Abstract
Trehalose is a disaccharide with potential applications in the biotechnology and food industries. We propose a method for industrial production of trehalose, based on improved strains of Corynebacterium glutamicum. This paper describes the heterologous expression of Escherichia coli trehalose-synthesizing enzymes trehalose-6-phosphate synthase (OtsA) and trehalose-6-phosphate phosphatase (OtsB) in C. glutamicum, as well as its impact on the trehalose biosynthetic rate and metabolic-flux distributions, during growth in a defined culture medium. The new recombinant strain showed a five- to sixfold increase in the activity of OtsAB pathway enzymes, compared to a control strain, as well as an almost fourfold increase in the trehalose excretion rate during the exponential growth phase and a twofold increase in the final titer of trehalose. The heterologous expression described resulted in a reduced specific glucose uptake rate and Krebs cycle flux, as well as reduced pentose pathway flux, a consequence of downregulated glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase. The results proved the suitability of using the heterologous expression of Ots proteins in C. glutamicum to increase the trehalose biosynthetic rate and yield and suggest critical points for further improvement of trehalose overproduction in C. glutamicum.
Trehalose (1-α-glucopyranosyl-1-α-glucopyranoside) is a nonreducing, particularly stable disaccharide formed by two glucose moieties (37). As a compatible osmolite (22) and protein stabilizer (39), trehalose shows a wide range of potential applications in biotechnology (increased stress tolerance of important crops, stability of recombinant proteins, etc.), as well as in the food industry (37).
In the past, trehalose was produced by using Saccharomyces cerevisiae (32). The high cost of this system and the promising applications of the disaccharide led to the development of a new trehalose production process based on the enzymatic biotransformation of maltodextrins (25, 26). The success of the enzymatic process has limited the interest in using microorganisms as alternative sources for trehalose synthesis. However, the recent development of metabolic engineering tools, allowing the rational design of microorganisms for metabolite production (27), prompted us to evaluate an alternative process for trehalose overproduction in the gram-positive bacterium Corynebacterium glutamicum (23). C. glutamicum was chosen for three major reasons: (i) it produces, and excretes, trehalose (15); (ii) it has a metabolic control architecture simpler than that of other microorganisms, maybe as a result of its comparatively small 3,500-kb genome size (10); and (iii) it is widely used in industrial biotechnological processes (10).
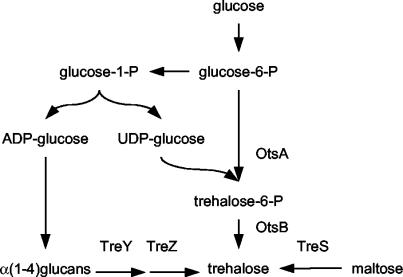
Three pathways for trehalose synthesis have been characterized in C. glutamicum (45), similarly to that found in Mycobacterium species (7) (Fig. 1). The first is the TreS pathway, in which trehalose is formed by maltose isomerization (7, 31). The second is the TreY-TreZ pathway, leading to trehalose from α(1-4)glucans through a two-step enzymatic catalysis. The first enzyme of this pathway (TreY) leads to glycosyl-trehalose formation from glycogen-like molecules; then, the second enzyme (TreZ) releases the trehalose residue from the oligosaccharide (9, 24). The third is the OtsA-OtsB pathway, in which trehalose synthesis starts from glucose-6-phosphate and UDP-glucose. It is also a two-step enzymatic catalysis, through the enzymes trehalose-6-phosphate synthase (OtsA) (5, 41) and trehalose-6-phosphate phosphatase (OtsB) (28, 41).
FIG. 1.
The three pathways for trehalose synthesis in C. glutamicum and M. tuberculosis. The otsBA pathway leads to trehalose from glucose-6-phosphate (glucose-6-P) through the intermediates UDP-glucose and trehalose-6-phosphate (trehalose-6-P). In the treYZ pathway, isomerization and hydrolysis of a α(1-4)glucan leads to trehalose. The synthesis of such glucans proceeds in a way fairly similar to the otsBA pathway, with ADP-glucose as the glucosyl donor. Finally, the treS pathway leads to trehalose after isomerization of maltose.
Although overexpression of homologous genes for trehalose production in C. glutamicum is an option, heterologous overexpression might be more efficient. For example, a foreign enzyme could be regulated in a different manner and therefore avoid the host regulatory network. Heterologous expression of E. coli genes has already been successfully used in C. glutamicum, such as for the expression of threonine dehydratase (17) and the lactose operon (3).
The trehalose synthesis pathway through OtsAB in E. coli has been thoroughly studied. In this bacterium, trehalose biosynthesis proceeds through the enzymes OtsA and OtsB, both of which are encoded by the otsBA operon (21). In this paper, we report the heterologous expression of the otsBA operon from E. coli in C. glutamicum 13032 ΔtreS—a mutant that lacks trehalose-maltose-isomerizing activity and hence has improved trehalose accumulation properties (45)—with the shuttle vector pXMJ19 (20). Upon induction with isopropyl-β-d-thiogalactopyranoside (IPTG), the enzyme activity of the OtsA and OtsB proteins in the C. glutamicum otsBA-expressing strain was quantified. Culture experiments were performed with the new strains, and the changes in the trehalose production rate and metabolic-flux distribution were determined.
