al., 2010) that are not found in miRBase are considered predicted miRNAs. The co-occurrence of sequence reads corresponding to miRNA, piRNA and those not mapping to known genomic elements with transcription start sites (TSS) and promoters was assessed using the RegionMiner tool from the Genomatix Software Suite v2.30426 (Genomatix Software, Munich, Germany).
Identification of miRNA targets retained in mature human spermatozoa
In order to identify likely targets of miRNAs classified as present in sperm (miRNAs with mapped reads from every sample—AS062, AS064, AS066), Diana MicroT prediction software was used (Maragkakis et al., 2009). For each of 35 individual miRNAs, all predicted targets were ranked based on calculated interaction score as a function of the weighted sum of binding and conservation levels for conserved and un-conserved miRNA recognition elements. Total number of target genes per single miRNA ranged from a maximum of 225 (mir-30) to 1 (mir-184), with interaction scores also ranging from 142 (top targets of MIR-LET-7) to a low threshold cutoff of ∼8. In order to limit the further analysis to most likely targeted genes, only transcripts with an interaction score in the top 10% for each miRNA were examined further. Parsed lists with still >100 transcripts were further reduced by taking only the top 50%. These high predictive value transcript lists were used for downstream Ingenuity IPA 9.0–3211 pathway analysis and correlation with gene expression in sperm and oocyte.
Identification of piRNAs and repetitive element associations
Candidate repetitive element piRNA targets were identified for the 1137 piRNA sequences represented in all three libraries using repeat consensus sequences (http://frodo.wi.mit.edu/cgi-bin/primer3/cat_humrep_and_simple.cgi) and a standalone version of the miRanda algorithm (John et al., 2004; Betel et al., 2007), with the default parameters available at the piRNABank database (http://pirnabank.ibab.ac.in/miranda/mir.html): gap-opening penalty = −8.0; gap-extending penalty = −2.0; score threshold = −50.0; energy threshold = −20.0 kcal/mol. The 113 repeat elements specified were grouped into 11 classes (7SK, MARINER, ALR, MIR, Tigger, ALU, MLT, LTR, L1, MER and Other). The relative affinity of each piRNA to target a repetitive element was calculated as [relative abundance of the repetitive element × (piRNA reads × target sites in repeat element) ÷ (length of the repetitive element)]. Accordingly, the number of reads representing each piRNA was standardized as a function of the size of the repetitive element and the relative abundance of the element in the human genome (rmsk.txt.gz).
Results
Although synchronously frozen in the cell cycle, semen contains a morphologically heterogeneous population of spermatozoa (Lewis, 2007) that can be thought of as reflecting various points of self-selection towards optimum reproductive fitness. Considering this source of variance, small RNAs of <200 bases were isolated from semen samples from three fertile donors. A 40 ng aliquot of the small RNA fraction from each donor was then used to generate three independent small RNA sequencing libraries. The sequences obtained by Illumina GAIIx sequencing were then mapped to the human genome (Hg19). Figure 1A and Table I highlight the global characteristics of the sequencing libraries while Supplementary data, Table S1 provides additional detail of the distribution of sequence reads. Sequences were eliminated from further consideration if their position could not be unequivocally assigned. This included those sequences that mapped to multiple genomic regions throughout the genome along with the fragmented rRNAs and remnants of other RNAs from sperm maturation. The majority of sequence reads that did not map back to the genome occurred once and were unique to each library. This was considered to represent the background distribution. Sequencing of the >200 nt RNA fraction did not show any evidence of this class of sequences, supporting the view that they represent degradation products or remnants. No in-depth analysis was undertaken. Of the reads that could be assigned as uniquely mapping to the human genome, >80% localized to intergenic and intronic regions.
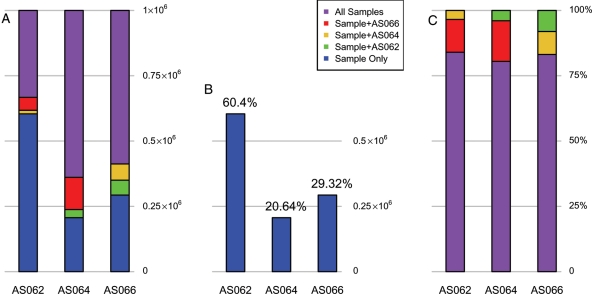
Figure 1.
