Abstract
Background
"Domestication" of transposable elements (TEs) led to evolutionary breakthroughs such as the origin of telomerase and the vertebrate adaptive immune system. These breakthroughs were accomplished by the adaptation of molecular functions essential for TEs, such as reverse transcription, DNA cutting and ligation or DNA binding. Cryptons represent a unique class of DNA transposons using tyrosine recombinase (YR) to cut and rejoin the recombining DNA molecules. Cryptons were originally identified in fungi and later in the sea anemone, sea urchin and insects.
Results
Herein we report new Cryptons from animals, fungi, oomycetes and diatom, as well as widely conserved genes derived from ancient Crypton domestication events. Phylogenetic analysis based on the YR sequences supports four deep divisions of Crypton elements. We found that the domain of unknown function 3504 (DUF3504) in eukaryotes is derived from Crypton YR. DUF3504 is similar to YR but lacks most of the residues of the catalytic tetrad (R-H-R-Y). Genes containing the DUF3504 domain are potassium channel tetramerization domain containing 1 (KCTD1), KIAA1958, zinc finger MYM type 2 (ZMYM2), ZMYM3, ZMYM4, glutamine-rich protein 1 (QRICH1) and "without children" (WOC). The DUF3504 genes are highly conserved and are found in almost all jawed vertebrates. The sequence, domain structure, intron positions and synteny blocks support the view that ZMYM2, ZMYM3, ZMYM4, and possibly QRICH1, were derived from WOC through two rounds of genome duplication in early vertebrate evolution. WOC is observed widely among bilaterians. There could be four independent events of Crypton domestication, and one of them, generating WOC/ZMYM, predated the birth of bilaterian animals. This is the third-oldest domestication event known to date, following the domestication generating telomerase reverse transcriptase (TERT) and Prp8. Many Crypton-derived genes are transcriptional regulators with additional DNA-binding domains, and the acquisition of the DUF3504 domain could have added new regulatory pathways via protein-DNA or protein-protein interactions.
Conclusions
Cryptons have contributed to animal evolution through domestication of their YR sequences. The DUF3504 domains are domesticated YRs of animal Crypton elements.
Keywords: tyrosine recombinase; Crypton; domestication, transposon; DUF3504
Background
The structural and mechanistic variety of transposable elements (TEs) is well-documented [1]. They encode proteins that include diverse functional domains involved in catalysis or interaction with DNA, RNA and other proteins. Because of this diverse repertoire, TEs can supply functional modules to generate new genes. "Molecular domestication" of transposable elements [2] led to evolutionary milestones such as the origin of telomerase and the vertebrate adaptive immune system. Telomerase reverse transcriptase (TERT) provides a solution for end replication problems accompanying linear chromosome replication and was derived from a reverse transcriptase (RT) related to Penelope-like elements in the very early stage of eukaryote evolution [3,4]. V(D)J recombination is a mechanism used in jawed vertebrates to generate a variety of immunoglobulins and T-cell receptors. It is catalyzed by the recombination activating gene 1 (RAG1) derived from a transposase encoded by the Transib family of DNA transposons [5]. Different kinds of transposon proteins were domesticated, including transposase, integrase, RT, envelope and gag proteins [6]. Herein we report in-depth studies of another type of transposon enzyme, tyrosine recombinase (YR), which was repeatedly domesticated in the history of animals.
To date four types of enzymes are known to catalyze DNA integration of eukaryotic transposons: DDE-transposase, YR, rolling-circle replication initiator and the combination of RT and endonuclease (EN) [7]. DDE-transposase is the most abundant gene in nature [8] and is carried by many DNA transposon superfamilies, self-synthesizing transposons (Polinton), as well as long terminal repeat (LTR) retrotransposons (Gypsy, Copia, BEL and endogenous retroviruses) [1,9-11]. They share three conserved amino acids (DDD or DDE) at their catalytic sites, which are separated by amino acid sequences of varying length. Some domesticated DDE-transposases became DNA-binding proteins, such as CENP-B in mammals and Daysleeper in Arabidopsis thaliana [12,13]. Non-LTR retrotransposons and Penelope-like elements use a combination of RT and EN in their transposition [14-17]. Helitron is the only group of eukaryotic transposons encoding rolling-circle replication initiator [9].
YR genes are ubiquitous in prokaryotes but rare in eukaryotes [18,19]. All YRs found in eukaryotes are encoded by mobile elements: yeast 2-micron circle plasmids [20], ciliate Euplotes crassus transposons (Tec1, Tec2 and Tec3) [21,22], three groups of retrotransposons (DIRS/Pat, Ngaro and VIPER) [23-25], and Cryptons [19]. The YR encoded by the yeast 2-micron plasmid, known as "flippase" (FLP), is widely used for site-specific recombination in the FLP-FRT system [26]. Tec1 and Tec2 transposons encode a DDE-transposase in addition to YR, and therefore the YR domains in these transposons are probably involved in resolving transposition intermediates. To date the only YR-encoding transposons found in the vertebrate genomes are DIRS and Ngaro retrotransposons. Cryptons were originally found in a basidiomycete Cryptococcus neoformans and several pathogenic fungi. Their boundaries are difficult to characterize because they have neither terminal inverted repeats (TIRs) nor long direct repeats. Instead they have short direct repeats at both termini. These 4- or 6-bp direct repeats are considered substrates for recombination. By analogy to prokaryotic YR-encoding transposons, Goodwin et al. [19] proposed that Cryptons are excised from the host genome as an extrachromosomal circular DNA and integrated at a different locus in the genome. YR typically recognizes recombination sites consisting of two inverted repeats that are 11 to 13 bp long and separated by a segment 6 to 8 bp long [27]. Recently, transposons encoding only a YR have been found in sea urchin, insects and cnidarians and classified as Cryptons [28,29]. YR contains four catalytically important residues (R-H-R-Y), but their overall sequence identity is very low among different genes and transposons [18,19]. The conserved tyrosine residue directly binds to DNA in the recombination reaction. In this paper, we report Cryptons from various species, including medaka fish, and six human genes originated from ancient domestication events of Crypton YRs.
Results
The diversity of Crypton elements in terms of their sequence and domain structure
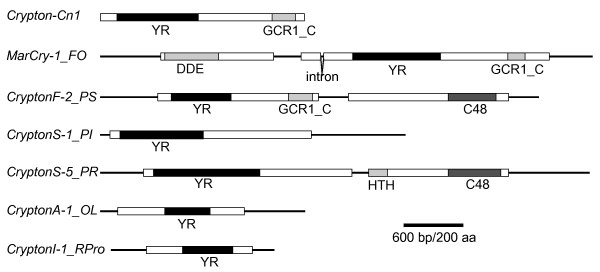
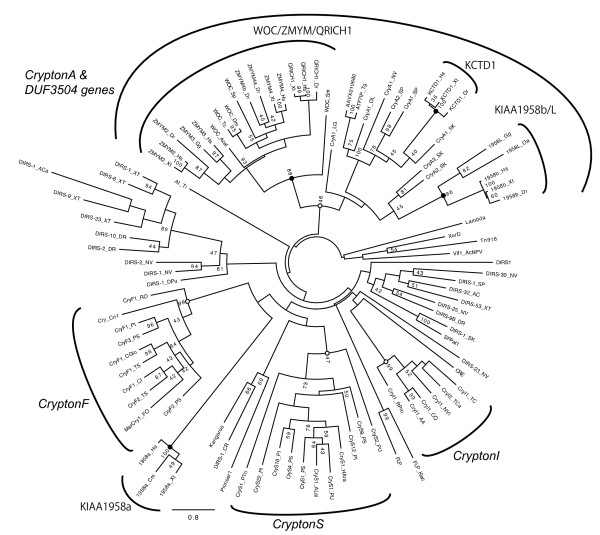
We identified 94 Crypton elements from 24 species representing animals, fungi and stramenopiles that include oomycetes and diatom (Figure 1, Table 1 and Additional file 1). Phylogenetic clustering of Cryptons on the basis of their YR domain sequences revealed four groups reflecting the systematics of their hosts (Figure 2, open circles), but two of them were not strongly supported phylogenetically because of the low bootstrap values. Herein we designate them as CryptonF, CryptonS, CryptonA and CryptonI to indicate their corresponding hosts: fungi, stramenopiles, animals and insects. CryptonA and CryptonI are structurally similar; however, CryptonF, CryptonS and CryptonA/CryptonI have distinct protein domain structures (see Figure 1 and detailed description in the next three sections). Because of the low resolution of the phylogenetic tree, we could not determine whether there is any relationship between these four Crypton groups and to other YR-encoding elements, and we cannot rule out the possibility that they have originated independently.