MATERIALS AND METHODS
Growth of bacterial strains.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium was used as the standard medium for E. coli DH5α-mcr (16) and strains derived from it. Tryptone soy broth (TSB) medium was used for C. glutamicum ΔtreS (Wolf et al., submitted) and strains derived from it. The defined medium used for C. glutamicum shake flask experiments (DMCG I) contained sodium citrate at 1.1 g/liter, NaCl at 1 g/liter, MgSO4 · 7H2O at 200 mg/ml, FeSO4 · 7H2O at 25 mg/liter, CaCl2 · 2H2O at 50 mg/liter, K2HPO4 at 8 g/liter, KH2PO4 at 1 g/liter, (NH4)2SO4 at 5 g/liter, MnSO4 at 2 mg/liter, Na2B4O7 · 10 H2O at 0.2 mg/liter, (NH4)6Mo7O24 · 4H2O at 0.1 mg/liter, FeCl3 · 6H2O at 2 mg/liter, ZnSO4 · 7H2O at 0.5 mg/liter, CuCl2 · 2H2O at 0.2 mg/liter, biotin at 1 mg/liter, thiamine hydrochloride at 1 mg/liter, and desferrioxamine mesylate at 3 mg/liter. Glucose (20 g/liter) was used as a carbon source. For batch bioreactor cultivation experiments, the defined medium (DMCG II) of Delaunay et al. (7) was used, with a few modifications. It contained nitrilotriacetic acid at 0.5 g/liter, NaCl at 2 g/liter, MgSO4 · 7H2O at 400 mg/liter, FeSO4 · 7H2O at 40 mg/liter, CaCl2 · 2H2O at 84 mg/liter, Na2HPO4 at 3 g/liter, KH2PO4 at 6 g/liter, (NH4)2SO4 at 8 g/liter, MnSO4 at 3.9 mg/liter, Na2B4O7 · 10H2O at 0.3 mg/liter, (NH4)6Mo7O24 · 4H2O at 0.1 mg/liter, FeCl3 · 6H2O at 3.9 mg/liter, ZnSO4 · 7H2O at 0.9 mg/liter, CuCl2 · 2H2O at 0.3 mg/liter, biotin at 4 mg/liter, thiamine hydrochloride at 20 mg/liter, desferrioxamine mesylate at 3 mg/liter, and polypropylene glycol (molecular weight, 2000) at 1 ml/liter. Glucose (100 g/liter) was used as a carbon source. Antibiotics were added to final concentrations of 50 (ampicillin) and 20 (chloramphenicol) μg/ml. IPTG was added to a final concentration of 1.0 mM.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype and/or description | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T | Ampr | Promega |
| pXMJ-19 | Cfr | 20 |
| pLPIotsBA00 | otsBA Ampr | This study |
| pLPIotsBA01 | ptac otsBA (direct insert) Cfr | This study |
| pLPIotsBA02 | ptac otsBA (inverted insert) Cfr | This study |
| Strains | ||
| E. coli DH5α-mcr | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA mcrA Δ(mrr-hsdRMS-mcrBC) | 16 |
| E. coli/pLPIotsBA00 | otsBA Ampr | This study |
| E. coli/pLPIotsBA01 | ptac otsBA (direct insert) Cfr | This study |
| E. coli/pLPIotsBA02 | ptac otsBA (inverted insert) Cfr | This study |
| C. glutamicum ΔtreS | ΔtreS derivative of C. glutamicum 13032 | 45 |
| C. glutamicum/pLPIotsBA01 | ΔtreS ptac otsBA (direct insert) Cfr | This study |
| C. glutamicum/pLPIotsBA02 | ΔtreS ptac otsBA (inverted insert) Cfr | This study |
DNA manipulations and bacterial transformation.
All DNA cloning procedures were performed in accordance with standard procedures (36). E. coli DH5α-mcr was transformed by a high-efficiency method (19). C. glutamicum ΔtreS was electroporated in accordance with a protocol published elsewhere (42).
Plasmid constructs.
The sequence of the E. coli otsBA operon was obtained from the GenBank database (accession no. X69160). The following oligodeoxynucleotides for PCR amplification were designed with Primer Premier software (Premier Biosoft International, Palo Alto, Calif.): OTSBA1 (5′GCG CGT CGA CAT AAG AAA AGA GAA GGA GG 3′) and OTSBA2 (5′TTC CAC TTA CGG TCG ACT AAC CGC TCC 3′). The underlined bases indicate SalI cut sites created in both primers for subsequent cloning steps.
The promoterless 2,263-bp fragment containing the coding sequence of the otsBA operon—flanked upstream to the start codon by a 37-bp sequence (harboring the ribosomal binding site of otsB) and downstream to the stop codon by a 27-bp sequence—was amplified by PCR from chromosomal DNA of E. coli JM109 (Promega Corporation, Madison, Wis.). The PCR product was ligated into the pGEM-T vector (Promega Corporation). The resulting construct (pLPIotsBA00) was transformed into E. coli DH5α-mcr, a DNA methylation mutant (16). The otsBA operon fragment was excised from plasmid pLPIotsBA00 with the restriction enzyme SalI. The excised fragment was filled with DNA polymerase I (Klenow fragment) and ligated into E. coli-C. glutamicum shuttle vector pXMJ19 (20), which had previously been digested with the restriction enzyme SmaI, 42 bp downstream of the tac promoter site (20). The resulting ligation mixture was used to transform E. coli DH5α-mcr. The transformants were selected by chloramphenicol resistance in LB plates. Two kinds of E. coli clones were obtained: pLPIotsBA01 clones, carrying the otsBA operon in the right orientation, and pLPIotsBA02 clones, carrying the operon in the inverted sense (used as a control construction). Plasmid DNA was extracted from both clones and used to electroporate C. glutamicum ΔtreS (Wolf et al., submitted). Positive clones were selected on chloramphenicol-LBHIS plates (42). In this way, we obtained C. glutamicum strains pLPIotsBA01 (otsBA+) and pLPIotsBA02 (otsBA inverted insert [control]). The resulting strains are summarized in Table 1.