Sequences shared among the three human sperm sncRNA libraries. The proportion of the total number of sequences that are unique to a library, shared with at least one other library or present in all three libraries (in common to all three libraries) are presented for each of the libraries. For each library, contigs were constructed according to information on chromosome, strand, start and end position, allowing new reads to join to a contig if there is overlap or if the read is less than or equal to four nucleotides away on the same chromosome/strand. A contig is synonymous with a unique locus. (A)–(C) Represent counts of reads in common between AS062, AS064 and AS066, based on these contigs. (A). For each library, reads were individually compared with the sets of contigs of the other libraries, resulting in numbers of reads that were unique to the library, or in common to one or both of the other libraries. The results were normalized to a million counts and plotted with a color code to show the percentage for each commonality subset, described in the legend, with purple representing reads common to all libraries and blue for reads that are donor-specific. (B). Identical to 1A, except showing only the reads that are only in one library, and the corresponding percentages. (C). This figure compares the relative percentages of shared reads—the percentages were calculated after the donor-specific reads were subtracted out.
Table I.
Total number of unique alignments obtained for each library prepared from human sperm obtained from three donors of proved fertility.
| Library | Reads mapping to a single location | Reads mapping to more than one location |
|---|---|---|
| AS062 | 492 096 | 901 807 |
| AS064 | 110 498 | 293 300 |
| AS066 | 276 923 | 688 881 |
Each sample was filtered for reads aligning to a unique position in the genome and these sequences were used for all subsequent analyses. The total number of reads for all samples is that which remains after removing reads containing primer sequences or failing Quality Control.
As summarized in Fig. 1B, between 20 and 60% of the mapped sequencing reads were donor-specific, reflecting the sample heterogeneity consistent with the distribution and depth of coverage of the known miRNAs and piRNAs within these libraries. The degree of sequences shared among the fertile donor sncRNAs was assessed by identifying those sequencing reads represented at least once in all libraries. One would expect that each sperm cell would contain ≈ 24 000 sncRNAs of a length of 20 nt, since each spermatozoon contains ∼0.3 fg of sncRNA. Sequence read density of each library is summarized in Table I and detailed in Supplementary data, Table S1 and illustrated in Fig. 1A. Accordingly, the coverage for all uniquely assigned sequences within the libraries exceeded 36-fold. Considering the heterogeneity of normal fertile ejaculate spermatozoa (Lalancette et al., 2009) the probability of the same read being present in all three individuals by chance within the bounds of the data is P< 0.0156. This is likely biologically significant. As shown in Fig. 1C, 95% of the mapped reads partitioned into two classes. Over 75% of the sncRNA sequence reads were shared between libraries. Of those remaining, at least 20% were common between two libraries. This supports the view that sequence saturation describing the population of sncRNAs within human sperm was obtained. The specifics of this group of sequences were considered further.
Identification of the sncRNAs in fertile donors
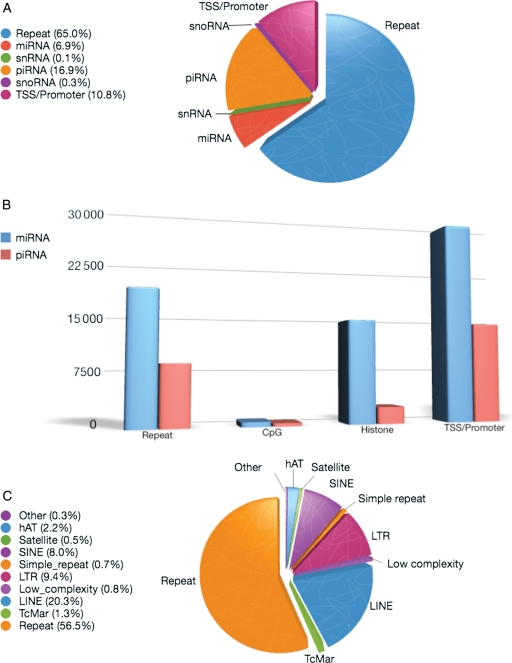
Speramtozoal sncRNA sequence content was initially assessed with respect to known BioMart classes of RNAs and is summarized in Supplementary data, Table S2. This summarizes all sequences that mapped to a single position on the human genome. Greater than 49% of the sncRNAs within the category designated as ‘other’ mapped to genomic locations where annotation has not been assigned. The remaining are residual products of spermiogenesis including fragments of coding, rRNAs and lincRNAs. As summarized in Fig. 2A, >95% of the sequences that mapped to known sncRNAs corresponded to two major classes: piRNAs (Supplementary data, Table S3) and miRNAs (Supplementary data, Table S4). These sncRNAs were nestled on a background of repetitive elements. A large portion of the miRNA sequence reads are associated with TSS/Promoter, histones and repetitive elements in contrast to the piRNAs that are not associated with histones and show less CpG enrichment compared with miRNAs (Fig. 2B). Other classes such as snRNA and snoRNA constituted a minor portion of the mapped sequence reads. Notably, the largest group of sequence reads corresponds to the repetitive elements as classified in Fig. 2C. SINES, LINES and LTR's are the most abundant of the well-described repetitive elements. This may be indicative of a physiologically significant role.