Figure 1.
Schematic structures of Cryptons. Crypton-Cn1 and MarCry-1_FO belong to the CryptonF group. YR = tyrosine recombinase; GCR1_C = DNA-binding domain; DDE = DDE-transposase; C48 = C48 peptidase; HTH = helix-turn-helix motif.
Table 1.
Distribution of Crypton elements
| Classification | Phylum or Class | Speciesa |
|---|---|---|
| Fungi | Basidiomycota | Cryptococcus neoformans [19] |
| Ascomycota | Coccidioides posadasii [19], Histoplasma capsulatum [19], Chaetomium globosum, Fusarium oxysporum, Ajellomyces capsulatus, Coccidioides immitis, Microsporan canis, Talaromyces stipitatus, Neosartorya fischeri | |
| Zygomycota | Rhizopus oryzae | |
| Animals | Chordata | Oryzias latipes |
| Echinodermata | Strongylocentrotus purpuratus [28] | |
| Hemichordata | Saccoglossus kowalevskii | |
| Mollusca | Lottia gigantea | |
| Arthropoda | Nasonia vitripennis [29], Tribolium castaneum [29], Rhodnius prolixus, Aedes aegypti, Culex quinquefasciatus | |
| Cnidaria | Nematostella vectensis [28] | |
| Stramenopiles | Oomycetes | Phytophthora infestans, Phytophthora sojae, Phytophthora ramorum, Pythium ultimum, Saprolegnia parasitica, Hyaloperonospora arabidopsidis, Albugo laibachii |
| Diatoms | Phaeodactylum tricornutum |
aCrypton elements from species without references are found in this study.
Figure 2.
Phylogeny of Cryptons, DUF3504 genes and other eukaryotic tyrosine recombinases. The numbers at nodes are bootstrap values over 40. Open circles indicate the clusters of Cryptons, and filled circles show the clusters of DUF3504 genes. YR = tyrosine recombinase. Prefixes of names are as follows. Cry = Crypton; 1958 = KIAA1958. Accession numbers of DUF3504 genes are shown in Additional file 5. Sequences of the transposable elements are deposited in Repbase http://www.girinst.org/repbase/. Other abbreviations and accession numbers are as follows. FLP = FLP recombinase of the 2-micron plasmid in Saccharomyces cerevisiae (NP_040488); FLP_Klac = FLP recombinase of the plasmid pKD1 in Kluyveromyces lactis (YP_355327); CRE = Cre recombinase of the enterobacteria phage P1 (YP_006472); Vlf1_AcNPV = very late expression factor 1 from the Autographa californica nucleopolyhedrovirus (NP_054107); Tn916 = Tn916 transposase from Enterococcus faecalis (NP_0687929); XerD = XerD from Escherichia coli (NP_417370); Lambda = lambda phage recombinase (NP_040609); At_Ti = recombinase from the Agrobacterium tumefaciens Ti plasmid (NP_059767); SpPat1 from Strongylocentrotus purpuratus (obtained at http://biocadmin.otago.ac.nz/fmi/xsl/retrobase/home.xsl). Suffixes for species names are as follows. Animals: Hs = human, Homo sapiens; Oa = platypus, Ornithorhynchus anatinus; Gg = chicken, Gallus gallus; Tg = zebra finch, Taeniopygia guttata; Ac/ACa = lizard, Anolis carolinensis; Xt/XT = frog, Xenopus tropicalis; Dr/DR = zebrafish, Danio rerio; OL = medaka, Oryzias latipes; Cm = chimaera, Callorhinchus milii; SP = sea urchin, Strongylocentrotus purpuratus; SK = acorn worm, Saccoglossus kowalevskii; Dm = fruit fly, Drosophila melanogaster; Tc/TC/TCa = beetle, Tribolium castaneum; NVi = parasitic wasp, Nasonia vitripennis; CQ = southern house mosquito, Culex quinquefasciatus; AA = yellow fever mosquito, Aedes aegypti; DPu = water flea, Daphnia pulex; Acal = sea hare, Aplysia californica; Sm = bloodfluke, Schistosoma mansoni; NV = sea anemone, Nematostella vectensis. Fungi: RO = Rhizopus oryzae; CGlo = Chaetomium globosum; TS = Talaromyces stipitatus; CI = Coccidioides immitis; FO = Fusarium oxysporum. Stramenopiles: PI = Phytophthora infestans; PS = Phytophthora sojae; PU = Pythium ultimum; HAra = Hyaloperonospora arabidopsidis; ALai = Albugo laibachii; PTri = Phaeodactylum tricornutum. Plants: CR = Chlamydomonas reinhardtii.
CryptonF elements from fungi and oomycetes, and CryptonF-derived genes
We identified CryptonF elements in nine species of fungi and four species of oomycetes (Table 1 and Additional file 1). These elements encode a protein that includes YR and GCR1_C DNA-binding domains (Figure 1). Most of the fungal Cryptons and the five oomycete Cryptons are associated with 6-bp terminal direct repeats, which are likely substrates for Crypton integration (Additional file 1). In Fusarium oxysporum, Crypton is fused with a Mariner-type DNA transposon and this composite transposon is hearafter named MarCry-1_FO (Figure 1). The analysis of four MarCry-1_FO copies with more than 97% identity to each other revealed the presence of 16-bp TIRs and target site duplications (TSDs) of the TA dinucleotide, indicating that their Mariner-type DDE-transposase is responsible for transposition. CryptonF-2_PS from Phytophthora sojae and related elements encode a C48 peptidase (Ulp1 protease) in addition to a YR (Figure 1). The oomycete CryptonF elements are nested in fungal CryptonF elements in the phylogenetic tree (Figure 2), indicating a horizontal transfer between fungi and oomycetes.
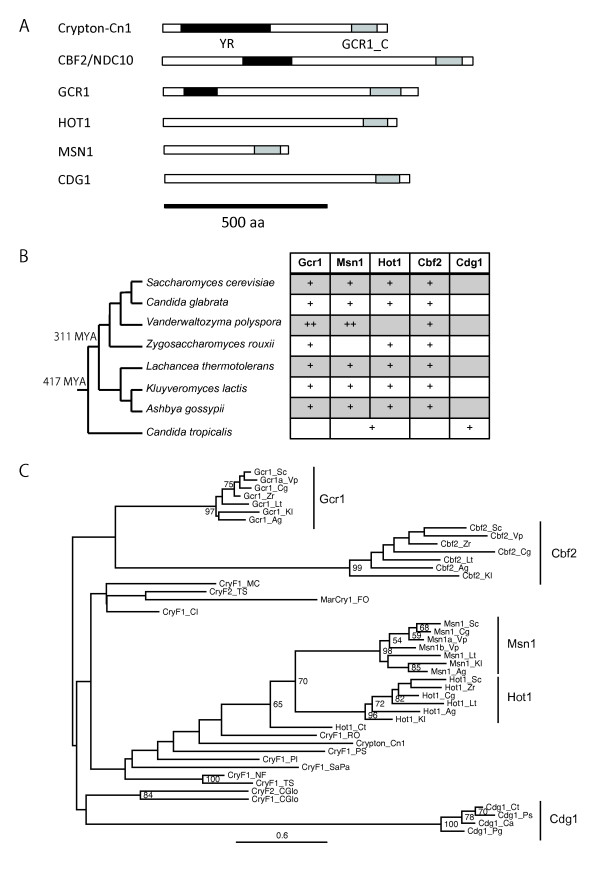
Four genes from Saccharomyces cerevisiae were derived from CryptonF elements (Figure 3 and Additional files 2 and 3). It was previously reported that the GCR1_C protein domain encoded by Gcr1, Msn1 and Hot1 genes is similar to the C-terminal part of fungal Cryptons [19]. In addition to these three genes, we found that Cbf2/Ndc10 contains a C-terminal domain similar to CryptonF proteins. The central portions of Cbf2 and Gcr1 are similar to CryptonF YR domains, but the catalytic site is not preserved (data not shown). Vanderwaltozyma polyspora carries two paralogous genes of Gcr1 and Msn1. Candida tropicalis and related species (Candida albicans, Pichia stipitis and Pichia guilliermondii) harbor another gene derived from a CryptonF element, represented by XP_002548716 in C. tropicalis. It is designated herein as Crypton-derived gene 1 (Cdg1) (Figure 3). The only domain shared by CryptonF elements and all Crypton-derived genes is the GCR1_C domain. The phylogenetic analysis of GCR1_C domains (Figure 3C) indicates that Hot1 and Msn1 are paralogous and that the gene related to Hot1/Msn1 in C. tropicalis represents an outgroup of both genes. Therefore, it is likely that four domestication events (for Hot1/Msn1, Gcr1, Cbf2 and Cdg1) occurred in this group.