Shake flask experiments.
For each induction experiment, 2 ml of a fresh overnight culture was inoculated into 100 ml of TSB. When the culture reached an absorbance at 600 nm of 0.3 to 0.4, IPTG was added to a final concentration of 1 mM. Samples were withdrawn at regular time intervals for metabolite and enzymatic analyses.
Bioreactor experiments.
For batch cultivation, a preinoculum of each strain was made in 100 ml of TSB medium. After overnight growth in a rotatory shaker at 30°C and 250 rpm, the cells were centrifuged and resuspended in 100 ml of defined medium. The cell suspension obtained was added to a 1-liter Bio-Flo IIc bioreactor (New Brunswick Scientific, Edison, N.J.) containing 900 ml of the defined medium. Cultures were run at 30°C with agitation at 700 rpm. The pH was kept at 7.0 with 6 M NaOH.
Sample preparation for metabolite measurement.
Ten-milliliter samples were withdrawn from the culture at regular time intervals. The samples were centrifuged at 4°C and 1,600 × g for 10 min immediately after collection. The supernatant was collected and used directly for high-performance liquid chromatography (HPLC) analysis of extracellular metabolites. For cytoplasmic metabolite analysis, the remaining pellet was resuspended in 2 ml of 35% perchloric acid at 4°C. The sample was neutralized by addition of 2.33 ml of cold 5 M KOH. The samples were centrifuged for 10 min in a bench centrifuge to remove cell debris and the KClO4 precipitate, and the supernatants were stored at −70°C. Intracellular metabolites were determined by HPLC analysis of the supernatant.
Quantitative HPLC analysis of carbohydrates was performed in a Merck-Hitachi L7100 pump system coupled to a Merck-Hitachi L7490 refraction index detector. An HPX-87H column (Bio-Rad Laboratories, Hercules, Calif.) was used with 5 mM sulfuric acid as the eluant (6). The column temperature was kept at 55°C in a Merck-Hitachi L7350 column oven.
Preparation of cell extracts and enzyme assays.
Cultures (100 ml) were harvested by centrifugation, washed twice in 20 ml of buffer containing Tris HCl at 100 mM (pH 7.5), KCl at 20 mM, MnSO4 at 5 mM, and dithiothreitol (DTT) at 1 mM. The cells were resuspended in 1 ml of the same buffer, mixed with 300 μl of 0.1-mm glass beads, and broken by six cycles of 20 s in a Mini Bead-beater apparatus (Biospec Products Inc., Bartlesville, Okla.). Total protein in the extracts was determined by the dye binding assay method (4) with bovine serum albumin as the standard.
OtsB activity was assayed by monitoring phosphate release from trehalose-6-phosphate (13). The reaction was carried out in a final volume of 1 ml containing 50 μmol of Tris HCl (pH 7.2), 5 μmol of MgCl2, and 1 μmol of trehalose-6-phosphate. Samples were withdrawn at regular time intervals and assayed for phosphate by the zinc acetate method (1) as follows: 300 μl of sample was mixed with 900 μl of a solution containing zinc acetate at 100 mM and ammonium molybdate at 15 mM (pH 5.0) and incubated in ice for exactly 1 min, and then the absorbance at 350 nm was quickly measured.
Since the enzyme activity of the OtsA protein is unstable during the crude-extract preparation procedure (21), we used a hybrid protocol to measure its activity. The cells were permeabilized in accordance with the method of Uy et al. (40), and then the activity of OtsA was measured in accordance with the protocol of Giæver et al. (12). Briefly, 100 ml of cells was harvested by centrifugation, washed twice with 20 ml of 50 mM NaCl, resuspended to an optical density at 570 nm of ∼150 in 100 mM HEPES buffer, pH 7.5, containing 20% (vol/vol) glycerol and then frozen at −20°C. For the permeabilization step, the frozen cells were slowly thawed on ice and then mixed with cetyltrimethylammonium bromide at a final concentration of 0.3% for exactly 1 min. The permeabilized cell suspension was immediately used to assay the OtsA-OtsB enzymatic system as follows. An amount of permeabilized cells containing about 3 mg of protein was added to a reaction mixture (final volume of 1 ml) containing 7.5 μmol of UDP-glucose, 2.5 μmol of MgCl2, 33 μmol of Tris HCl (pH 7.5), and 250 μmol of KCl. The reaction mixture was placed in a 30°C water bath, and the reaction was started by addition of 15 μmol of glucose-6-phosphate. Samples were taken at regular time intervals for 15 min, boiled to stop the reaction, and analyzed for trehalose content as already described.