Figure 2.
Classification of all the sequences found in the three human sperm sncRNA libraries. (A) Sequences mapping to known genomic elements, TSS and promoters. The distribution of sequences mapping to miRNA, piRNA, snoRNA, snRNA and repeats are shown here. In addition, those sequences that did not map to known genomic elements were analyzed for TSS and promoter association. (B) Sequences mapping to miRNA or piRNA as well as repeats, CpG islands, histones and TSS or promoters. All sequences associated with miRNA (blue) or piRNA (red) were further analyzed to determine if they also map to repeats, CpG islands, histones or TSS/promoters. (C) Sequences mapping to known repeat classes. This figure shows the majority of sequences associated with an undefined repeat category. Of the remaining categories, LINE, LTR and SINE are highly represented.
microRNA
Several miRNA family members have been identified in mammalian testis (Ro et al., 2007a,c; Yan et al., 2007, 2009; Mishima et al., 2008). As summarized in Table II, (Supplementary data, Table S5) a comparatively small fraction are retained in mature spermatozoa. They are distributed across most chromosomes with the exception of chromosomes 4, 5, 10, 13, 14, 20 and Y. These known miRNAs only represent a minor fraction of the male sncRNA contribution. Less than one-half of the miRNAs have been previously identified in mouse (Amanai et al., 2006) and greater than one-half have also been detected in human testis (Landgraf et al., 2007).
Table II.
Associations of the miRNAs.
| miRNA | Sequences per library (reads per million) |
Association with genomic features |
Previously identified # |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AS062 | AS064 | AS066 | Average | Epi-regulated | Epi-miRNA | TSS Promoter | CpG Island | Histones | Sperm* | Testis | Ovary | Zygote* | |
| Has-miR-34c | 908.36 | 4371.12 | 12234.45 | 5837.98 | √ | – | √ | – | – | √ | √ | – | √ |
| hsa-miR-320a | 63.00 | 506.80 | 895.56 | 488.45 | – | – | √ | √ | √ | √2 | – | √2 | √ |
| hsa-miR-let-7b | 95.51 | 271.50 | 826.94 | 397.98 | – | – | √ | – | √ | √ | √ | √ | √ |
| hsa-miR-22 | 471.45 | 117.65 | 805.28 | 464.79 | – | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-122 | 93.48 | 63.35 | 790.83 | 315.89 | – | – | √ | – | – | – | – | √ | √ |
| hsa-miR-423 | 52.84 | 144.80 | 747.50 | 315.04 | – | – | √ | – | √ | – | √ | √ | √ |
| hsa-miR-375 | 20.32 | 253.40 | 682.50 | 318.74 | – | – | √ | √ | √ | √ | √ | – | √ |
| hsa-miR-let-7c | 36.58 | 325.80 | 574.17 | 312.18 | – | – | √ | – | – | √1 | √ | √ | √ |
| hsa-miR-140 | 16.26 | 99.55 | 411.67 | 175.82 | – | √ | √ | – | – | √ | √ | √ | √ |
| hsa-miR-21 | 48.77 | 99.55 | 292.50 | 146.94 | – | √ | √ | – | – | – | √ | √ | √ |
| hsa-miR-152 | 4.06 | 63.35 | 281.67 | 116.36 | √ | √ | √ | – | – | – | – | – | √ |
| hsa-miR-30a | 40.64 | 108.60 | 270.83 | 140.03 | – | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-148a | 8.13 | 54.30 | 220.28 | 94.24 | √ | √ | √ | – | – | – | – | – | √ |
| hsa-miR-let-7g | 36.58 | 144.80 | 220.28 | 133.89 | – | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-192 | 16.26 | 108.60 | 180.56 | 101.80 | – | – | √ | – | – | – | – | – | √ |
| hsa-miR-184 | 56.90 | 570.15 | 180.56 | 269.20 | – | – | √ | – | √ | √ | √ | – | √ |
| hsa-miR-10a | 63.00 | 27.15 | 166.11 | 85.42 | √ | – | √ | – | – | √ | √ | – | √ |
| hsa-miR-335 | 50.80 | 9.05 | 144.44 | 68.10 | – | – | √ | – | – | – | – | – | √ |
| hsa-miR-1323 | 6.10 | 18.10 | 122.78 | 48.99 | – | – | √ | – | – | – | – | – | – |
| hsa-miR-191 | 8.13 | 27.15 | 104.72 | 46.67 | – | – | √ | – | √ | √ | √ | √ | √ |
| hsa-miR-25 | 24.39 | 18.10 | 90.28 | 44.25 | – | – | √ | – | – | √ | – | – | √ |
| hsa-miR-99a | 2.03 | 36.20 | 57.78 | 32.00 | – | – | √ | – | – | √ | √ | √ | √ |
| hsa-miR-34b | 6.10 | 36.20 | 54.17 | 32.15 | √ | – | √ | √ | √ | – | – | – | √ |
| hsa-miR-221 | 52.84 | 18.10 | 54.17 | 41.70 | √ | – | √ | – | – | √ | – | √ | √ |
| hsa-miR-let-7e | 12.19 | 9.05 | 50.56 | 23.93 | – | – | √ | – | √ | √ | √ | √ | √ |
| hsa-miR-363 | 2.03 | 18.10 | 50.56 | 23.56 | – | – | √ | – | – | – | – | – | – |
| hsa-miR-30e | 8.13 | 9.05 | 43.33 | 20.17 | – | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-23a | 2.03 | 18.10 | 43.33 | 21.16 | – | – | √ | – | – | √ | √ | √ | √ |
| hsa-miR-100 | 38.61 | 90.50 | 39.72 | 56.28 | √ | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-509-3 | 2.03 | 9.05 | 32.50 | 14.53 | – | – | √ | – | – | – | √3 | – | – |