Figure 3.
Distribution and schematic structures of Crypton-derived genes in Saccharomycetaceae fungi. (A) Schematic protein structures encoded by Crypton-derived genes and Cryptons. (B) Distribution of Crypton-derived genes. Each gene identified in the haploid genome is represented by a plus symbol. (C) The phylogeny of Crypton-derived genes and Cryptons using the GCR1_C domain sequences. The numbers at nodes are bootstrap values over 50. Accession numbers of genes are shown in Additional file 2. "Cry" stands for Crypton. Suffixes for species names are as follows. Sc = Saccharomyces cerevisiae; Cg = Candida glabrata; Vp = Vanderwaltozyma polyspora; Zr = Zygosaccharomyces rouxii; Lt = Lachancea thermotolerans; Kl = Kluyveromyces lactis; Ag = Ashbya gossypii; Ct = Candida tropicalis; Ca = Candida albicans; Ps = Pichia stipitis; Pg = Pichia guilliermondii.
We could not find any Crypton insertions in the subphylum Saccharomycotina (including S. cerevisiae, C. tropicalis and related species). The distribution of Crypton-derived genes indicates that Crypton was active in the past and that the DNA-binding domain GCR1_C was most likely derived from Cryptons.
CryptonS, a new group of Cryptons from oomycetes and diatom
We found CryptonS elements in seven oomycete and one diatom species (Figure 1, Table 1 and Additional file 1). CryptonS elements do not encode any GCR1_C domain, but the C-terminal region is conserved among CryptonS elements. CryptonS elements are associated with 5- or 6-bp terminal direct repeats. The majority of CryptonS elements share TATGG termini. Some CryptonS elements encode an additional protein containing a C48 peptidase domain. The peptidases encoded by CryptonS and CryptonF elements in oomycetes belong to the same family and are related to the Ulp1 protease family. Domain shuffling between two groups of Crypton elements could explain the similarity, but more data are needed to determine the relationship between these peptidases and other cellular peptidases.
Cryptons in animals (CryptonA and CryptonI groups)
We identified Cryptons in seven metazoan animals belonging to five phyla (Table 1 and Additional file 1). CryptonI elements were found only in insects, whereas CryptonA elements were found in various animals, including cnidarians. Animal Cryptons (both CryptonA and CryptonI) have no C-terminal domain (Figure 1). We did not find any terminal repeats in animal Cryptons. CryptonI-1_RPro from Rhodnius prolixus hosts a non-autonomous derivative family, CryptonI-1N1_RPro, in which 5' 438 bp and 3' 260 bp are 98% identical to those of CryptonI-1_RPro. This is the first report of non-autonomous Crypton elements. Comparison of 50 copies of CryptonI-1_RPro and CryptonI-1N1_RPro revealed no terminal repeats (neither direct nor inverted). In medaka, we also found two families of non-autonomous derivatives (CryptonA-1N1_OL and CryptonA-1N2_OL) of CryptonA-1_OL. As in the case of other DNA transposons, Crypton non-autonomous elements are much more abundant than their autonomous counterparts.
We can safely rule out the theoretically possible contamination of the genomic sequences from medaka used in this study. First, we identified more than 2,700 copies of autonomous and non-autonomous Crypton elements with DNA sequence identities to consensus ranging from 59% to 98%. The nucleotide diversity of Cryptons from medaka is consistent with their long-term presence in the medaka lineage. Second, we found many Crypton sequences in the database of expressed sequence tags (ESTs) from three different medaka strains: Hd-rR, CAB and HNI (data not shown). We also found several Cryptons with inserted medaka-specific transposons such as piggyBac-N1_OL and RTE-1_OL (Table 2).
Table 2.
Crypton copies containing insertions of other transposable elements
| Chromosome | Starta | Enda | Element | Startb | Endb | Directionc | Identityd |
|---|---|---|---|---|---|---|---|
| chr1 | 35100344 | 35100443 | Crypton-1N1_OL | 470 | 573 | c | 0.8614 |
| 35100444 | 35100648 | piggyBAC-N1_OL | 1 | 204 | d | 0.9512 | |
| 35100649 | 35101118 | Crypton-1N1_OL | 1 | 474 | c | 0.8728 | |
| chr23 | 4844226 | 4844361 | Crypton-1N1_OL | 1 | 147 | d | 0.9489 |
| 4844362 | 4844566 | piggyBAC-N1_OL | 1 | 205 | d | 0.9854 | |
| 4844567 | 4844980 | Crypton-1N1_OL | 144 | 570 | d | 0.9294 | |
| chr23 | 7640079 | 7640510 | Crypton-1N1_OL | 1 | 441 | d | 0.8918 |
| 7640511 | 7640716 | piggyBAC-N1_OL | 1 | 205 | d | 0.9466 | |
| 7640717 | 7640832 | Crypton-1N1_OL | 438 | 573 | d | 0.8814 | |
| chr5 | 2033028 | 2033143 | Crypton-1N1_OL | 3 | 116 | d | 0.8966 |
| 2033144 | 2033348 | piggyBAC-N1_OL | 1 | 205 | c | 0.9659 | |
| 2033349 | 2033803 | Crypton-1N1_OL | 113 | 573 | d | 0.9132 | |
| chr5 | 22193852 | 22194261 | Crypton-1N1_OL | 144 | 573 | c | 0.9030 |
| 22194262 | 22194466 | piggyBAC-N1_OL | 1 | 205 | d | 0.9707 | |
| 22194467 | 22194605 | Crypton-1N1_OL | 1 | 147 | c | 0.9078 | |
| chr8 | 7666866 | 7667209 | Crypton-1N1_OL | 21 | 376 | d | 0.9075 |
| 7667210 | 7667896 | RTE-1_OL | 2,666 | 3,352 | c | 0.9796 | |
| 7667897 | 7668065 | Crypton-1N1_OL | 372 | 550 | d | 0.8667 |
aSequence coordinates in the corresponding chromosome. bCorresponding coordinates of the consensus sequences of repeat elements from Repbase. cSequence orientation relative to consensus: (d)irect and (c)omplementary orientation. dIdentity to the consensus sequences.
Crypton-derived sequences in the ATF7IP gene
Identification of Cryptons in three deuterostome species (medaka, sea urchin and acorn worm) prompted us to extend analysis of Cryptons in chordates, including four sequenced actinopterygian species (Fugu rubripes, Tetraodon nigroviridis, Gasterosteus aculeatus and Danio rerio). Although multiple copies of Crypton elements were found only in medaka, sequences similar to Cryptons were found in various chordate species (Table 3). Most of them do not encode any functional recombinases, owing to frameshifts, deletions and substitutions at catalytically essential residues.
Table 3.
Molecular fossils of Cryptons in chordates
| Species | Accession numbers |
|---|---|
| Danio rerio | BX530066* |
| Xenopus tropicalis | NP_001120376, AAMC01135377*, AAMC01082917* |
| Callorhinchus milii | AAVX01521991*, AAVX01068049*, AAVX01132927* |
| Ciona intestinalis | XP_002124034, XP_002125964 |
| Ciona savignyi | AACT01002283*, AACT01041791* |
| Halocynthia roretzi | BAB40645 |
| Oikopleura dioica | CBY34656 |
| Branchiostoma floridae | XP_00260067, XP_002595788, XP_002613958, XP_002613959, XP_002587732, XP_002607491 |
*Nucleotide sequences including Crypton fragments.