Measurement of catabolic enzymes was performed in accordance with previously published methods, with modifications. Glucose-6-phosphate dehydrogenase and gluconate-6-phosphate dehydrogenase were measured as described by Moritz et al. (29), in a mixture containing Tris HCl buffer (pH 7.5) at 100 mM, DTT at 1 mM, MgCl2 at 5 mM, NADP at 1 mM, glucose-6-phosphate at 2 mM, and cell extract. The reaction was started by addition of glucose-6-phosphate. Phosphoglucoisomerase was assayed as described by Schray et al. (38), in a mixture containing Tris HCl buffer (pH 7.5) at 100 mM, DTT at 1 mM, MgCl2 at 5 mM, NADP at 1 mM, fructose-6-phosphate at 5 mM, glucose-6-phosphate dehydrogenase at 2 U/ml, and cell extract. The reaction was started by addition of fructose-6-phosphate. Pyruvate kinase was assayed as described by Cocaign-Bousquet et al. (6), in a mixture containing Tris HCl buffer (pH 7.5) at 100 mM, DTT at 1 mM, MnSO4 at 5 mM, KCl at 100 mM, ADP at 10 mM, NADH at 3 mM, fructose-6-phosphate at 5 mM, lactate dehydrogenase at 40 U/ml, phosphoenolpyruvate at 2 mM, and cell extract. The reaction was started by addition of phosphoenolpyruvate. Isocitrate dehydrogenase was measured as described by Eikmanns et al. (11), in a mixture containing Tris HCl buffer (pH 7.5) at 100 mM, DTT at 1 mM, MnSO4 at 5 mM, NADP at 0.2 mM, threo-(DSLS)-isocitrate at 1.6 mM, and cell extract. The reaction was started by addition of isocitrate. The malate dehydrogenase reverse reaction was assayed in a mixture containing CHES [2-(N-cyclohexamino)ethanesulfonic acid] buffer (pH 10) at 100 mM, NADH at 3 mM, oxaloacetate at 2 mM, and cell extract. The reaction was started by addition of oxaloacetate.
Metabolic-flux analysis.
Metabolic fluxes were calculated by using a stoichiometric metabolic model, described elsewhere (8, 43), that was adapted to the C. glutamicum metabolic network. Four rate measurements are required for flux calculation (glucose consumption, trehalose, biomass, and lactate production). Since the CO2 rate measurement is redundant, it was used to check the consistency of the results (9). Prior to the flux analysis, we verified that carbon balances accounted for 95 to 100% in each batch culture performed.
RESULTS
Heterologous expression of the otsBA operon.
The first goal of this work was to obtain the functional expression of ots genes from E. coli in C. glutamicum ΔtreS. To check the accomplishment of this goal, we tested the enzyme activity of Ots proteins in crude extracts or permeabilized cells from the new strains obtained in this work. otsBA-overexpressing strain C. glutamicum ΔtreS pLPIotsBA01 showed a sixfold increase in OtsB activity in crude extracts (956 pkat/mg of protein) relative to the control strain harboring the pLPIotsBA02 plasmid (145 pkat/mg of protein). Unfortunately, OtsA activity was not detected in crude extracts as a consequence of its instability, as previously reported by Kaasen et al. (21). Hence, it was assayed in permeabilized cells, with UDP-glucose and glucose-6-phosphate as substrates. Since the trehalose-6-phosphate formed in this reaction is dephosphorylated to trehalose by the OtsB protein also present in the preparation, this assay measures the combined activity of OtsA and OtsB when the trehalose formed is measured. By this method, the otsBA-overexpressing strain showed a fivefold increase in activity (151 pkat/mg of protein) compared to the control strain (28 pkat/mg of protein). The observed differences in enzymatic activity between the OtsB and OtsA-OtsB assays can be ascribed to (i) differences in the level of expression of the otsA and otsB genes (OtsA could be the rate-limiting enzyme in the assay) and (ii) differences in enzyme activity recovery in the crude extract and in permeabilized cells. The results demonstrated the functional expression of the E. coli OtsA and OtsB proteins in C. glutamicum.
Macroscopic response to otsBA heterologous expression.
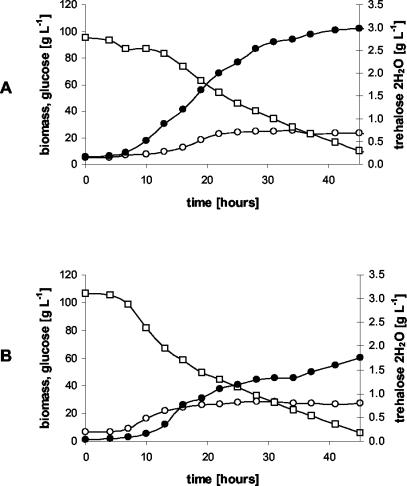
Our second goal was to test the behavior of the new recombinant strain under controlled culture conditions—high cell density and glucose concentrations (100 g/liter) with proper aeration and pH control—during batch cultures in a 1-liter bioreactor. The temporal profiles of glucose, biomass, and trehalose during the cultures of the otsBA-expressing and control strains are shown in Fig. 2.
FIG. 2.
Batch cultures of C. glutamicum strains pLPIotsBA01 (A) and pLPIotsBA02 (B) in DMCG II (see Materials and Methods). The temporal profiles of glucose (□), biomass (○), and trehalose (•) are shown. The culture medium used was supplemented with chloramphenicol at 20 mg/liter to avoid plasmid loss, and IPTG (inducer) was added at 4 h of cultivation.
Kinetic parameters and fermentation yields of C. glutamicum changed significantly upon expression of the E. coli otsBA operon. During the exponential growth phase, the specific growth rate showed a reduction from 0.15 h−1 in the control strain to 0.082 h−1 in the pLPIotsBA01 strain. This change was accompanied by the following changes in the specific rates of the otsBA-expressing and control strains, respectively: glucose consumption, 1.3 and 1.9 mmol g (dry cell weight [DCW])−1 h−1; trehalose excretion, 0.021 and 0.005 mmol g (DCW)−1 h−1; biomass production, 1.1 and 1.8 mmol g (DCW)−1 h−1; CO2 production, 3.1 and 3.9 mmol g (DCW)−1 h−1.