| hsa-miR-135a-2 | 2.03 | 18.10 | 28.89 | 16.34 | – | – | √ | – | – | – | – | – | √4 |
| hsa-miR-200a | 6.10 | 9.05 | 25.28 | 13.47 | √ | – | √ | – | √ | – | √ | – | √ |
| hsa-miR-31 | 4.06 | 36.20 | 7.22 | 15.83 | √ | – | √ | – | – | – | √ | – | √ |
| hsa-miR-27b | 2.03 | 9.05 | 7.22 | 6.10 | – | – | √ | – | – | – | √ | √ | √ |
| hsa-miR-185 | 2.03 | 18.10 | 7.22 | 9.12 | – | – | √ | – | √ | – | √ | √ | √ |
The biological and genomic context of the miRNAs within all libraries is presented. These include, designation as an epi-miRNA, promoter, TSS, CpG island or association with a histone-enriched region. The miRNAs previously identified in testis, ovary, sperm (mouse) or zygote (mouse) are noted.
Information from miR-let-7c-11, miR-3202, miR-5093, miR-135a4 as indicated.
*Mouse data.
#Hairpin origin was not considered.
Association with epigenetic machinery: Valeri et al. (2009), Iorio et al. (2010), Pan et al. (2010), Sato et al. (2011), Han et al. (2007).
Genomic expression profiles from: Amanai et al. (2006), Landgraf et al. (2007), Tang et al. (2007).
Translational repression is a characteristic of late spermatogenesis and miRNAs interacting at 3′UTRs can either direct their targets towards degradation or translational repression (Bushati and Cohen, 2007). Numerous software tools are available for the computational prediction of miRNA-UTR targets. The significance of the prediction is considered as a function of the thermodynamic stability of a miRNA–mRNA hybrid, and conservation of the proposed site of interaction across multiple species (Min and Yoon, 2010). Predictions vary widely among different tools, even within this common framework. For example, Microcosm (Tombol et al., 2010) prediction suggests 1167 targets with predictive score above the default threshold while TargetScan (Lewis et al., 2005; Liang, 2008) yields 84 has-mir-34c-5p targets. Diana MicroT (Maragkakis et al., 2009) was recently developed using a comparatively large set of biologically validated targets (Satoh and Tabunoki, 2011). The results are comparatively robust enabling one to select and/or rank likely targets and were thus employed in this analysis.
A total of 579 high-confidence and unique mRNA targets were identified by taking the top 10% Diana MicroT predicted scores of targets for each miRNA. A search with Genomatix GePS tool revealed that the majority of these targets were expressed in a wide range of tissues. Approximately 94% (547/579) of potential targets were represented on the Illumina microarray platform previously used to describe the composition of the normal fertile male (Lalancette et al., 2009). Of 2575 transcripts measured as present (P< 0.01 in 21 samples), only 46 were identified as targeted genes. This is significantly less (P= 2.7e−5) than would be expected and suggests that miRNA targets are likely absent in sperm. This is consistent with a potential role of miRNAs in translational suppression and/or degradation. Transcript profiling by RNA-seq (data not shown) showed that miRNA-targeted genes were subject to preferential 3′ degradation. For example, of the abundant miRNAs observed in spermatozoa, miR-122 has been described to participate in the post-transcriptional down-regulation of the transition protein 2 (TNP2) during spermatogenesis (Yu et al., 2005). TNP2, together with transition protein 1 (TNP1) transitionally substitute for some of the histones during spermiogenesis. This intermediate step precedes and facilitates protamine replacement that is required to compact the paternal genome into the relatively small sperm head [reviewed in (Johnson et al., 2011b)]. As we have shown (Johnson et al., 2011a) the corresponding transcripts must be eliminated once their mission has been accomplished. It is likely that the elevated quantities of this miRNA mark this and other transcripts for degradation. Perhaps they provide a rheostat for the maternally stored RNAs. The unfertilized human oocyte (GEO GDS 3256) and fertilized (GSE18290) human oocyte were interrogated to identity those transcripts that may be degraded upon fertilization. Similar to the above, ∼90% (521/579) of the miRNA targets identified by the sperm miRNAs were represented within these datasets. However, most showed no significant up- or down-regulation when compared with the unfertilized oocyte. This suggests no immediate effect, consistent with recent reports that miRNA activity is suppressed in mouse until after zygotic genome activation (Ma et al., 2010; Suh et al., 2010).