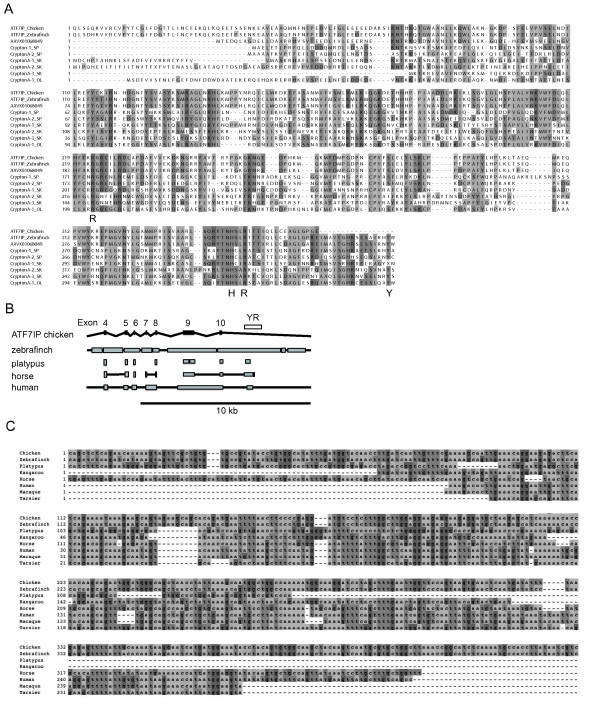
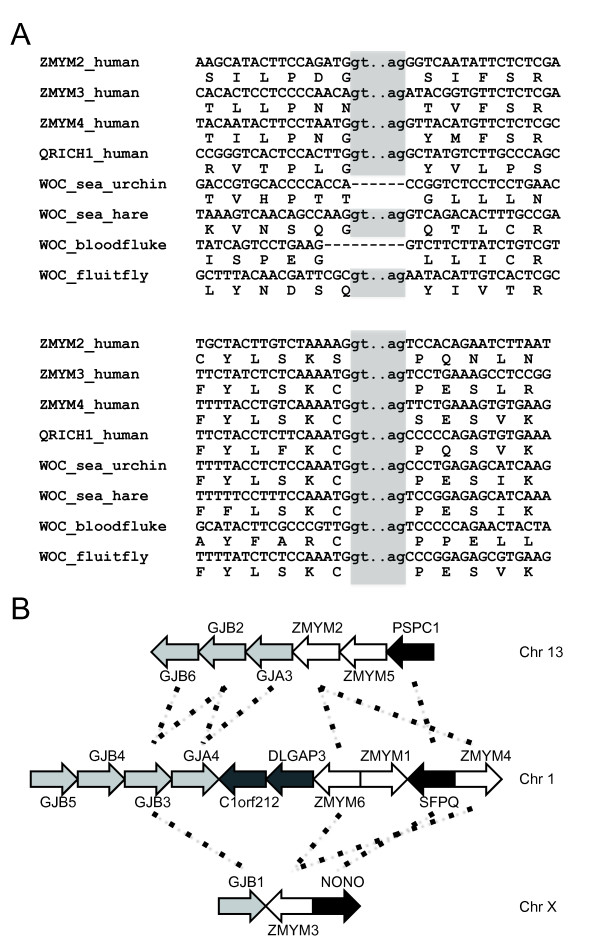
However, two similar sequences (ABQF01015803 from the zebra finch Taeniopygia guttata and AAVX01068049 from the chimaera (elephant shark) Callorhinchus milii) include an intact open reading frame of YR (Figure 4A). We did not further analyze the sequence from chimaera, because the sequenced region was only 2,661 bp in length. The Crypton-like sequence in zebra finch is inside an intron of a gene coding for activating transcription factor 7 interacting protein (ATF7IP) (Figure 4B). There is a YR sequence at the orthologous locus of chicken Gallus gallus, which encodes a protein 97% identical to that of zebra finch, but it contains a frameshift inside the YR region. The orthologous YR sequence from the turkey Meleagris gallopavo contains a frameshift at the same position (data not shown). Because the divergence between chicken and zebra finch occurred some 107 million years ago (MYA) [30], this unusually high similarity indicates a strong selection operating on these YR sequences. An exon-intron prediction program would predict alternative splicing in the ATF7IP gene from zebra finch, although at present there are no mRNA or ESTs corresponding to the fusion transcript. It is possible that the YR is translated as part of the ATF7IP protein and retains catalytic activity in some birds.
Figure 4.
Crypton-derived sequence in an intron of ATF7IP gene. (A) Alignment of proteins coded by deuterostome Cryptons and Crypton-derived sequences. Catalytically essential residues are shown below the alignment. (B) Illustration of the conservation of ATF7IP loci. The position of the YR sequence is indicated by the open box. Black boxes represent exons of the chicken ATF7IP gene. Gray boxes indicate conserved blocks between chicken and respective species based on the Net Tracks of the UCSC Genome Browser http://genome.ucsc.edu/. Lines between gray boxes indicate that boxes are connected by unalignable sequences. (C) Alignment of nucleotide sequences of Crypton-derived sequences.
Using the University of California Santa Cruz (UCSC) Genome Browser http://genome.ucsc.edu/, we found that there are partial Crypton sequences at the orthologous positions of the ATF7IP gene from the human, horse, kangaroo and platypus genomes (Figures 4B and 4C). There are also closely related sequences present in the genomes of rhesus macaque and tarsier. Therefore, the insertion of Crypton in the ATF7IP gene must have occurred in the common ancestor of amniotes more than 325 MYA [30]. None of the mammalian orthologous sequences encode intact YR proteins, and many mammalian species are missing the YR sequence. This indicates only a slight, if any, selective pressure on this sequence in mammals.
Ancient domestication of Cryptons in animals
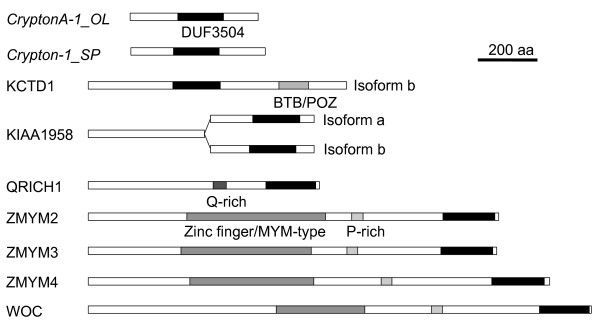
Most vertebrate genes similar to Crypton code for proteins (Additional file 4). In the human genome, there are seven proteins similar to Crypton YRs, which are annotated as parts of six genes (Figure 5 and Additional file 5). The KIAA1958 gene contains two isoforms, both of which include YR-derived sequences. The other genes are potassium channel tetramerization domain containing 1 (KCTD1), zinc finger, myeloproliferative and mental retardation type 2 (ZMYM2)/zinc finger protein 198 (ZNF198), ZMYM3/ZNF261, ZMYM4/ZNF262 and glutamine-rich protein 1 (QRICH1) (Figure 5). A PSI-BLAST search of these proteins against the National Center for Biotechnology Information (NCBI) conserved domain database (CDD) revealed that they share a domain of unknown function (DUF3504 superfamily; E-value ≤ 1e-29). The six genes are widespread among vertebrates (Figure 6) and are highly conserved among phylogenetically distant species (Table 4). The phylogenetic relationship of each gene agreed with that of species (data not shown). The nucleotide sequences corresponding to all seven DUF3504 domains were present in the NCBI EST database, indicating their expression. The data clearly show that they are neither pseudogenes nor defective Cryptons (see the accession numbers of DUF3504 genes in Additional file 5). However, none of them preserve the YR catalytic site. All of them lost the catalytic tyrosine and the second conserved arginine, and all but KCTD1 also lost the conserved histidine.
Figure 5.
Schematic structures of DUF3504 proteins. KIAA1958 gene has two isoforms, each of which encodes a DUF3504 domain. The structures of KCTD1, KIAA1958, QRICH1, ZMYM2, ZMYM3 and ZMYM4 are from humans. The structure of WOC is from Drosophila melanogaster.
Figure 6.