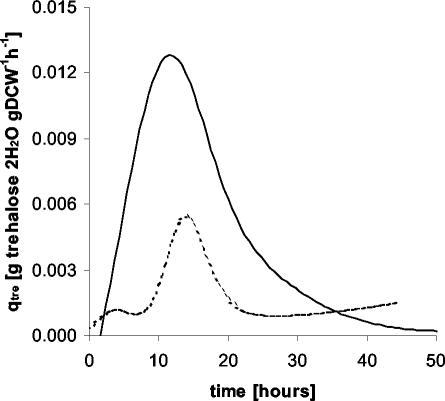
From these data, the metabolic yields, which are a measure of the process performance of the cells during cultivation, were calculated. The biomass-to-glucose yield was reduced from 0.54 g (DCW) g of glucose−1 to 0.42 g (DCW) g of glucose−1. The main product during cultivation was trehalose, which reached a maximal external concentration of 3.0 g · liter−1 in the otsBA-expressing strain and 1.7 g liter−1 in the control strain (Fig. 2). Although trehalose excretion in the otsBA-expressing strain continued during the whole cultivation period, it was higher in the exponential growth phase. The trehalose-on-glucose yield was 50 mg of trehalose g of glucose−1 during the exponential growth phase and 10 mg of trehalose g of glucose−1 at stationary phase. In the control strain, the trehalose-on-glucose yield was constant at around 10 mg of trehalose · g of glucose−1. In the otsBA-expressing strain, the maximal trehalose specific productivity reached 13 mg of trehalose · g (DCW)−1 h−1 (Fig. 3), almost threefold higher than in the control strain (5 mg of trehalose · g [DCW]−1 h−1). Neither lactate nor acetate production was detected during the exponential growth phase, indicating that oxygen was not limiting in this stage of cultivation.
FIG. 3.
Specific productivity of trehalose (in grams of trehalose per gram [DCW] per hour) during batch culturing of C. glutamicum strains pLPIotsBA01 (continuous line) and pLPIotsBA02 (dashed line) in DMCG II. The curves were obtained after curve fitting of biomass and trehalose data (see Materials and Methods).
Metabolic-flux response to heterologous otsBA expression.
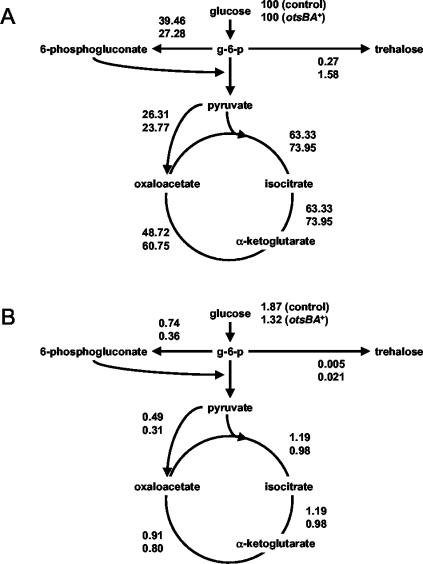
The third goal of this work was to obtain a picture of the changes in the central metabolism upon otsBA expression, reflected in metabolic fluxes. These fluxes were calculated with a stoichiometric metabolic model based on a previously reported metabolic network of C. glutamicum (43). To avoid singularities in the stoichiometric matrix, pyruvate carboxylase was assumed to be the only operative anaplerotic reaction (34), and only one pathway for trehalose synthesis was included. Although they are biochemically different, the OtsA-OtsB and TreY-TreZ pathways are stoichiometrically indistinguishable.
Flux distributions during the exponential growth of C. glutamicum ΔtreS strains pLPIotsBA01 and pLPIotsBA02—with the glucose consumption, trehalose, biomass, and lactate production rates as the inputs for calculations—are illustrated in Fig. 4. Fluxes were normalized to the glucose uptake rate (millimoles per millimole of glucose) to obtain a relative distribution of carbon utilization. A fivefold increase in trehalose excretion flux was observed upon expression of the OtsA and OtsB enzymes, along with increases of 35 and 21% in the fluxes through glycolysis and the Krebs cycle, respectively. Concomitantly, a 31% reduction in the flux through the pentose phosphate pathway (PPP) was determined.
FIG. 4.
Relative (A) and specific (B) fluxes during exponential growth in batch bioreactor cultures of C. glutamicum strains. Metabolic fluxes were calculated with a stoichiometric metabolic model adapted for the C. glutamicum metabolic network (see Materials and Methods).
Carbon utilization inside the cell was calculated through the specific flux rates (expressed in millimoles of metabolite per gram [DCW] per hour) (Fig. 4). In this case, the fluxes through glycolysis and the Krebs cycle decreased by 18 and 15%, respectively, the opposite of that observed for the relative fluxes discussed above. The reduction of specific flux through the pentose phosphate shunt was decreased to 51%, which is significantly more pronounced than that observed for normalized fluxes and by far the most significant flux change obtained.
It is interesting that the intracellular trehalose concentration did not show a significant differences upon expression of the otsBA operon, e.g., 290 μmol/g (DCW) for the otsBA-expressing strain and 280 μmol/g (DCW) for the control strain.
Effect of heterologous expression of otsBA on the synthesis of important catabolic enzymes.