piRNAs and repeat-associated small RNAs
Since the discovery of piRNAs in developing mouse male germ cells, they have been thought to be absent from mature spermatozoa (Girard et al., 2006). However, sequence reads corresponding to a total of 1137 piRNAs (Supplementary data, Table S3) were shared among the mature human spermatozoa sncRNAs libraries and this certainly cannot be the case. The majority of the sequence reads mapping to piRNAs aligned to piRNA clusters located on several chromosomes with the exception of 4, 13 and X. Of the 1137 piRNAs identified chromosome 15 contained the greatest concentration of piRNAs (338) followed by chromosome 6 (224) and chromosome 19 (156).
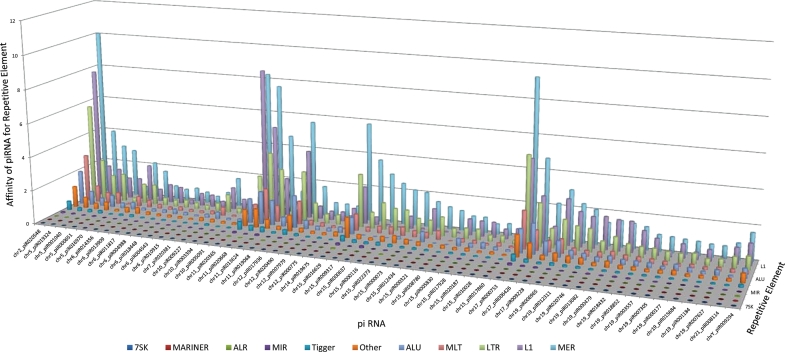
It has been proposed that piRNAs protect the genome from the deleterious effects of invasive elements [reviewed in (O'Donnell and Boeke, 2007)] and/or perhaps the means to achieve confrontation and consolidation. It is known that simple repeat sequence SINEs comprised of short sequence repeats and LINEs that harbor low-complexity repeats can provide a platform for reverse transcription that is requisite for retrotransposition (Weiner, 2006). Whether the binding of piRNAs to these repetitive elements is the mechanism by which they protect the genome from retrotransposition and/or a component of confrontation and consolidation, it is reasonable to assume that this may be mediated by targeting repetitive elements. To test this hypothesis, the potential of the identified piRNAs to bind to repeat sequences was assessed. Potential piRNA targets were identified by miRanda (John et al., 2004) with conditions optimized for piRNAs (piRNA database) and using repeat consensus sequences (http://frodo.wi.mit.edu/cgi-bin/primer3/cat_humrep_and_simple.cgi). As summarized in Fig. 3 (Supplementary data, Table S6), the principal classes targeted by spermatozoal piRNAs are MER, L1 and LTR elements. The number of sequence reads associated with the piRNA-targeting repeats was standardized as a function of the number of locations the piRNA targets a given repeat, the length of the repetitive element and frequency within the genome. Of these, hsapiR020548 is the most abundant, preferentially targeting the MER family, a class of retrotransposons that appears to represent a fossil in the human genome (Smit and Riggs, 1996). This apparent paradox raises the issue of the proposed silent nature.
Figure 3.
piRNA-targeting repetitive elements. Distribution of repetitive elements targeted by piRNAs. For each repeat consensus the relative affinity of the piRNA in each category was determined. The value for each piRNA in each repeat class is standardized to account for the number of positions within the repeat a piRNA targets, the length of the repeat and its frequency in the genome. Only piRNA with a maximum relative affinity in the top 5% are illustrated.