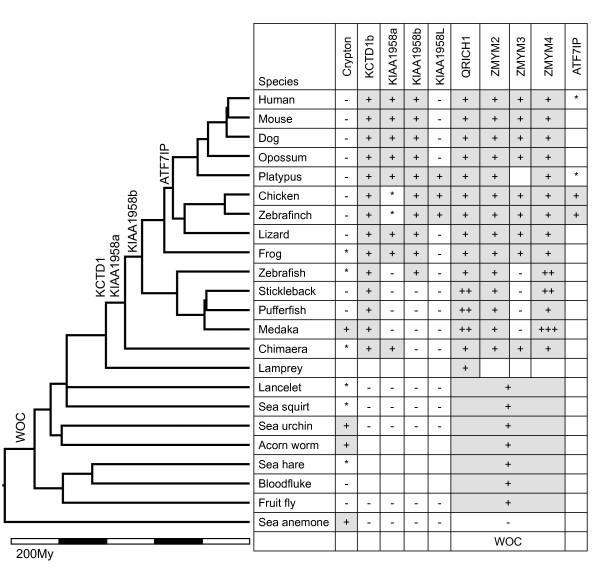
Distribution of Cryptons and Crypton-derived genes. Each gene identified in the haploid genome is represented by a plus symbol. Minus symbols indicate the absence of Cryptons or Crypton-derived genes. Asterisks indicate the presence of their disrupted fragments. The branch ages are based on TimeTree [30]. The unit of time is indicated. Crypton-derived genes listed at nodes of the tree indicate the times of their domestication based on their distribution in different species. KIAA1958L, QRICH1, ZMYM2, ZMYM3 and ZMYM4 are not shown, because they were likely derived by gene duplications.
Table 4.
Protein identities between DUF3504 domains in two species
| Comparison | KCTD1 | KIAA1958b | QRICH1 | ZMYM2 | ZMYM3 | ZMYM4 |
|---|---|---|---|---|---|---|
| Human-chicken | 98% (268 of 274) | 99% (282 of 287) | 97% (299 of 309) | 92% (287 of 313) | 85% (265 of 315) | 88% (302 of 347) |
| Human-zebrafish | 90% (273 of 304) | 86% (245 of 287) | 75% (229 of 306) | 47% (145 of 310) | - | 52% (ZMYM4a) (176 of 345) 52% (ZMYM4b) (169 of 331) |
Numbers in parentheses represent the number of identical residues (left) and the aligned domain length (right). Gaps are excluded in the comparison.
Although the resolution is low because of high divergence and the short length of the YR sequence, animal DUF3504 genes tend to cocluster with animal Cryptons (CryptonA) in the YR phylogenetic tree (Figure 2). There are four independent clusters of DUF3504 genes: KCTD1, KIAA1958a, KIAA1958b/KIAA1958L and WOC/ZMYM/QRICH1 (Figure 2, filled circles). KCTD1 coclusters with several animal Cryptons, and the clustering is supported by 100% bootstrap value. Cryptons form a paraphyletic cluster, which indicates that the DUF3504 domain of KCTD1 was derived from a Crypton YR. The position of KIAA1958a is distinct from either CryptonA or CryptonI, and WOC/ZMYM/QRICH1 is clustered as a sister group of all animal CryptonA elements. Therefore, phylogeny alone does not support the domestication of animal Cryptons leading to WOC/ZMYM/QRICH1 and KIAA1958a.
The DUF3504 domain was derived from YR, not vice versa, because DUF3504 lacks the complete catalytic tetrad essential for YR activity. YR is essential for transposition, and repeated generation of active YRs from defective YRs is highly improbable. The distributions of WOC/ZMYM/QRICH1 and KIAA1958a are restricted to bilaterians and jawed vertebrates, respectively. Apart from Cryptons, the only other possible sources of YRs in animal genomes are the retrotransposon families DIRS and Ngaro [23,24]. However, all searches of the YRs from CryptonA-1_OL, Crypton-1_SP and CryptonA-1_SK matched the DUF3504 sequence with E-values ≤ 8e-12, whereas YRs of DIRS and Ngaro did not match the DUF3504 sequence at all (even when the threshold E-value was set at 100). Several representatives of DUF3504 are actually Crypton sequences; for example, XP_001639277 is the protein coded by Crypton-1_NV. The patchy distribution of Cryptons and the inconsistency between Crypton and host phylogenies indicate ancient amplification and extinction events in Crypton evolution. The ancient amplifications would have generated many lineages of Cryptons, and it is likely that WOC/ZMYM/QRICH1 and KIAA1958a derived from lineages of Cryptons that are now extinct. We cannot completely rule out the possibility that the two genes and CryptonA elements were independently derived from DIRS-like retrotransposons or some as yet uncharacterized types of mobile elements, but this implies independent origins of CryptonA and other Crypton groups (CryptonF, CryptonS and CryptonI). Therefore, four independent domestication events of animal Cryptons are the most parsimonious explanation for the origins of animal DUF3504 genes.
A representative of DUF3504 from Halocynthia roretzi (BAB40645) has orthologs in other tunicates: Ciona intestinalis (XP_002125964), Ciona savignyi (AACT01002283 and AACT10141791) and Oikopleura dioica (CBY34656). They could also represent a domestication event of Crypton. Another representative (YP_025778) is coded in the mitochondrion of the green alga Pseudendoclonium akinetum. It could also be a candidate Crypton-derived gene; owing to the lack of related sequences, however, we did not analyze it further.
All DUF3504 genes encode much longer proteins than their DUF3504 domains, and it is possible that the preexisting genes captured entire Crypton protein-coding sequences. However, the only recognizable domain encoded by animal Cryptons is YR (DUF3504), and there is little sequence similarity beyond YRs among Cryptons themselves. Therefore, it is unlikely that any significant sequence similarity was preserved beyond the DUF3504 domains between the DUF3504 and Crypton proteins.
KCTD1 gene
The KCTD1 gene contains the DUF3504 domain confined within a single exon. Among the vertebrate genes, the KCTD1 DUF3504 domain is the closest to the Crypton YRs in terms of protein sequence similarity. The sequence identity between the KCTD1 DUF3504 domain and the YR of Crypton-1_SP is 32%, which exceeds the analogous identity among different lineages of Cryptons. For example, Crypton-1_SP in the CryptonA lineage and Crypton-1_TC in the CryptonI lineage show less than 30% sequence identity to each other. KCTD1 encodes two protein isoforms of different lengths. The longer isoform (isoform b) contains both an N-terminal DUF3504 domain and a C-terminal BTB/POZ (Broad-complex, Tramtrack and Bric-a-brac/poxvirus and zinc finger) domain (Figure 5), whereas the shorter one (isoform a) contains only the BTB/POZ domain. The shorter isoform is approximately 80% identical to the KCTD15 gene at the protein level, and related genes are found in various organisms, including lancelet, sea urchin and insects (Additional file 6). The KCTD15 gene does not have any DUF3504 domain and is found in gnathostomes from mammals to chimaera. We infer that KCTD1 and KCTD15 were duplicated from a single gene in the early evolution of vertebrates and after that a Crypton copy was inserted upstream of the KCTD1 gene, which generated a new transcriptional variant encoding the isoform b. This insertion should have occurred before the branching of the Chondrichthyes (sharks, rays, skates and chimaeras) about 527 MYA [30].
KIAA1958 gene
The KIAA1958 gene, of unknown function, is present in two isoforms (a and b) which contain different DUF3504 domains encoded by different exons (Figure 5). DUF3504 domains in isoforms a and b are only 26% identical to each other in the human genome. Although alternative splicing has not been confirmed experimentally, the high conservation of both DUF3504-coding sequences indicates that both encode functional proteins (Table 4 and Additional file 4). Neither of the two DUF3504-coding sequences is interrupted by introns. We found both isoforms in a wide range of tetrapods (Figure 6). Zebrafish lacks isoform a, whereas Chondrichthyes (chimaera) lack isoform b. Some Actinopterygii (medaka, stickleback, fugu and pufferfish) lack both isoforms. Chicken, zebra finch and platypus have an additional gene similar to KIAA1958 isoform b, designated KIAA1958L. The exons encoding isoform-specific regions of KIAA1958b are positioned upstream from those of KIAA1958a. The KIAA1958L gene likely originated from a duplication of the segment including KIAA1958b-specific exons but not including KIAA1958a-specific exons, and this duplication event predated the branching between mammals and birds. KIAA1958L is less conserved than KIAA1958 isoforms a and b. The DUF3504 domains of KIAA1958L proteins from platypus and chicken are only 44% identical, whereas those of the KIAA1958b are 97% identical between the species. KIAA1958a is nonfunctional in chicken and zebra finch, but it is intact in lizard. Isoform b was not found in Chondrichthyes, and it is possible that it originated later in the lineages which branched off Chondrichthyes. KIAA1958a might have originated in the common ancestor of gnathostomes. Another possibility is that both isoforms were acquired in the common ancestor of gnathostomes and isoforms a and b had been lost in the lineages of Actinopterygii and Chondrichthyes, respectively.