To investigate possible reasons for the observed flux variations, additional enzymatic measurements were performed to assess the effect of otsBA expression on the synthesis of enzymes involved in central catabolic pathways of C. glutamicum (Table 2). The activities tested were glucose-6-phosphate dehydrogenase-gluconate-6-phosphate dehydrogenase (pentose phosphate shunt), phosphoglucoisomerase (glycolysis), pyruvate kinase (glycolysis), isocitrate dehydrogenase (Krebs cycle), and malate dehydrogenase (Krebs cycle). Pyruvate kinase and malate dehydrogenase activities did not show significant differences in the otsBA-expressing strain compared to the control strain. Phosphoglucoisomerase and isocitrate dehydrogenase showed variations (a 27% increase and a 17% decrease, respectively), but taking into account the high specific activities of both enzymes, it may be doubted whether there was an effect of such variations on metabolic fluxes. The most significant change was observed for the glucose-6-phosphate dehydrogenase-gluconate-6-phosphate dehydrogenase system, which showed a 77% decrease. This reduction is in accordance with the strong reduction observed in the specific flux through the pentose phosphate shunt (51%) upon expression of the otsBA operon.
TABLE 2.
Specific activities of some enzymes of the central metabolism in crude extracts from C. glutamicum after induction with 1 mM IPTGb
| Enzyme | Sp act (pkat/mg of protein)a
|
||
|---|---|---|---|
| ΔtreS | ΔtreS pLPIotsBA01 | ΔtreS pLPIotsBA02 | |
| G-6-phosphate-gluconate- 6-phosphate dehydrogenase | 192 | 39 | 173 |
| Phosphoglucoisomerase | 12,524 | 17,891 | 14,038 |
| Pyruvate kinase | 7,119 | 5,716 | 6,014 |
| Isocitrate dehydrogenase | 37,124 | 31,303 | 38,016 |
| Malate dehydrogenase | 22,018 | 22,198 | 21,629 |
Tabulated activities are the means of at least two independent measurements, and the standard deviation was always less than 10%.
Extracts prepared from cells grown in shake flasks containing DMCG I medium and induced for 2h with 1 mM IPTG. The inducer was added when the culture reached an optical density at 600 nm of 0.3 to 0.4.
DISCUSSION
Overexpression of the otsBA operon in C. glutamicum pLPIotsBA01 resulted in a five- to sixfold increase in the enzymatic activity of the OtsA-OtsB system during the exponential growth phase, compared to the C. glutamicum pLPIotsBA02 control strain. The major effect of the heterologous expression of otsBA in C. glutamicum, when evaluated in batch cultures, was the significant increase in excreted trehalose (a 70% increase). This rise in the trehalose titer was accompanied by a lower glucose specific uptake rate (a 29% reduction) and a lower specific growth rate (a 38% reduction), as well.
The maximum trehalose specific productivity of both strains coincides with the exponential growth phase. This could possibly be explained by a shortage of the UDP-glucose required for trehalose synthesis through the otsBA pathway. During exponential growth, bacteria synthesize more UDP-glucose for cell wall formation (14). Evidence from Mycobacterium tuberculosis (44) suggests that UDP-glucose is required as a precursor of UDP-galactose, the source of the galactosyl residues of the arabinogalactan complex of the cell wall in mycobacteria and corynebacteria (35). Insufficient UDP-glucose synthesis through the corresponding enzyme, UTP:glucose-1-phosphate uridylyl-transferase, might be the bottleneck in this pathway. During the exponential growth phase in shake flask experiments, the specific activity of that enzyme was 14 pkat/mg of protein (data not shown), which is very similar to the calculated trehalose synthesis specific flux during exponential phase (15 pkat/mg). It is worth noting that these values are 1 order of magnitude lower than the measured specific activity of OtsB and of the OtsA-OtsB combination, as measured in permeabilized cells of the otsBA-expressing strain. A similar situation was observed in the gram-positive bacterium Lactococcus lactis, where exopolysaccharide synthesis is limited by the UDP-glucose supply (2). Since the trehalose excretion rate seems to be coupled to the flux through the OtsAB pathway, the pattern for the trehalose specific productivity of both strains is consistent with the hypothesis of a UDP-glucose shortage. The greater amount of the OtsA and OtsB enzymes in the otsBA-expressing strain leads to the withdrawal of more UDP-glucose for trehalose synthesis during exponential growth. Improvement of the UDP-glucose supply for trehalose synthesis is a current focus of research in our laboratory.
The basal activity of the Ots enzymes in the C. glutamicum control strain should be ascribed to the chromosomal homologues of the heterologously expressed ots genes (45), which are not involved in osmotic tolerance, in contrast to the situation in E. coli (45). Their function in C. glutamicum is unclear, although their involvement in the synthesis of corynomycolic acids, key components of the cell wall of corynebacteria, has been recently suggested (35). In the closely related bacterium Mycobacterium smegmatis, which also synthesizes mycolic acids, the activity of the OtsAB enzymatic pathway was 158 pkat/mg (9), in close agreement with our results for OtsB activity in crude extracts of the C. glutamicum control strain (145 pkat/mg).
Surprisingly, the internal concentration of trehalose was similar for the otsBA-expressing and control strains, suggesting the existence of a mechanism for maintaining a constant internal concentration of trehalose, possibly by activating an export carrier.
Metabolite production requires the understanding of central metabolism fluxes. We calculated metabolic fluxes relative to both glucose uptake and biomass (specific fluxes). The former is suitable for assessment of carbon utilization in both strains. The increase in trehalose synthesis flux observed upon expression of the otsBA operon was accompanied by significant perturbations in the central metabolism: reduced flux through the PPP, along with increased glycolytic and Krebs cycle fluxes. The reduction in glucose-6-phosphate consumption through the PPP (12 mmol of glucose-6-phosphate/mmol of glucose) was more pronounced than the increase in the OtsAB pathway flux (2.6 mmol of glucose-6-phosphate/mmol of glucose). The reduction in PPP relative flux was consistent with the increase in the Krebs cycle flux, as well as the glycolytic flux responsible for fueling it. Since a PPP flux decrease results in an NADPH shortage, the Krebs cycle flux should be increased in order to replenish the NADPH pool through isocitrate dehydrogenase.