Discussion
The complexity of the population of mature human spermatozoal RNAs has now been extended to include a host of sncRNAs for which we may begin to suggest function. Given that the elevated quantities of miRNAs likely mark some paternal transcripts for degradation, could the remaining paternal miRNAs provide additional function? Others have shown that exogenously added miRNAs can markedly affect fetal development and impact subsequent development (Grandjean et al., 2009). To identify those paternal miRNAs that may impact early development, a comparison of the most abundant miRNAs identified in human sperm with those of the mouse oocyte and 1-cell zygote was undertaken. As presented in Table II, several paternal effect candidates were identified, four of which are not found in ovary. These include hsa-mir-34c, hsa-mir-375, hsa-mir-252 and hsa-mir-25. Interestingly, Ingenuity Pathway Analysis defined the top functions of the upper 10% of the Diana MicroT targets as Embryonic Development, Organismal Development, Tissue Development, Organ Development, Cellular Growth and Proliferation and Organism survival (P< 10−25). Among the abundant miRNAs, hsa-mir-34c is of particular note. This miRNA belongs to a family of highly conserved miRNA (miR-34a, miR-34b and miR-34c). It has recently been described as playing an important role in promoting the germinal phenotype during male gametogenesis. In this germinal lineage, expression seems to be p53-independent in contrast to somatic cells (Bouhallier et al., 2010). The top predicted targets for this miRNA included DLL1 and NOTCH1. These two genes have been described to play an important role in segmentation and somite formation in vertebrates (Zhang et al., 2002; Krebs et al., 2003). Further, Ingenuity Pathway analysis annotation by function infers embryo segmentation (P= 0.0000196), specification of midline axis (P= 0.000537) and development of the embryonic node (P= 0.000869). Other morphogenic miRNA targets include OTX1 and MAFB (McKay et al., 1994; Acampora et al., 1998; Li and Joyner, 2001; Martinez-Morales et al., 2001; Theil et al., 2002).
After fertilization, the highly methylated paternal genome is actively demethylated before establishing the new epigenetic marks necessary for early embryonic development [reviewed in (Morgan et al., 2005; Palini et al., 2011)]. Passive and progressive demethylation is facilitated by the exclusion of Dnmt1 during these steps (Carlson et al., 1992; Bestor, 2000) and accompanies replication to enable reprogramming and imprinting (Monk et al., 1987). This methyltransferase is the most abundant in mammalian cells and plays a key role in the maintenance of DNA methylation although it is also active in de novo methylation (Bestor, 2000). In this context, miRNAs transmitted by the male gamete would immediately inhibit epigenetic marking. Inhibition could then be released and the paternal contribution marked at the appropriate developmental time. This view is supported by the observation that among the most abundant miRNAs detected in human spermatozoa (Table II), four epi-miRNAs repress the expression of transcripts encoding proteins involved in the epigenetic machinery: hsa-miR-140 (Tuddenham et al., 2006), hsa-miR-21 (Pan et al., 2010), hsa-miR-152 (Braconi et al., 2010) and hsa-miR-148a (Duursma et al., 2008). For example, miR-21 down-regulates the expression of DNA methyltransferase 1 (DNMT1) in an indirect manner (Pan et al., 2010) by targeting RASGRP1 (RAS guanyl-releasing protein 1), which is an upstream critical regulator of the Ras-MAPK signaling cascade of DNMT1 (MacLeod et al., 1995). In contrast, miR-152 together with hsa-miR-148a, directly target DNMT1 (Braconi et al., 2010). In comparison, hsa-miR-148a down-regulates DNA methyltransferase 3b (DNMT3b) by recognizing an evolutionary conserved coding sequence (Duursma et al., 2008). This methyltransferase is thought to function in de novo methylation during early development and gametogenesis (Okano et al., 1999). Its expression is reduced in somatic cells but it is highly expressed in undifferentiated embryonic stem cells. Inactivation is associated with embryonic lethality and DNMT3a knock-out mice die shortly after birth (Okano et al., 1999).
miRNAs can also target structures beyond the 3′UTRs and regulate gene expression at the transcriptional level (Kim et al., 2008; Place et al., 2008; Khraiwesh et al., 2010). It has been suggested that miRNAs retained in mature spermatozoa are associated with the histone-enriched regions (Hammoud et al., 2009) and may act to modulate the bipotential state of the male chromatin. Most of the spermatozoal miRNAs identified were derived from promoter regions. The correlation of these miRNAs was considered in relation to the proximity of the TSS/promoters of their target genes and histones. As summarized in Table II, while only 10 of the 35 miRNAs found in the libraries were derived from a histone-enriched region in human spermatozoa (Hammoud et al., 2009) all were found associated with TSS/promoter sites.