ZMYM2, ZMYM3, ZMYM4 and QRICH1 genes
The ZMYM2, ZMYM3 and ZMYM4 genes are present in diverse gnathostomes from human to chimaera (Figure 6). ZMYM2, ZMYM3 and ZMYM4 are similar to arthropod WOC in terms of their sequence and structure [31,32]. The DUF3504 domains from the Drosophila melanogaster WOC gene and the human ZMYM2 gene are 41% identical at the protein level. Some introns are also positioned at the corresponding sites of ZMYM2 and WOC (Figure 7A). In addition to chordates and arthropods, we found sequence fragments related to WOC in echinoderms (Strongylocentrotus purpuratus), hemichordates (Saccoglossus kowalevskii), mollusks (Pinctada maxima and Aplysia californica) and platyhelminthes (Schistosoma mansoni, S. japonicum and Schmidtea mediterranea) (Additional file 5). There is no evidence that WOC forms multiple gene families in invertebrates. The ZMYM2, ZMYM3 and ZMYM4 genes are listed in the data set of ohnologs reported recently by Makino and McLysaght [33], which means that they were duplicated from a single gene during two rounds of whole-genome duplication in the early evolution of vertebrates before the split between jawed vertebrates and agnathans [34,35]. The synteny blocks of ZMYM2, ZMYM3 and ZMYM4 share several genes in addition to ZMYM genes (Figure 7B). The most parsimonious scenario is that the WOC/ZMYM gene family originated from the domestication of Crypton in the common ancestor of bilaterians.
Figure 7.
Paralogous relationships of WOC/ZMYM/QRICH1 genes. (A) Two conserved intron positions among WOC, ZMYM2, ZMYM3, ZMYM4 and QRICH1. Introns are printed in lowercase letters and shaded. Protein sequences are shown below nucleotide sequences. The upper and lower intron positions correspond to the 20th and 22nd introns of human ZMYM2, respectively. (B) The synteny blocks of ZMYM2, ZMYM3 and ZMYM4. Ohnologous relationships reported by Makino and McLysaght [33] are indicated by dotted lines. GJB = gap junction protein β; GJA = gap junction protein α; DLGAP3 = discs large homolog-associated protein 3; C1orf212 = chromosome 1 open reading frame 212. Other gene names are described in the text.
There are three other ZMYM genes (ZMYM1, ZMYM5 and ZMYM6) in the synteny blocks (Figure 7B), but they have no DUF3504 domain. The N-terminal part of ZMYM5 is similar to that of ZMYM2, whereas those of ZMYM1 and ZMYM6 are similar to that of ZMYM4. These three genes are present only among eutherian mammals. These data support independent gene duplication events inside each synteny block. It is noteworthy that the C-terminal parts of ZMYM1, ZMYM5 and ZMYM6 derived from transposases of hAT-type DNA transposons, but these hAT-derived sequences are not close to each other. The C-terminal part of ZMYM6 is close to Charlie elements in the human genome, whereas that of ZMYM1 is closer to plant hAT elements such as HAT-1_Mad from apple (data not shown).
The QRICH1 gene was found in diverse vertebrates, including lamprey (Figure 6). The DUF3504 domain in QRICH1 is quite similar to those of ZMYM2, ZMYM3 and ZMYM4. Besides, five of eight introns of QRICH1 were at the sites corresponding to those of ZMYM2, ZMYM3 and ZMYM4 (Figure 7A and data not shown). The high structural and sequence similarity between WOC, ZMYM2/3/4 and QRICH1 indicates that QRICH1 originated from either WOC or ZMYM genes. In the neighborhood of QRICH1, we could not find any genes paralogous to genes in the synteny blocks of ZMYM2, ZMYM3 and ZMYM4. However, because QRICH1 is present in the lamprey genome, it must have originated at the time close to the whole-genome duplication events.
Discussion
Evolution of WOC: the third-oldest event of transposon domestication
The most ancient transposon-derived gene known to date is TERT, which was generated by the domestication of a Penelope-like retroelement [4], and Prp8, a spliceosomal component derived from a retrointron (group II self-splicing intron) [36]. TERT retains the catalytic activity of RT, but Prp8 does not. These two genes are shared by almost all eukaryotes. Another example of an ancient domestication event is the RAG1 gene [5]. It is distributed widely among gnathostomes, but no RAG1 ortholog was found in agnathans, including lamprey and hagfish. Given that agnathans have a different type of adaptive immune system called "variable lymphocyte receptors" [37], the domestication of RAG1 likely occurred in the last common ancestor of gnathostomes after their branching from agnathans. Other transposons domesticated in the distant past are in HARBI1 and PBDG5 genes, both of which are present in vertebrates from humans to actinopterygian fish [38,39]. The KCTD1b, KIAA1958a and KIAA1958b genes are as old as or older than the HARBI1 and PBDG5 genes (Figure 6). A transposon-derived CENP-B, a highly conserved mammalian centromere, and three CENP-B-like proteins (Abp1, Cbh1 and Cbh2) in fission yeast resemble each other in terms of their sequences and functions, but they derived independently from different pogo-like transposases [40]. The human genome harbors a significant number of genes derived from transposons [6]. Some of them were domesticated in the distant past, and there are no traces of related repetitive sequences or TEs from which they were derived. For example, the HARBI1 gene was derived from PIF/Harbinger and PHSA (THAP domain-containing protein 9, or THAP9) from a P-like element [38,41]. Both HARBI1 and PHSA were found by screening mammalian genes against DNA transposons from zebrafish. Similarly, the key to our findings of Crypton-derived genes was screening of genes against Cryptons preserved in medaka, because there are only a few remnants of Cryptons left in vertebrate genomes sequenced to date, except in medaka.
The ancestral gene for WOC/ZMYM probably originated in the common ancestor of all bilaterians more than 910 MYA [30]. This is the third-oldest transposon domestication event known to date, following the two domestication events of RT [4,36]. Our study indicates that domestication of Crypton-like elements in eukaryotes was relatively common in the distant past. This implies that Cryptons are very ancient and, given their rare occurrence in the genomic fossil record and their great diversity, they were probably much more active in the distant past than in more recent evolutionary history.
Functional implications for domesticated Crypton YRs
No function of DUF3504 domains has been reported to date. Even so, the YR origin of DUF3504 domains implies their functions to some extent. YR forms a multimer when it binds substrate DNA during recombination [42]. On that basis, we can envision two possible functions derived from YRs: DNA binding and protein-protein interaction. There are several indications for functions of domesticated YRs. First, many genes derived from YRs are transcriptional regulators. Gcr1, KCTD1, WOC, ZMYM2, ZMYM3 ZMYM4, and ATF7IP are either transcriptional activators or repressors [43-48]. Cbf2 acts as a centromeric protein directly binding to centromere-specific sequences and is essential for spindle pole body formation [49,50]. Although these proteins usually contain a DNA-binding domain other than DUF3504, exemplified by the GCR1_C domain of Gcr1 and Cbf2, the DUF3504 domain could also work as a DNA-binding domain. Second, there is an interesting resemblance between functions of domesticated DDE-transposases and YRs. Daysleeper is a transcription factor derived from a hAT DDE-transposase and binds a specific motif for transcription regulation [12]. CENP-B is a centromere protein derived from the DDE-transposase of a pogo-like transposon [13]. In these genes, transposase-derived domains act as DNA-binding domains. Third, a large family of prokaryotic transcriptional activators, AraC/XylS, shows structural similarity to YRs. The overall fold of the 129-amino acid protein MarA, a member of the AraC/XylS family, almost entirely recapitulates the YR domain of Cre recombinase [51]. MarA can simultaneously bind RNA polymerase II and DNA to form a ternary complex [52]. These data support the putative function of DUF3504 to be DNA or protein binding.