Since the calculation of relative fluxes does not take into account the biomass content, we also calculated specific fluxes. A slight decrease in the glycolytic and Krebs cycle fluxes was found, probably as a consequence of the lower glucose uptake rate in the otsBA-expressing strain. This result was expected because the specific growth rate was found to be reduced upon expression of the otsBA operon, which leads to lower requirements for biomass precursors (30). Again, a decrease in PPP flux was observed (0.38 mmol of glucose-6-phosphate/g [DCW]), and it was more pronounced than the increase in the glucose-6-phosphate flux for trehalose synthesis (0.032 mmol of glucose-6-phosphate/g [DCW]). The measurements of enzyme activities in crude extracts showed a 77% reduction in the activity of the glucose-6-phosphate dehydrogenase-gluconate-6-phosphate dehydrogenase system. Thus, the observed reduction in PPP flux seems to be a synergistic effect of glucose-6-phosphate withdrawal, mediated by OtsA, and reduced synthesis of glucose-6-phosphate dehydrogenase and/or gluconate-6-phosphate dehydrogenase. Since the internal trehalose concentration did not change upon otsBA expression, a putative perturbation in the internal pool of trehalose-6-phosphate could be related to the reduced activity of the enzymes mentioned above. In E. coli, for example, trehalose-6-phosphate regulates the expression of the treB and treC genes (18). We speculate that this compound might also exert some kind of regulation in C. glutamicum.
The consequences of a reduced growth rate in C. glutamicum are reduced glucose catabolism (30) and reduced glucose uptake by the phosphotransferase system, a process tightly associated with glycolytic flux (33). The reduced growth rate observed upon otsBA expression is a desirable feature for trehalose production, but the associated reduced glucose uptake might limit the efficiency of the process. Consequently, further research into trehalose production by C. glutamicum will be aimed at increasing the UDP-glucose supply and uncoupling glucose uptake from glycolytic flux, in order to favor trehalose overflow metabolism.
In conclusion, in this study we obtained a C. glutamicum strain successfully expressing the otsA and otsB genes from E. coli. As a consequence, redirection of carbon flux toward trehalose biosynthesis was achieved. Flux estimations obtained from a stoichiometric model allowed us to determine the impact of otsA and otsB expression on central metabolic pathways. The new strain, C. glutamicum pLPIotsBA01, is a basis for further metabolic engineering studies of trehalose overproduction by this microorganism.
Acknowledgments
We thank Andreas Wolf and Andreas Burkovski for providing the ΔtreS mutant and plasmid pXMJ19. The help of Héctor Soto in bioreactor experiments is also acknowledged.
This research was supported by Fondo Nacional para el Desarrollo Científico y Tecnológico de Chile (FONDECYT) grant 2000063 and the Volkswagen Foundation. Leandro Padilla was supported by a doctoral fellowship from the Consejo Nacional de Ciencia y Tecnología de Chile (CONICYT).
REFERENCES
- 1.Bencini, D. A., J. R. Wild, and G. A. O'Donovan. 1983. Linear one-step assay for the determination of orthophosphate. Anal. Biochem. 132:254-258. [DOI] [PubMed] [Google Scholar]
- 2.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brabetz, W., W. Liebl, and K. H. Schleifen. 1991. Studies on the utilization of lactose by Corynebacterium glutamicum, bearing the lactose operon of Escherichia coli. Arch. Microbiol. 155:607-612. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cabib, E., and L. F. Leloir. 1958. The biosynthesis of trehalose phosphate. J. Biol. Chem. 231:259-275. [PubMed] [Google Scholar]
- 6.Cocaign-Bousquet, M., A. Guyonvarach, and N. D. Lindley. 1996. Growth rate dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose limited chemostat cultures of Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaunay, S., D. Uy, M. F. Baucher, J. M. Engasser, A. Guyonvarch, and J. L. Goergen. 1999. Importance of phosphoenolpyruvate carboxylase of Corynebacterium glutamicum during the temperature triggered glutamic acid fermentation. Metab. Eng. 1:334-343. [DOI] [PubMed] [Google Scholar]
- 8.Delgado, J., and J. C. Liao. 1997. Inverse flux analysis for reduction of acetate excretion in Escherichia coli. Biotechnol. Prog. 13:361-367. [DOI] [PubMed] [Google Scholar]
- 9.De Smet, K. A. L., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Eggeling, L., H. Sahm, and A. A. de Graaf. 1996. Quantifying and directing metabolite flux: application to amino acid overproduction. Adv. Biochem. Eng. Biotechnol. 54:1-30. [Google Scholar]
- 11.Eikmanns, B. J., D. Rittmann, and H. Sahm. 1995. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giæver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goddijn, O. J. M., T. C. Verwoerd, E. Voogd, R. W. H. H. Krutwagen, P. T. H. M. de Graaf, J. Poelos, K. van Dun, A. S. Ponstein, B. Damm, and J. Pen. 1997. Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol. 113:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk, G. 1986. Bacterial metabolism, 2nd ed. Springer Verlag, New York, N.Y.