Greater than 10% of the sequence reads that uniquely mapped to the genome were assigned to the TSS/promoter. They form 4549 clusters that are not miRNA, piRNA, snRNA or snoRNA. They may appear similar to the tiRNAs found in association with TSSs (Ambros et al., 2003; Taft et al., 2009) but are not GC rich nor are they correlated with the presence of modified histones (Hammoud et al., 2009; Brykczynska et al., 2010; Johnson et al., 2011b; Vavouri and Lehner, 2011). On one hand, unlike the majority of TSSs in sperm (Hammoud et al., 2009; Johnson et al., 2011b), <20% of this group correspond to promoters or TSSs that exhibit significant histone enrichment (Supplementary data, Fig. S1). This small fraction of the TSS/promoter RNAs frame the histone regions, sharing similar spatial properties reminiscent of a barrier function. These novel RNAs are denoted as quiescent RNAs (qRNAs). This small group segregates into similar ontological classes of early development as described previously (Hammoud et al., 2009). On the other hand, ∼80% of the qRNAs are not associated with histones and are ontologically non-descript. Perhaps they mark enhancer sites like the recently described eRNA, i.e. enhancer RNA (Wang et al., 2011). Though this novel class of sperm non-coding RNA remains to be further defined, the abundance of these transcripts is consistent with the view that they may be of developmental consequence (Wu et al., 2011). Although these insights suggest the existence of a potential role of these paternal transcripts during early embryonic development it should be noted that recent reports have suggested that miRNA activity is suppressed in mouse during this period (Ma et al., 2010; Suh et al., 2010). Nevertheless it is possible that miRNAs delivered by human sperm do play a functional role in the oocyte, given the delay in zygotic genome activation.
It is intriguing to suggest that the contribution of this group of paternal piRNAs, miRNAs qRNAs and repeat-associated RNAs at fertilization may constitute some of the mechanistic members of the confrontation–consolidation pathway (Bourc'his and Voinnet, 2010). During confrontation they may be used to assure genome compatibility and fitness. For example, this could be accomplished through the pairing of oocyte and sperm RNAs to prevent or activate a response, or by silencing the hypomethylated paternal genome while guiding recombination or through their direct interaction of the repeat-associated RNAs with the maternal complement of repetitive elements. Through consolidation, these paternal sncRNAs may again act as a guide, perhaps to heterochromatization. Interestingly, both recombination and heterochromatization are known to be facilitated by their interaction with the nuclear matrix, a source rich in sperm RNAs (Lalancette et al., 2008).
Using deep sequencing, the complexity of the population of mature human spermatozoal RNAs has now been extended to include sncRNAs. In addition to the miRNA class previously detected in mature mouse porcine and human spermatozoa (Ostermeier et al., 2005; Watanabe et al., 2006; Ro et al., 2007b; Mishima et al., 2008; Luo et al., 2010) for the first time piRNAs were also observed. This work represents the most extensive survey of sncRNAs in mammalian spermatozoa to date. The results of this study show that the spermatozoal RNA contribution extends beyond the presence of mRNA and miRNA populations. Their possible role in early embryo development and as fertility markers is intriguing.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
A.K., E.A., S.D. and R.T. analyzed the data and helped to prepare the manuscript. M.D. provided the samples. S.A.K. participated in the experimental design, analysis and wrote the manuscript with C.L. who provided some of the initial drafts and created the sequencing libraries and the analysis of the data.
Funding
This work is supported in part by the Presidential Research Enhancement Program in Computational Biology, the Charlotte B. Failing Professorship and NIH grant HD36512 to S.A.K. This work was also supported in part by a Michigan Core Technology grant from the State of Michigan's 21st Century Fund Program to the Wayne State University Applied Genomics Technology Center. E.A. is recipient of a mobility grant (JC 2010-0247) from the Ministerio de Educación (Gobierno de España).
Supplementary Material
Acknowledgements
The authors would like to thank Adrian E. Platts who assisted with some of the initial analysis, Shihong Mao and Edward Sendler for their assistance in data analysis along with Graham Johnson for his review of the manuscript.