To date relatively little is known about Cryptons. There have been no studies of their transposition, transcription, translation or regulation. The sequence similarity between Cryptons is very low, especially in their non-protein-coding regions. We compared DNA sequences of Cryptons from different species, but we could not find any conserved nucleotide sequences among them. Furthermore, all Crypton domestication events are very old. Therefore, it is very difficult to propose any specific functions of DUF3504 domains. Instead, herein we propose potential pathways in which DUF3504 domains could be involved.
KCTD1 and KCTD15 are paralogs that have diverged during the early evolution of vertebrates (Additional file 6). KCTD1 isoform b, generated by an insertion of Crypton upstream of the original KCTD1 gene, is widely conserved among jawed vertebrates (Figure 6), although it is unclear whether the agnathans carry the KCTD1b gene. The high conservation of KCTD1b (Table 4) indicates its essential function shared among jawed vertebrates. KCTD1 represses the activity of the AP-2α transcription factor, and the BTB/POZ domain is responsible for the interaction [46]. AP-2α plays an essential role in neural border (NB) and neural crest (NC) formations during embryonic development [53]. NB is the precursor of NC. KCTD15 is expressed in NB and inhibits NC induction [54]. The NC cells are a transient, multipotent, migratory cell population unique to vertebrates. They give rise to diverse cell lineages. We can speculate that by adding a new protein-protein or protein-DNA interaction KCTD1b can contribute to the network of NC formation through the regulation of AP-2α.
Among DUF3504 genes, the function of WOC/ZMYM is of special interest because two of the genes in this group, ZMYM2 and ZMYM3, are linked to human diseases. A chromosomal translocation between ZMYM2 and fibroblast growth factor receptor 1 (FGFR1) causes lymphoblastic lymphoma and a myeloproliferative disorder [55]. A translocation involving ZMYM3 is associated with X-linked mental retardation [56]. Mutations of their ortholog, WOC, cause larval lethality in D. melanogaster [31].
WOC/ZMYM gene-encoded proteins are involved in various processes, including transcription, DNA repair and splicing. WOC is a transcriptional regulator that colocalizes with the initiating forms of RNA polymerase II [31,32]. The WOC proteins also colocalize with all telomeres, and mutants of WOC are associated with frequent telomeric fusions [31,32]. ZMYM2, ZMYM3 and ZMYM4 are components of a multiprotein corepressor complex, including histone deacetylase 1 (HDAC1) and HDAC2 [47,48]. ZMYM2 binds to various transcriptional regulators including Smad proteins [57]. It also binds to proteins involved in homologous recombination, such as RAD18, HHR6A and HHR6B, which are human orthologs of the yeast RAD proteins [58], and to spliceosomal components including SFPQ (splicing factor, proline- and glutamine-rich) [59].
Interestingly, the SFPQ gene is a component of the syntenic cluster of ZMYM4 (Figure 7B). The paralog of SFPQ in the cluster of ZMYM3 is NONO (non-POU domain-containing, octamer-binding), which is a partner of SFPQ in heteromers [60]. PSPC1 (paraspeckle component 1) present in the cluster of ZMYM2 also shows similarity to SFPQ and NONO genes. In addition to their involvement in splicing, the SFPQ proteins contribute to DNA repair by interacting with RAD51 [61]. They are also recruiting HDAC1 to the STAT6 transcription complex [62]. Therefore, it is likely that WOC/ZMYM and SFPQ/NONO/PSPC1 proteins cooperatively act in transcription regulation, splicing and DNA repair, and that they have coevolved by maintaining their functional relationships. Their DUF3504 domains may contribute to some of the protein-protein interactions.
Evolution of Cryptons
To date Cryptons have been identified in a limited number of fungi and animal species. Herein we report the presence of Cryptons in new species, but information regarding their overall distribution continues to be patchy (Table 1). CryptonF elements are present in three phyla of fungi (Ascomycota, Basidiomycota and Zygomycota) and two orders of oomycetes (Peronosporales and Saprolegniales). Our phylogenetic analysis supports the horizontal transfer of CryptonF elements between fungi and oomycetes (Figure 2), which is consistent with frequent horizontal transfer of genes between them [63]. CryptonS elements are also present in two oomycete orders and one species of diatoms. Both oomycetes and diatoms are lineages of stramenopiles, and the origin of CryptonS elements could date back to their common ancestor.
Animal Cryptons (CryptonA and CryptonI) were found in six phyla: Chordata, Echinodermata, Hemichordata, Arthropoda, Mollusca and Cnidaria. CryptonI elements have the same overall structure as CryptonA elements and were observed only in insect genomes. It is possible that CryptonI elements constitute a branch of CryptonA but they have evolved more rapidly in insects. The overall distribution in fungi, oomycetes and animals indicates that Cryptons were long present in these three eukaryotic groups, probably with some contribution of a horizontal transfer. It is likely that Cryptons originated in the common ancestor of these three groups, although because of the low resolution of the YR phylogeny, we cannot rule out the possibility of their independent origins.
The identification of Crypton elements in medaka is surprising. The nucleotide diversity of Cryptons in the medaka genome clearly shows that Cryptons were maintained in the lineage leading to medaka for a long time. It is possible that Cryptons invaded the medaka population after the split of medaka from the three actinopterygian fish species (Gasterosteus, Takifugu and Tetraodon), whose genomes have been sequenced. The vertical transfer of Cryptons in the lineage leading to medaka is a preferable scenario because of the domestication of Crypton in the common ancestor of bilaterian animals, which led to the origin of WOC genes. In most identified host organisms, Cryptons are preserved in very low copy numbers (Additional file 1). We found several fragments of Cryptons in various vertebrates, including zebrafish (Table 3). The origin of Crypton-derived genes took place at different times during the evolution of vertebrates (Figure 6). This is consistent with the hypothesis that Cryptons continued to maintain very low copy numbers in the vertebrate genomes and were occasionally amplified in certain lineages.
Conclusions
This study has revealed the diversity of a unique class of DNA transposons, Cryptons, and their repeated domestication events. The DUF3504 domains are domesticated YRs of animal Crypton elements. Our findings add a new repertoire of domesticated proteins and provide further evidence for an important role of transposable elements as a reservoir for new cellular functions.
Methods
Data source
Genome sequences of various species were obtained mostly from GenBank, and sequences of known Cryptons, DIRS and Ngaro were obtained from Repbase http://www.girinst.org/repbase/. All characterized Cryptons have been deposited in Repbase.
Sequence analysis
Characterization of new Cryptons was achieved by repeated BLAST [64] and CENSOR [65] searches using genome sequences of various species with Cryptons as queries. All analyses were done with default settings. The consensus sequences of elements were derived using the majority rule applied to the corresponding sets of multiple aligned copies of Cryptons. Alignment gaps were manually adjusted to maximize similarity to other related elements. Characterization of DUF3504 genes was performed by BLAST searches against both protein and nucleotide databases with known DUF3504 genes as queries. We predicted exon-intron boundaries with the aid of SoftBerry FGENESH:
http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind and manually adjusted them through the comparison to orthologous sequences in other species.
Sequence alignment and phylogenetic analysis
We used MAFFT [66] with the linsi option or MUSCLE [67] with default settings to align both nucleotide and protein sequences of various Cryptons and Crypton-derived proteins. We constructed maximum likelihood trees by using PhyML [68,69] with 100 bootstrap replicates [70] for the amino acid substitution model LG. We also constructed trees with other substitution models, WAG, RtREV and DCMut, and with the Neighbor-joining method, but the resolution did not improve. The tree topology search method was Nearest Neighbor Interchange (NNI), and the initial tree was BIONJ. The phylogenetic trees were drawn with FigTree 1.3.1 software http://tree.bio.ed.ac.uk/software/figtree/.