- 15.Gourdon, P., and N. D. Lindley. 1999. Metabolic analysis of glutamate production by Corynebacterium glutamicum. Metab. Eng. 1:224-231. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli K-12. J. Bacteriol. 166:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillouet, S., A. A. Rodal, G. An, P. A. Lessard, and A. J. Sinskey. 1999. Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl. Environ. Microbiol. 65:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horlacher, R., and W. Boos. 1997. Characterization of treR, the major regulator of the Escherichia coli trehalose system. J. Biol. Chem. 272:13026-13032. [DOI] [PubMed] [Google Scholar]
- 19.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 20.Jakoby, M., C-E. Ngouoto-Nkili, and A. Burkovski. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Tech. 13:437-441. [Google Scholar]
- 21.Kaasen, I., P. Falkenberg, O. B. Styrvold, and A. R. Strøm. 1992. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by KatF (AppR). J. Bacteriol. 174:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on the amino acid fermentation. Part I: production of l-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 24.Kobayashi, K., M. Kato, Y. Miura, M. Kettoku, T. Komeda, and A. Iwamatsu. 1996. Gene cloning and expression of new trehalose-producing enzymes from the hyperthermophilic archaeum Sulfolobus solfataricus KM1. Biosci. Biotechnol. Biochem. 60:1882-1885. [DOI] [PubMed] [Google Scholar]
- 25.Kubota, M., K. Maruta, T. Miyake, and T. Sugimoto.December 1994. Trehalose-releasing enzyme, and its preparation and uses. European patent EP0628630.
- 26.Kubota, M., T. Miyake, K. Maruta, and T. Sugimoto.July 1994. Non-reducing saccharide-forming enzyme, and its preparation and uses. European patent EP0606753.
- 27.Lee, S. Y., and E. T. Papoutsakis. 1999. The challenges and promise of metabolic engineering, p. 1-12. In S. Y. Lee and E. T. Papoutsakis (ed.), Metabolic engineering, 1st ed. Marcel Dekker Inc., New York, N.Y.
- 28.Matula, M., M. Mitchell, and A. D. Elbein. 1971. Partial purification and properties of a highly specific trehalose phosphate from Mycobacterium smegmatis. J. Bacteriol. 107:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moritz, B., K. Striegel, A. A. de Graaf, and H. Sahm. 2000. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway in vivo. Eur. J. Biochem. 267:3442-3452. [DOI] [PubMed] [Google Scholar]
- 30.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates Inc., Sunderland, Mass.
- 31.Nishimoto, T., H. Chaen, T. Sugimoto, and T. Miyake.June 1998. Trehalose and its production and use. U.S. patent 5,759,610.
- 32.Panek, A. D., V. M. F. Paschoalin, and C. R. M. Meleiro.August 1993. Processo para la obtenção de trehalose de Saccharomyces cerevisiae. Patent PI9303490, INPI, Brazil.
- 33.Parche, S., A. Burkovski, G. A. Sprenger, B. Weil, R. Krämer, and F. Titgemeyer. 2001. Corynebacterium glutamicum: a dissection of PTS. J. Mol. Microbiol. Biotechnol. 3:423-428. [PubMed] [Google Scholar]
- 34.Peters-Wendisch, P. G., C. Kreutzer, J. Kalinowski, M. Patek, H. Sahm, and B. J. Eikmanns. 1998. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915-927. [DOI] [PubMed] [Google Scholar]
- 35.Puech, V., M. Chami, A. Lemassu, M. A. Laneelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffe. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147:1365-1382. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schiraldi, C., I. Di Lernia, and M. De Rosa. 2002. Trehalose production: exploiting novel approaches. Trends Biotechnol. 20:420-425. [DOI] [PubMed] [Google Scholar]
- 38.Schray, K. J., S. J. Benkovic, P. A. Benkovic, and I. A. Rose. 1973. Catalytic reactions of phosphoglucoisomerase with cyclic forms of glucose-6-phosphate and fructose-6-phosphate. J. Biol. Chem. 248:2219-2224. [PubMed] [Google Scholar]
- 39.Singer, M. A., and S. Lindquist. 1998. Thermotolerance in Saccharomyces cerevisiae: the yin and yang of trehalose. Trends Biotechnol. 16:460-467. [DOI] [PubMed] [Google Scholar]
- 40.Uy, D., S. Delaunay, J. Engasser, and J. Goergen. 1999. A method for the determination of pyruvate carboxylase activity during the glutamic acid fermentation with Corynebacterium glutamicum. J. Microbiol. Methods 39:91-96. [DOI] [PubMed] [Google Scholar]
- 41.Vandercammen, A., J. F. Hers, and H. G. Hers. 1989. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase of Saccharomyces cerevisiae. Eur. J. Biochem. 182:613-620. [DOI] [PubMed] [Google Scholar]
- 42.Van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 43.Varela, C., E. Agosin, M. Baez, M. Klapa, and G. Stephanopoulos. 2003. Metabolic flux redistribution in Corynebacterium glutamicum in response to osmotic stress. Appl. Microbiol. Biotechnol. 60:547-555. [DOI] [PubMed] [Google Scholar]
- 44.Weston, A., R. J. Stern, R. E. Lee, P. M. Nassau, D. Monsey, S. L. Martin, M. S. Scherman, G. S. Besra, K. Duncan, and M. R. McNeil. 1997. Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber. Lung Dis. 78:123-131. [DOI] [PubMed] [Google Scholar]
- 45.Wolf, A., R. Krämer, and S. Morbach. 2003. Three different pathways for trehalose synthesis in Corynebacterium glutamicum and their significance in response to osmotic stress. Mol. Microbiol. 49:1119-1134. [DOI] [PubMed] [Google Scholar]