References
- Acampora D, Avantaggiato V, Tuorto F, Briata P, Corte G, Simeone A. Visceral endoderm-restricted translation of Otx1 mediates recovery of Otx2 requirements for specification of anterior neural plate and normal gastrulation. Development. 1998;125:5091–5104. doi: 10.1242/dev.125.24.5091. [DOI] [PubMed] [Google Scholar]
- Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Betel D, Sheridan R, Marks DS, Sander C. Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol. 2007;3:e222. doi: 10.1371/journal.pcbi.0030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16:720–731. doi: 10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourchis D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Curry E, Ellis SE, Pratt SL. Detection of porcine sperm microRNAs using a heterologous microRNA microarray and reverse transcriptase polymerase chain reaction. Mol Reprod Dev. 2009;76:218–219. doi: 10.1002/mrd.20980. [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic acids research. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53:161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol. Therapy. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: An intricate network. Biochim Biophys Acta-Gene Regul Mech. 2010;1799:694–701. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Sendler E, Lalancette C, Hauser R, Diamond MP, Krawetz SA. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol Hum Reprod. 2011a doi: 10.1093/molehr/gar054. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011b;141:21–36. doi: 10.1530/REP-10-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics. 2010;11:77. doi: 10.1186/1471-2164-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nature reviews. Genetics. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O'Brien TP, Hamada H, Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17:1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: new functional insights for spermatozoal RNA. J Cell Biochem. 2008;104:1570–1579. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalancette C, Platts AE, Johnson GD, Emery BR, Carrell DT, Krawetz SA. Identification of human sperm transcripts as candidate markers of male fertility. J Mol Med. 2009;87:735–748. doi: 10.1007/s00109-009-0485-9. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE. Is sperm evaluation useful in predicting human fertility? Reproduction. 2007;134:31–40. doi: 10.1530/REP-07-0152. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Lian J, Zhang X, Tian H, Liang N, Wang Y, Liang C, Li X, Sun F. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC Med Genomics. 2008;1:61. doi: 10.1186/1755-8794-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Ye L, Liu G, Shao G, Zheng R, Ren Z, Zuo B, Xu D, Lei M, Jiang S, et al. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS One. 2010;5:e11744. doi: 10.1371/journal.pone.0011744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–11337. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Morales JR, Signore M, Acampora D, Simeone A, Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- McKay IJ, Muchamore I, Krumlauf R, Maden M, Lumsden A, Lewis J. The kreisler mouse: a hindbrain segmentation mutant that lacks two rhombomeres. Development. 1994;120:2199–2211. doi: 10.1242/dev.120.8.2199. [DOI] [PubMed] [Google Scholar]
- Min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med. 2010;42:233–244. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y, Ishikawa T, Mori M, Kanda T, Goto T. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ-cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, et al. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J Androl. 2005;26:70–74. [PubMed] [Google Scholar]
- Palini S, De Stefani S, Scala V, Dusi L, Bulletti C. Epigenetic regulatory mechanisms during preimplantation embryo development. Ann N Y Acad Sci. 2011;1221:54–60. doi: 10.1111/j.1749-6632.2010.05937.x. [DOI] [PubMed] [Google Scholar]
- Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4(+) T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007a;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007b;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Song R, Nguyen D, Jin J, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007c;13:1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- Satoh J, Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Mining. 2011;4:17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Hawkins PG, Mattick JS, Morris KV. The relationship between tiRNAs and CTCF localization. Epigenetics Chromatin. 2011;4:13. doi: 10.1186/1756-8935-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Ariza-McNaughton L, Manzanares M, Brodie J, Krumlauf R, Wilkinson DG. Requirement for downregulation of kreisler during late patterning of the hindbrain. Development. 2002;129:1477–1485. doi: 10.1242/dev.129.6.1477. [DOI] [PubMed] [Google Scholar]
- Tombol Z, Eder K, Kovacs A, Szabo PM, Kulka J, Liko I, Zalatnai A, Racz G, Toth M, Patocs A, et al. MicroRNA expression profiling in benign (sporadic and hereditary) and recurring adrenal pheochromocytomas. Modern Pathol. 2010;23:1583–1595. doi: 10.1038/modpathol.2010.164. [DOI] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet 7. 2011:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeri N, Vannini I, Fanini F, Calore F, Adair B, Fabbri M. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation. Mamm Genome. 2009;20:573–580. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM. SINEs and LINEs: troublemakers, saboteurs, benefactors, ancestors. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2006. pp. 507–533. [Google Scholar]
- World Health Organization. WHO Laboratory Manual for Examination of Human Semen. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, Ma Y. A microarray for microRNA profiling in mouse testis tissues. Reproduction. 2007;134:73–79. doi: 10.1530/REP-07-0056. [DOI] [PubMed] [Google Scholar]
- Yan W, Morozumi K, Zhang J, Ro S, Park C, Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and MicroRNAs in the sperm nuclei. Biol Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- Yan N, Lu Y, Sun H, Qiu W, Tao D, Liu Y, Chen H, Yang Y, Zhang S, Li X, et al. Microarray profiling of microRNAs expressed in testis tissues of developing primates. J Assist Reprod Genet. 2009;26:179–186. doi: 10.1007/s10815-009-9305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- Zhang N, Norton CR, Gridley T. Segmentation defects of Notch pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double mutant mice. Genesis. 2002;33:21–28. doi: 10.1002/gene.10081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.