Abbreviations
bp: base pair; EN: endonuclease; MYA: million years ago; RT: reverse transcriptase; TE: transposable element; TERT: telomerase reverse transcriptase; TIR: terminal inverted repeat; TSD: target site duplication; YR: tyrosine recombinase.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KKK initiated the research. KKK and JJ performed the research and wrote the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
PDF file listing Crypton elements found in this study.
PDF file listing Crypton-derived genes in fungi.
PDF file showing alignment of Cryptons and Crypton-derived genes in Saccharomycetaceae fungi in fasta format.
PDF file showing alignment of Cryptons and Crypton-derived genes in animals in fasta format.
PDF file listing the accession numbers for DUF3504 genes.
PDF file showing alignment of KCTD1, KCTD15 and related protein sequences in fasta format.
Contributor Information
Kenji K Kojima, Email: kojima@girinst.org.
Jerzy Jurka, Email: jurka@girinst.org.
Acknowledgements
This work was supported by National Institutes of Health grant 5 P41 LM006252. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
References
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–414. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Miller WJ, Hagemann S, Reiter E, Pinsker W. P-element homologous sequences are tandemly repeated in the genome of Drosophila guanche. Proc Natl Acad Sci USA. 1992;89:4018–4022. doi: 10.1073/pnas.89.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Derbyshire KM. The outs and ins of transposition: from Mu to kangaroo. Nat Rev Mol Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci USA. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 2009;26:983–993. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P. An Arabidopsis hAT-like transposase is essential for plant development. Nature. 2005;436:282–284. doi: 10.1038/nature03667. [DOI] [PubMed] [Google Scholar]
- Tudor M, Lobocka M, Goodell M, Pettitt J, O'Hare K. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet. 1992;232:126–134. doi: 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. An extraordinary retrotransposon family encoding dual endonucleases. Genome Res. 2005;15:1106–1117. doi: 10.1101/gr.3271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Malik HS, Eickbush TH. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc Natl Acad Sci USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatkov KI, Arkhipova IR, Malkova NV, Finnegan DJ, Evgenev MB. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc Natl Acad Sci USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Butler MI, Poulter RT. Cryptons: a group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology. 2003;149:3099–3109. doi: 10.1099/mic.0.26529-0. [DOI] [PubMed] [Google Scholar]
- Broach JR, Hicks JB. Replication and recombination functions associated with the yeast plasmid, 2 μ circle. Cell. 1980;21:501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Doak TG, Witherspoon DJ, Jahn CL, Herrick G. Selection on the genes of Euplotes crassus Tec1 and Tec2 transposons: evolutionary appearance of a programmed frameshift in a Tec2 gene encoding a tyrosine family site-specific recombinase. Eukaryot Cell. 2003;2:95–102. doi: 10.1128/EC.2.1.95-102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs ME, Sánchez-Blanco A, Katz LA, Klobutcher LA. Tec3, a new developmentally eliminated DNA element in Euplotes crassus. Eukaryot Cell. 2003;2:103–114. doi: 10.1128/EC.2.1.103-114.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol. 2004;21:746–759. doi: 10.1093/molbev/msh072. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. The DIRS1 group of retrotransposons. Mol Biol Evol. 2001;18:2067–2082. doi: 10.1093/oxfordjournals.molbev.a003748. [DOI] [PubMed] [Google Scholar]
- Lorenzi HA, Robledo G, Levin MJ. The VIPER elements of trypanosomes constitute a novel group of tyrosine recombinase-enconding retrotransposons. Mol Biochem Parasitol. 2006;145:184–194. doi: 10.1016/j.molbiopara.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- van Duyne GD. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editor. Washington, DC: American Society of Microbiology Press; 2002. A structural view of tyrosine recombinase site-speciic recombination; pp. 93–117. [Google Scholar]
- Jurka J, Kapitonov VV. First Cryptons from invertebrates. Repbase Rep. 2008;8:232–233. [Google Scholar]
- Jurka J. First Cryptons from insects. Repbase Rep. 2009;9:468–480. [Google Scholar]
- Hedges SB, Kumar S. The TimeTree of Life. New York: Oxford University Press; 2009. [Google Scholar]
- Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. The putative Drosophila transcription factor Woc is required to prevent telomeric fusions. Mol Cell. 2005;20:821–831. doi: 10.1016/j.molcel.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Font-Burgada J, Rossell D, Auer H, Azorín F. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 2008;22:3007–3023. doi: 10.1101/gad.481408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Dlakić M, Mushegian A. Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA. 2011;17:799–808. doi: 10.1261/rna.2396011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–324. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM, Collins FH. Molecular evolutionary analysis of the widespread piggyBac transposon family and related "domesticated" sequences. Mol Genet Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer SE, Strehl S, Hagemann S. Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol Biol Evol. 2005;22:833–844. doi: 10.1093/molbev/msi068. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- Holland MJ, Yokoi T, Holland JP, Myambo K, Innis MA. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:813–820. doi: 10.1128/mcb.7.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ishihara K, Ichimura T, Fujita N, Hino S, Tomita S, Watanabe S, Saitoh N, Ito T, Nakao M. MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem. 2009;284:5165–5174. doi: 10.1074/jbc.M807098200. [DOI] [PubMed] [Google Scholar]
- Ding X, Luo C, Zhou J, Zhong Y, Hu X, Zhou F, Ren K, Gan L, He A, Zhu J, Gao X, Zhang J. The interaction of KCTD1 with transcription factor AP-2α inhibits its transactivation. J Cell Biochem. 2009;106:285–295. doi: 10.1002/jcb.22002. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette WK, Martin RG, Rosner JL. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J Mol Biol. 2000;299:1245–1255. doi: 10.1006/jmbi.2000.3827. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- de Crozé N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci USA. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Dawid IB. Kctd15 inhibits neural crest formation by attenuating Wnt/β-catenin signaling output. Development. 2010;137:3013–3018. doi: 10.1242/dev.047548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ, Fletcher JA. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–87. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- van der Maarel SM, Scholten IHJM, Huber I, Philippe C, Suijkerbuijk RF, Gilgenkrantz S, Kere J, Cremers FPM, Ropers HH. Cloning and characterization of DXS6673E, a candidate gene for X-linked mental retardation in Xq13.1. Hum Mol Genet. 1996;5:887–897. doi: 10.1093/hmg/5.7.887. [DOI] [PubMed] [Google Scholar]
- Warner DR, Roberts EA, Greene RM, Pisano MM. Identification of novel Smad binding proteins. Biochem Biophys Res Commun. 2003;312:1185–1190. doi: 10.1016/j.bbrc.2003.11.049. [DOI] [PubMed] [Google Scholar]
- Kunapuli P, Somerville R, Still IH, Cowell JK. ZNF198 protein, involved in rearrangement in myeloproliferative disease, forms complexes with the DNA repair-associated HHR6A/6B and RAD18 proteins. Oncogene. 2003;22:3417–3423. doi: 10.1038/sj.onc.1206408. [DOI] [PubMed] [Google Scholar]
- Kasyapa CS, Kunapuli P, Cowell JK. Mass spectroscopy identifies the splicing-associated proteins, PSF, hnRNP H3, hnRNP A2/B1, and TLS/FUS as interacting partners of the ZNF198 protein associated with rearrangement in myeloproliferative disease. Exp Cell Res. 2005;309:78–85. doi: 10.1016/j.yexcr.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54nrb/NonO: multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/S0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Rajesh C, Baker DK, Pierce AJ, Pittman DL. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Zhang X, Fu X, Zhang X, Gao X, Zhu M, Wang X, Yang Z, Jensen ON, Saarikettu J, Yao Z, Silvennoinen O, Yang J. PTB-associated splicing factor (PSF) functions as a repressor of STAT6-mediated Igε gene transcription by recruitment of HDAC1. J Biol Chem. 2011;286:3451–3459. doi: 10.1074/jbc.M110.168377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005. pp. W557–W559. [DOI] [PMC free article] [PubMed]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDF file listing Crypton elements found in this study.
PDF file listing Crypton-derived genes in fungi.
PDF file showing alignment of Cryptons and Crypton-derived genes in Saccharomycetaceae fungi in fasta format.
PDF file showing alignment of Cryptons and Crypton-derived genes in animals in fasta format.
PDF file listing the accession numbers for DUF3504 genes.
PDF file showing alignment of KCTD1, KCTD15 and related protein sequences in fasta format.